Abstract
Superinfection exclusion is the phenomenon whereby a virus prevents the subsequent infection of an already infected host cell. The Pekin duck hepatitis B virus (DHBV) model was used to investigate superinfection exclusion in hepadnavirus infections. Superinfection exclusion was shown to occur both in vivo and in vitro with a genetically marked DHBV, DHBV-ClaI, which was unable to establish an infection in either DHBV-infected ducklings or DHBV-infected primary duck hepatocytes (PDHs). In addition, exclusion occurred in vivo even when the second virus had a replicative advantage. Superinfection exclusion appears to be restricted to DHBV, as adenovirus, herpes simplex virus type 1, and vesicular stomatitis virus were all capable of efficiently infecting DHBV-infected PDHs. Exclusion was dependent on gene expression by the original infecting virus, since UV-irradiated DHBV was unable to mediate the exclusion of DHBV-ClaI. Using recombinant adenoviruses expressing DHBV proteins, we determined that the large surface antigen mediated exclusion. The large surface antigen is known to cause down-regulation of a DHBV receptor, carboxypeptidase D (CPD). Receptor down-regulation is a mechanism of superinfection exclusion seen in other viral infections, and so it was investigated as a possible mechanism of DHBV-mediated exclusion. However, a mutant large surface antigen which did not down-regulate CPD was still capable of inhibiting DHBV infection of PDHs. In addition, exclusion of DHBV-ClaI did not correlate with a decrease in CPD levels. Finally, virus binding assays and confocal microscopy analysis of infected PDHs indicated that the block in infection occurs after internalization of the second virus. We suggest that superinfection exclusion may result from the role of the L surface antigen as a regulator of intracellular trafficking.
Hepadnaviruses are a family of enveloped, hepatotropic viruses with small (3.0- to 3.2-kb), partially double-stranded DNA genomes (28). The family includes viruses infecting the woodchuck, ground squirrel, grey heron, snow goose, and Pekin duck (duck hepatitis B virus [DHBV]) as well as the medically important human hepatitis B virus (HBV).
The virion is an icosahedral capsid made up of a core protein surrounded by a lipid bilayer that contains the viral envelope proteins. Contained within the capsid is the viral genome with the polymerase protein covalently attached to the 5′ terminus of the minus strand. The hepadnavirus genome is organized into overlapping reading frames that encode the precore, core, polymerase, and surface proteins. The mammalian hepadnaviruses, as well as the majority of the avian hepadnaviruses, contain an additional open reading frame that encodes the X protein (10). Infection is initiated by the interaction of the virus with a receptor present on the surface of hepatocytes. Carboxypeptidase D (CPD) has been identified as a receptor for DHBV, although it appears that additional coreceptors are required (4, 7, 35). Following attachment, the virus enters the cell, likely by endocytosis, and nucleocapsids are released into the cytoplasm (28). Transport of the nucleocapsids to the nuclear membrane is mediated by a nuclear localization signal present in the core protein (6, 20, 39). Disassembly of the nucleocapsids occurs either in the cytoplasm or at the nuclear membrane and is followed by release of the viral DNA into the nucleus. The relaxed circular genome then is converted into covalently closed circular DNA (cccDNA), which serves as the template for virus transcription. The viral genome is transcribed by host RNA polymerase II, and the transcripts are transported to the cytoplasm (10, 28). Following translation of the viral gene products in the cytoplasm, the pregenomic RNA is packaged along with the viral polymerase into the nucleocapsids, where DNA synthesis occurs (31). Reverse transcription of the pregenomic RNA followed by DNA-dependent DNA polymerization results in the relaxed circular, partially double-stranded genome. At this point, the nucleocapsids either are targeted to the nucleus or, alternatively, bud into the endoplasmic reticulum (ER) lumen and exit the cell through the secretory pathway as enveloped, infectious virions (37). This process is partially regulated by the level of expression of the large (L) surface antigen (19, 32, 33). Early in infection, when there is minimal surface antigen expression, the mature nucleocapsids are directed to the nucleus to contribute to the amplification of cccDNA. Later, when a threshold level of surface antigen expression is achieved, the mature nucleocapsids attach to ER membranes containing surface antigen, resulting in envelopment and secretion (19, 32, 33).
Superinfection exclusion is a phenomenon whereby a cell infected with a virus is resistant to superinfection by the same virus. Superinfection exclusion is observed during infections by a broad range of viruses, including human immunodeficiency virus (HIV) (17), vesicular stomatitis virus (VSV) (38), vaccinia virus (5), and alphavirus (14). There is some evidence to suggest that superinfection exclusion occurs in hepadnavirus infections as well. When liver transplant and nontransplant patients with chronic HBV infections were treated with lamivudine, differences in the patterns of development of lamivudine resistance could be seen. The average duration of lamivudine monotherapy before resistant HBV variants emerged was longer in nontransplant patients than in transplant patients, 562 days versus 371 days, respectively (11). In addition, the rates of resistance appeared to be higher in transplant patients than in nontransplant patients (8, 16, 23). These results suggest that lamivudine-resistant HBV establishes an infection more readily in an uninfected liver than in an HBV-infected liver.
Recent studies with the DHBV animal model have also suggested that superinfection exclusion occurs in avian hepadnavirus infections. Studies of viral kinetics in ducks have shown that enrichment of wild-type DHBV over replication-defective mutants is rapid during the initial spread of infection. Thereafter, the enrichment rate is slower and appears to be dependent on the generation of new, uninfected hepatocytes (40, 41). In a similar study, the emergence of wild-type DHBV in competition with either cytopathic or noncytopathic DHBV was found to be dependent on cell death caused by the cytopathic virus, because the wild-type virus did not emerge in competition studies with the noncytopathic variant (18).
In this study, the DHBV animal model was used to investigate superinfection exclusion in hepadnavirus infections. The results show that superinfection exclusion occurs in DHBV infection, is mediated by the L surface antigen, and does not involve the down-regulation of CPD, a known receptor for DHBV.
MATERIALS AND METHODS
Animals and virus stocks.
Lamivudine was obtained from GlaxoSmithKline, Research Triangle Park, N.C. Newborn Pekin ducklings, either congenitally infected with DHBV type 16 or uninfected, were obtained from the University of Alberta, Edmonton, Alberta, Canada, and were maintained according to the regulations of the Canadian Council on Animal Care. All animals were screened for the presence of DHBV infection by dot blotting prior to use in these studies. Wild-type DHBV was obtained from the serum of congenitally infected ducks. The mutant virus, DHBV-ClaI, was made as previously described (9). DHBV-ClaI contains a point mutation at nucleotide 1858 which introduces a ClaI restriction site without altering any amino acid sequence. All nucleotide nomenclature in this article is based on the numbering system used by Mandart et al. for the DHBV genome (21). Serum containing DHBV-ClaI or DHBV-M512V (9) was passaged several times in ducks to obtain high-titer serum. When DHBV-M512V was passaged, animals were maintained on lamivudine therapy at 40 mg/kg of body weight intramuscularly (i.m.) twice daily to prevent reversion to the wild-type virus. Viral titers were quantitated by dot blotting with plasmid standards and are expressed as viral genome equivalents (VGE). All animals were infected by i.m. injections.
For studies involving UV-inactivated virus, 100 μl of high-titer DHBV-positive serum was irradiated for 1 h at 4°C by using a Southern New England UV Co. RPM200 UV box (reactor wavelength, 350 nm) and then used immediately.
Preparation and infection of PDHs.
Primary duck hepatocytes (PDHs) from 14- to 21-day-old ducklings were prepared by using collagenase as previously described (34). The resulting PDHs were plated at 750,000 cells per well in six-well plates and cultured at 37°C in Leibovitz 15 medium supplemented with 1.2 μg of insulin/ml, 1.7 μg of glucose/ml, 11 μM hydrocortisone hemisuccinate, 15 mM HEPES, 5% fetal bovine serum, 50 IU of penicillin/ml, 10 μg of streptomycin/ml, and 25 μg of nystatin/ml (PDH medium). At 1 day postplating, the medium was replaced with serum-free medium, and the cells were cultured for an additional 2 days. At 3 days postplating, the cells were infected with DHBV-positive serum at a multiplicity of infection (MOI) of 100 to 200 in medium containing 1.5% dimethyl sulfoxide overnight at 37°C. The medium was replaced with fresh serum-free medium every second day.
Extraction of extracellular viral DNA from serum.
Twenty microliters of serum was added to 80 μl of 50 mM Tris-HCl (pH 8)-150 mM NaCl- 10 mM EDTA-0.1% sodium dodecyl sulfate (SDS)-800 μg of proteinase K/ml and incubated at 42°C for a minimum of 4 h. The sample was extracted with an equal volume of phenol-chloroform. DNA was precipitated by adding a 0.10 volume of 3 M sodium acetate, 10 μg of yeast tRNA, and 2 volumes of 95% ethanol. The DNA was resuspended in 20 μl of water. Ten microliters was used for a subsequent PCR.
Isolation of intracellular viral DNA from PDHs.
Monolayers of PDHs were rinsed with phosphate-buffered saline (PBS) and lysed in 10 mM Tris-HCl (pH 7.5)-50 mM NaCl- 1 mM EDTA- 0.25% NP-40- 8% sucrose. The nuclei and cellular debris were pelleted by centrifugation at 13,000 × g for 4 min and discarded. To digest cellular nucleic acids, 6 mM MgCl2, 100 μg of DNase I/ml, and 10 μg of RNase A/ml were added to the lysates, and the mixtures were incubated at 37°C for 30 min. The samples were centrifuged as described above, and the virus was precipitated from the supernatants by the addition of 0.3 volume of 26% polyethylene glycol 6000- 1.4 M NaCl- 25 mM EDTA and incubation overnight at 4°C, followed by centrifugation at 13,000 × g for 15 min at 4°C. The pellets, containing the virus, were resuspended in 50 mM Tris-HCl (pH 8)-150 mM NaCl- 10 mM EDTA. To digest the capsid and polymerase, 800 μg of proteinase K/ml and 0.1% SDS were added, and the mixture was incubated at 42°C overnight. The samples were extracted with phenol-chloroform. Ten micrograms of yeast tRNA was added as a carrier, and DNA was precipitated as described above and resuspended in 20 μl of water. Five to 10 μl was used for subsequent PCRs or Southern blots.
Analysis of viral DNA.
The extracted viral DNA was amplified by PCR with Taq polymerase (Gibco BRL) according to the manufacturer's specifications, 1.5 mM MgCl2, and the following primers at 0.25 μM: 5′-CTCAAGAGATTCCTCAGCC-3′ and 5′-GTCATACCATTCTCCTACT-3′. Cycling conditions were as follows: 95°C for 4 min; 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min; and 72°C for 7 min. To distinguish between wild-type DHBV and DHBV-ClaI, the PCR products were digested with restriction enzyme ClaI at 37°C for at least 1 h. The digestion products were separated on 1.3% agarose gels and visualized with ethidium bromide. To distinguish between wild-type DHBV and DHBV-M512V, the PCR products were sequenced.
Cell sorting and single-cell PCR.
Primary cells growing in cultures were washed twice with PBS, treated with glycine buffer (50 mM glycine, 150 mM NaCl [pH 2.2]) for 1.5 min to remove bound virus (3), and washed twice more with PBS. The cells then were removed from the culture dish by trypsin digestion, pelleted by centrifugation, and washed twice more with PBS. The cells were counted and checked for viability by trypan blue exclusion. Single-cell PCR was performed by using a modified version of a previously described protocol (36). The cells were sorted into 0.2-ml PCR tubes containing 10 μl of lysis solution (200 mM KOH, 50 mM dithiothreitol), heated at 65°C for 10 min, and cooled briefly, and the KOH was neutralized with 5 μl of 0.2 N HCl and 5 μl of 400 mM Tris (pH 8.3). The cells then were heated at 93°C for 15 min and cooled briefly, and a PCR mixture was added to a final volume of 100 μl. The PCR and subsequent analysis were performed as described above, except that 40 cycles were used for the single-cell PCR.
Virus stocks and infections.
John Elliot (University of Alberta) kindly provided a recombinant adenovirus which expresses a β-galactosidase with a nuclear localization signal. PDHs were infected with this adenovirus at an MOI of approximately 1 at 3 to 5 days postplating. PDHs were washed with PBS and incubated with 0.5 to 1 ml of adenovirus for 1 h at 37°C. The adenovirus then was removed and replaced with PDH medium. The number of adenovirus-infected cells was determined 24 to 48 h postinfection by staining for β-galactosidase activity. Briefly, the cells were washed with PBS and fixed for 5 min at 4°C with 0.25% glutaraldehyde. The cells were washed three times with PBS and incubated with X-Gal solution (1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]/ml, 2 mM MgCl2, 100 mM each potassium ferricyanide and potassium ferrocyanide in PBS) overnight at 37°C. The cells were washed twice with PBS, and the number of β-galactosidase-positive cells was counted.
James Smiley (University of Alberta) kindly provided VSV. For infection of PDHs, cell monolayers were incubated with culture medium containing VSV at an MOI of 0.1 to 0.15 at 37°C overnight. VSV then was removed and replaced with fresh medium. Cells were monitored daily for cytopathic effects. When the majority of cells were dead, the cells were washed with PBS and fixed and stained with Wright's solution.
Herpes simplex virus type 1 (HSV-1) subtype KOS 1.1 was also provided by James Smiley and used in infection studies. PDHs were infected at 3 days postplating with HSV-1 at an MOI of 10 and were monitored daily for cytopathic effects.
Generation of recombinant adenoviruses.
Recombinant adenoviruses were generated by using the AdEasy system. Bert Vogelstein (Johns Hopkins Oncology Center) kindly provided vectors pAdtrack-CMV and pAdEasy-1 and Escherichia coli strain BJ5183.
The nucleotide sequences encoding the L surface antigen, the small (S) surface antigen, and the core genes of DHBV were first amplified by using an Expand high-fidelity PCR system (Roche, Laval, Quebec, Canada) according to the manufacturer's specifications. The primers used were as follows: for the L surface antigen, 5′-CAGATATCACCATGGGGCAACATCCAGCAAAATCAATGG-3′ and 5′-CAGATATCCTAACTCTTGTAAAAAAGAGC-3′; for the S surface antigen, 5′-CAGATATCACCATGTCTGGTACCTTCGGG-3′ and 5′-CAGATATCCTAACTCTTGTAAAAAAGAGC-3′; and for the core, 5′-CTTGGGATCCGATGGATATCAATGCTTCTAGAGC-3′ and 5′-GCAAAGCTTTTATTTCCTAGGCGAGGGAG-3′. The L surface antigen PCR product was digested with EcoRV and cloned directly into EcoRV-digested pAdtrack-CMV to generate Adtrack-CMV-LsAg. The S surface antigen PCR product was first subcloned into pCR2.1 (Invitrogen, Carlsbad, Calif.), which was then digested with EcoRV; the resulting fragments were cloned into EcoRV-digested pAdtrack-CMV to generate pAdtrack-CMV-SAg. The core PCR product was blunt ended by filling in with T4 DNA polymerase (Invitrogen) and cloned into EcoRV-digested pAdtrack to generate pAdtrack-CMV-core. To generate an L surface antigen with a deletion of amino acids 83 to 109 (L surface antigen Δ83-109), the overlap extension method was used (13). The primers used were as follows: the 5′-flanking primer was 5′-CAGATATCACCATGGGGCAACATCCAGCAAAATCAATGG-3′, the 3′-flanking primer was 5′-CAGATATCCTAACTCTTGTAAAAAAGAGC-3′, and the internal primers were 5′-CTCTTGAGGAGTCGGATTTGATAATCC-3′ and 5′-CGACTCCTCAAGAGGAAACCACCACCATTCCTCCGTCTTCC-3′. The resulting PCR products were treated in the same manner as the S surface antigen PCR product to generate Adtrack-CMV-LsAgΔ83-109. The integrity of the sequences generated by PCR amplification was confirmed by DNA sequencing. Recombinant adenoviruses Ad-LsAg, Ad-LsAgΔ83-109, Ad-SsAg, Ad-core, and Ad-GFP were generated as previously described (12).
Infection of PDHs with recombinant adenoviruses.
Two-day-old cultures of PDHs were incubated with recombinant adenoviruses at an MOI of 50 at 37°C overnight. The cells then were washed once with PBS, and fresh medium was added. At 4 days postinfection, the efficiency of infection was estimated by using fluorescence microscopy to detect green fluorescent protein (GFP)-expressing cells. At 4 days after adenovirus infection, DHBV antigen expression within the PDHs was examined by Western blotting, and the PDHs were infected with DHBV as previously described. One week later, intracellular virus was harvested and analyzed by Southern blotting.
Fluorescent labeling of DHBV stocks.
DHBV virions from duck serum were first partially purified on a 20% sucrose cushion. Approximately 20 to 30 ml of DHBV-positive serum was layered over 5 to 6 ml of 20% sucrose and centrifuged at 76,000 × g for 18 h at 4°C. The resulting pellet was resuspended in 500 μl of PBS, followed by the addition of an equal volume of 400 mM Na2BO3 (pH 8.5) and 0.04 mg of 5 (and 6)-carboxy-X-rhodamine (succinimidyl ester)/ml. The mixture was incubated at room temperature for 1 h. Unreacted rhodamine label was removed by using a PD-10 gel filtration column (Bio-Rad, Mississauga, Ontario, Canada). To remove any contaminating labeled serum albumin, rhodamine-labeled DHBV (approximately 1.5 ml) was incubated at 4°C with 250 μl of Affi-Gel Blue (Bio-Rad) for 2 h. The sample then was centrifuged briefly to remove the Affi-Gel Blue beads. Serum from an uninfected animal was labeled in the same manner as a control.
Binding studies and confocal microscopy.
PDHs from uninfected and congenitally DHBV-infected ducks were prepared as described above. The cells were incubated with rhodamine-labeled DHBV either at room temperature or at 4°C for 5 to 6 h, washed a minimum of six times with PBS to remove unbound virus, and analyzed by fluorescence microscopy. For blocking experiments, PDHs were incubated with labeled DHBV preincubated with monoclonal antibodies specific for either the L or the S surface antigen. Alternatively, the cells were incubated with labeled DHBV in the presence of 25 μl of concentrated subviral particles. The subviral particles were obtained from 50 ml of culture supernatant from Huh-7 cells that were infected with both Ad-LsAg and Ad-SsAg; the supernatant was concentrated to 3 ml by using a Millipore 50K MWCO Centriplus concentrator. Serum albumin was removed by incubation with 500 μl of Affi-Gel Blue as described above. For confocal microscopy, PDHs were grown on glass coverslips, incubated with rhodamine-labeled DHBV for 18 h at 37°C, and washed six times with PBS. The nuclei were stained with Hoechst 33342 dye (Molecular Probes Inc.) (500 ng/ml) for 5 min at room temperature. The coverslips were mounted on slides with 50% glycerol, and the cells were examined with a Zeiss LSM 5 confocal microscope.
SDS-PAGE and Western blot analysis of viral antigens and CPD.
The cells were washed with PBS, harvested from 12-well cell culture dishes by using a cell scraper, and resuspended in 100 μl of 6× loading buffer (125 mM Tris [pH 6.8], 5% SDS, 10% β-mercaptoethanol, 15% glycerol, 0.1% bromophenol blue). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with 8 or 10% polyacrylamide. The separated proteins were transferred to nitrocellulose (Hybond ECL; Amersham Biosciences, Little Chalfont, United Kingdom), processed according to the manufacturer's specifications, and visualized by chemiluminescence. Rabbit anti-CPD antisera were kindly provided by Heinz Schaller (University of Heidelberg), rabbit anti-core antisera were provided by Jesse Summers (University of New Mexico), and monoclonal antibodies to L and S surface antigens were obtained from Pat Nakajima (Fox Chase Institute, Philadelphia, Pa.). Horseradish peroxidase- goat-anti-mouse and horseradish peroxidase- goat-anti-rabbit antibodies were obtained from Cappel Rockland Inc., Gilbertsville, Pa.
Nucleotide sequence accession number.
The sequence of the Alberta strain of DHBV type 16 has been deposited in GenBank under accession number AF047045.
RESULTS
DHBV-ClaI can establish an infection in uninfected but not congenitally DHBV-infected animals.
To determine whether superinfection exclusion occurs in hepadnavirus infections, the ability of a second DHBV to establish an infection in congenitally infected ducklings was examined. A genetically tagged virus, DHBV-ClaI, was used in these studies so it could be distinguished from the endogenous DHBV present in the animals. DHBV-ClaI contains a single nucleotide change which introduces a ClaI restriction site into the DHBV genome without altering the amino acid sequences of any viral proteins. To determine whether an infection with DHBV-ClaI could be established in an animal already infected with DHBV, serum containing 2 × 1010 VGE of DHBV-ClaI was used to inoculate five congenitally infected newborn ducks. Five uninfected animals of the same age were similarly infected as controls. Serum samples were taken weekly to monitor infection. Viral DNA was extracted from serum, amplified by PCR, and digested with ClaI. The full-length wild-type PCR product is 906 bp. ClaI digestion of the PCR product from DHBV-ClaI reduces the size of the product to 820 bp.
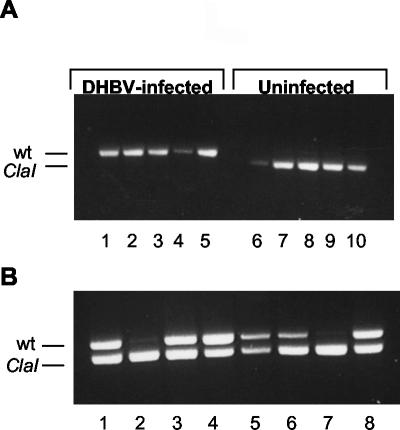
Serum from the congenitally infected animals (Fig. 1A, lanes 1 to 5) showed no evidence of DHBV-ClaI superinfection at 14 days postinfection, since no 820-bp product was detected. However, the virus present in the serum from uninfected animals was entirely DHBV-ClaI (Fig. 1A, lanes 6 to 10), indicating that the DHBV-ClaI stock was infectious and that the ClaI digestion was complete. Analysis of serum taken at all other time points postinfection yielded the same results (data not shown). In total, 13 congenitally infected and 12 uninfected animals were studied. All 12 uninfected animals were infected with DHBV-ClaI, whereas none of the 13 congenitally infected animals showed DHBV-ClaI infection. Four of the congenitally infected ducklings were monitored for 12 weeks, with no evidence of DHBV-ClaI infection. The PCR assay was able to detect 2% of DHBV-ClaI in a background of wild-type DHBV (data not shown). These results suggest that the preexisting infection in these animals prevented superinfection by DHBV-ClaI.
FIG. 1.
Infection of uninfected and congenitally infected ducklings with DHBV-ClaI. Ducklings were inoculated i.m. with 2 × 1010 VGE of either DHBV-ClaI alone (A) or a mixture of DHBV-ClaI and DHBV (B). Serum viral DNA was extracted and amplified by PCR. The PCR products were digested with ClaI and analyzed on agarose gels. (A) Five newborn congenitally infected ducklings (lanes 1 to 5) and five uninfected ducklings (lanes 6 to 10) were infected with DHBV-ClaI. At 14 days postinfection, serum viral DNA was analyzed as described above. (B) Eight newborn uninfected ducklings were infected with an equal mixture of DHBV-ClaI and DHBV. Results represent an analysis of serum viral DNA 7 days postinfection. wt, wild type.
To determine whether DHBV and DHBV-ClaI could establish a coinfection, a mixture of sera containing equivalent amounts of the two viruses was used to infect uninfected newborn ducklings. Eight 1-day-old ducklings were inoculated i.m. with 2 × 1010 VGE of a 1:1 mixture of DHBV and DHBV-ClaI. Serum viral DNA was analyzed weekly as described above, and the results obtained at 7 days postinfection are shown in Fig. 1B. Six of eight ducks showed a mixture of both viruses in their sera. The remaining two ducks showed predominantly DHBV-ClaI (Fig. 1B, lanes 2 and 7). The results obtained at 14 and 21 days were the same, and the ratios of DHBV and DHBV-ClaI in each duck remained consistent at the different time points (data not shown). In total, 13 of 18 ducks studied showed coinfection with both viruses. The remaining five had predominantly either DHBV or DHBV-ClaI. The reason for the predominance of one virus or the other is not clear. The establishment of a coinfection in the majority of the animals indicates that the simultaneous introduction of two viruses can result in the establishment of a dual infection. Thus, the lack of DHBV-ClaI in the congenitally infected ducklings in the previous experiment was not due to a replicative advantage of DHBV over DHBV-ClaI.
Single-cell PCR was performed to determine whether individual hepatocytes were dually infected with both viruses or whether DHBV and DHBV-ClaI replicated exclusively in separate cells. Newborn ducklings were infected with serum containing an approximately 1:1 ratio of DHBV to DHBV-ClaI and were monitored for coinfection by PCR of serum viral DNA. At 2 weeks postinfection, the liver of a coinfected duckling was perfused, and the hepatocytes were cultured for 1 week. The cells then were sorted into single cells by fluorescence-activated cell sorting, and viral DNA was amplified and analyzed as for previous experiments.
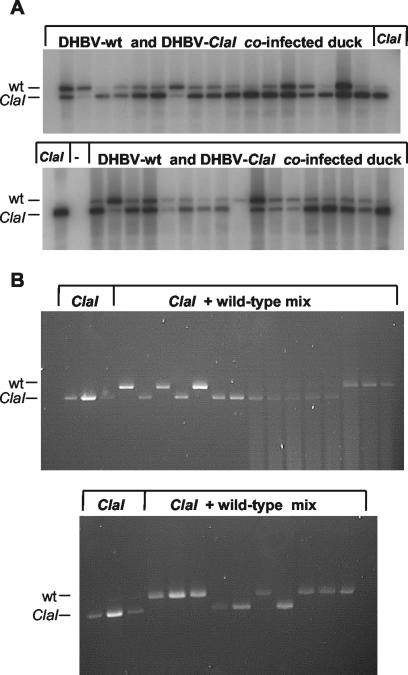
Some of the results of the single-cell PCR analysis for the coinfected duckling are shown in Fig. 2A. In total, 89 of 105 cells showed the presence of both viruses. Five cells were infected exclusively with DHBV, while 11 cells contained only DHBV-ClaI. Interestingly, analysis of serum viral DNA from this animal also showed slightly more DHBV-ClaI than DHBV. These results indicate that the majority of the cells contained both viruses. When cells infected with wild-type DHBV and with DHBV-ClaI were prepared separately and mixed prior to cell sorting, only one virus was detected in each well (Fig. 2B). In total, 48 of 50 positive cells showed the presence of either DHBV or DHBV-ClaI. These results confirm that cells were sorted into single cells which were not contaminated by extracellular virus. Therefore, the presence of both DHBV and DHBV-ClaI in the hepatocytes of the coinfected duckling was due to a dual infection.
FIG. 2.
Single-cell PCR analysis of the viral population within DHBV-infected hepatocytes. (A) PCR analysis of individual hepatocytes obtained from a duck coinfected with DHBV and DHBV-ClaI. PCR products were separated by size on 1.2% agarose gels, transferred to nylon membranes by Southern blotting, and probed with a 32P-labeled plasmid containing DHBV sequences. ClaI lanes represent ClaI-digested controls; − lanes represent negative controls (no cells). (B) PCR analysis of individual hepatocytes obtained from a DHBV-infected duck and a DHBV-ClaI-infected duck and mixed prior to sorting into single cells. PCR products were separated by size on 1.2% agarose gels and visualized by ethidium bromide staining. wt, wild type.
A lamivudine-resistant virus, DHBV-M512V, is unable to efficiently establish an infection in congenitally infected animals despite having a selective advantage.
To assess whether a virus with a replicative advantage over the endogenous virus could establish an infection, DHBV-M512V was administered to congenitally infected ducklings that had been treated with lamivudine. The mutation of methionine to valine in the YMDD motif of polymerase confers resistance to lamivudine, so that DHBV-M512V has a selective advantage over endogenous virus in lamivudine-treated, congenitally infected ducks. Fourteen congenitally infected newborn ducks were treated for 1 week with 40 mg of lamivudine/kg i.m. twice daily to suppress endogenous DHBV replication. Suppression of viremia by lamivudine was confirmed by dot blotting (data not shown). On day 8, these ducks were infected with 5 × 109 VGE of DHBV-M512V and were maintained on lamivudine treatment. Serum samples were taken weekly, viral DNA was isolated and amplified by PCR, and the YMDD motif was analyzed by DNA sequencing. As a control, 10 uninfected newborn ducks were treated in the same manner. The results for samples analyzed at 4 weeks postinfection are depicted in Table 1. Nine of the 10 uninfected animals that were infected with DHBV-M512V were found to be positive for DHBV-M512V exclusively, indicating that the inoculum was infectious and that there was no reversion to wild-type DHBV. Conversely, 12 of the 14 congenitally infected animals showed no evidence of DHBV-M512V superinfection. One of the 14 congenitally infected ducklings had only DHBV-M512V in its serum, and a second had a mixture of DHBV and DHBV-M512V. The presence of DHBV-M512V in two congenitally infected ducklings likely was the result of the DHBV-M512V inoculation, because DHBV-M512V does not spontaneously arise in wild-type DHBV-infected ducks maintained on long-term lamivudine therapy, even after 2 years of continuous therapy (unpublished results). These results indicate that despite a selective advantage, DHBV-M512V was unable to establish an infection in the majority of congenitally infected animals.
TABLE 1.
Inoculation of congenitally DHBV-infected and uninfected ducklings with lamivudine-resistant DHBV-M512Va
| Sequence determined for codon 512 of DHBV polymerase | No. of the following ducklings with the indicated sequence:
|
|
|---|---|---|
| Congenitally infected | Uninfected | |
| ATG (wild type) | 12 | 0 |
| GTG (M512V) | 1 | 9 |
| A/GTG (wild type + M512V) | 1 | 0 |
Fourteen newborn congenitally DHBV-infected ducklings were treated for 1 week with 40 mg of lamivudine/kg twice daily and then inoculated i.m. with DHBV-M512V. Ten uninfected ducklings of the same age were similarly infected. All ducks were maintained on lamivudine therapy after inoculation with DHBV-M512V. Serum samples were taken weekly, and viral DNA was extracted and amplified by PCR. The PCR products were sequenced to determine the sequence at codon 512 of the polymerase. Results represent weekly analysis of serum viral DNA for 4 weeks after DHBV-M512V inoculation.
Exclusion of DHBV-ClaI in PDHs.
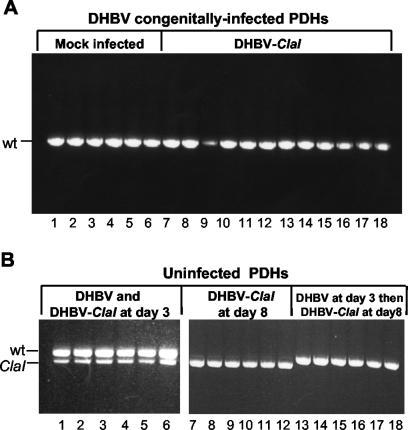
To determine whether superinfection exclusion could be observed in vitro, the ability of DHBV-ClaI to superinfect PDHs from a congenitally infected duckling was examined. PDHs were prepared and, at 3 days postplating, either mock infected (Fig. 3A, lanes 1 to 6) or infected with DHBV-ClaI (lanes 7 to 18) at an MOI of approximately 100 to 200. At 15 days postplating, intracellular viral DNA was harvested, amplified by PCR, and analyzed for the presence of DHBV-ClaI. As shown in Fig. 3A, no evidence of DHBV-ClaI was observed. To determine whether the lack of DHBV-ClaI superinfection was true exclusion or simply was due to inefficient infection of the PDHs, uninfected PDHs either were coinfected with a 1:1 mixture of DHBV and DHBV-ClaI or were infected with wild-type DHBV first and then with DHBV-ClaI 5 days later (Fig. 3B). Uninfected cells cultured for the same amount of time (8 days) were also infected with DHBV-ClaI to ensure that the PDHs were still susceptible to infection. DHBV-ClaI was detected in cells with no prior infection (Fig. 3B, lanes 1 to 12) but not in cells which had previously been infected with wild-type DHBV (lanes 13 to 18). The virus detected in these cells was the result of replicating virus, because a Southern blot of intracellular virus from these samples demonstrated the presence of the single-stranded form of viral DNA (data not shown). An initial infection with wild-type DHBV was found to exclude a subsequent infection with DHBV-ClaI at between 5 and 7 days after the initial infection, depending on the preparation of PDHs (data not shown, but see Fig. 5). Interestingly, the ratio of DHBV to DHBV-ClaI remained constant in cell cultures (Fig. 3B, lanes 1 to 6), unlike what was seen in ducklings coinfected with the two viruses. This result indicates that the variation seen in animals likely involves factors other than the actual infection of the hepatocytes. The results of these experiments indicate that PDHs are equally susceptible to infection with both viruses and that the lack of DHBV-ClaI in both the congenitally infected PDHs and the naive PDHs infected with DHBV prior to DHBV-ClaI is due to superinfection exclusion.
FIG. 3.
Exclusion of DHBV-ClaI in PDHs. PDHs from congenitally DHBV-infected or uninfected ducklings were harvested and plated as described in Materials and Methods. (A) PDHs from a congenitally DHBV-infected duckling were either mock infected (lanes 1 to 6) or infected with DHBV-ClaI (lanes 7 to 18) at 3 days postplating. Intracellular viral DNA was harvested and analyzed for the presence of DHBV-ClaI. Each lane represents intracellular viral DNA harvested from cells in one well of a six-well culture plate. (B) PDHs from an uninfected duckling were infected with DHBV and DHBV-ClaI simultaneously at 3 days postplating (lanes 1 to 6), infected with DHBV-ClaI alone at 8 days postplating (lanes 7 to 12), or infected with DHBV at 3 days postplating and then infected with DHBV-ClaI at 8 days postplating (lanes 13 to 18). Intracellular viral DNA was harvested and analyzed for the presence of DHBV-ClaI. wt, wild type.
FIG. 5.
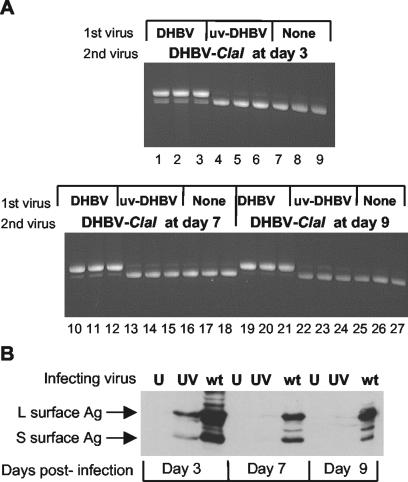
Superinfection exclusion of DHBV-ClaI is dependent on viral gene expression during the initial DHBV infection. PDHs were first infected with DHBV or UV-treated DHBV (uv-DHBV) or mock infected. They were then infected with DHBV-ClaI either 3, 7, or 9 days later. (A) Intracellular virus was harvested 1 week later and analyzed for the presence of DHBV-ClaI. (B) Uninfected cells (U) or cells infected with either UV-irradiated DHBV (UV) or DHBV (wt) were harvested 3, 7, and 9 days postinfection and analyzed by Western blotting for the expression of L and S surface antigens (Ag).
Uninfected and DHBV-infected PDHs are equally susceptible to infection with adenovirus, HSV-1, and VSV.
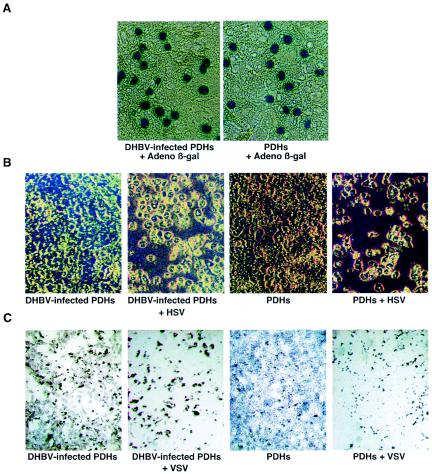
To determine whether the superinfection exclusion was specific for DHBV, we compared the susceptibilities of both naive and DHBV-infected PDHs to adenovirus, HSV-1, and VSV infections. Uninfected and congenitally infected PDHs were infected with an adenovirus that expresses β-galactosidase with a nuclear localization signal at an MOI of approximately 1. At 2 days postinfection, the cells were stained for β-galactosidase activity. Figure 4A shows that there was no significant difference in the numbers of adenovirus-infected cells between uninfected and DHBV-infected hepatocytes. The same experiment was done with HSV-1. PDHs were incubated overnight with HSV-1 at an MOI of 10. Cytopathic effects, characterized by rounding of the majority of cells, were observed at 24 h postinfection, followed by cell death at 48 h postinfection. Again, there was no visible difference between cytopathic effects observed in DHBV-infected and uninfected hepatocytes (Fig. 4B). These experiments indicate that DHBV does not affect the ability of adenovirus or HSV-1 to infect PDHs.
FIG. 4.
Superinfection exclusion is limited to DHBV. Uninfected and DHBV-infected PDHs were prepared as described in Materials and Methods. (A) Cells were infected with a supernatant containing a recombinant adenovirus (Adeno) expressing β-galactosidase (β-gal). At 2 days postinfection, cells were fixed and stained for β-galactosidase activity. (B) Cells were infected with HSV-1 at an MOI of 10 and monitored for cytopathic effects. Cells are shown 2 days after HSV-1 infection. (C) Cells were infected with VSV at an MOI of 0.1 and monitored for cytopathic effects. Cells are shown 4 days after VSV infection.
Studies with HBV-transgenic mice have shown higher levels of HBV in gamma interferon (IFN-γ) and IFN-α/β receptor knockout mice (22). This finding suggests that these cytokines inhibit HBV replication to some extent. In addition, DHBV replication has been shown to be inhibited by both IFN-α/β and IFN-γ (26, 27). However, the effect is greatest when IFN is present at or before the time of infection. It is possible that infection with DHBV induces a low level of IFN expression which does not affect the replication of the established DHBV infection but which inhibits the establishment of a second infection, in this case, a DHBV-ClaI infection. The response of VSV to DHBV infection was used to test this hypothesis. VSV is extremely sensitive to both duck IFN-α/β and duck IFN-γ (26, 27). If a low level of IFN-α/β is expressed in DHBV-infected hepatocytes, then they should be protected against VSV-mediated lysis. Both DHBV-infected and uninfected PDHs were infected with VSV at an MOI of 0.1 and monitored daily for cytopathic effects. The majority of both uninfected and DHBV-infected hepatocytes were killed by day 4 after VSV infection, indicating that they were equally susceptible to VSV infection (Fig. 4C). This result suggests that the mechanism of DHBV exclusion is unlikely to be mediated by IFN. The results from the adenovirus superinfection experiment support this conclusion, as the entry and possibly the gene expression of recombinant adenoviruses have also been shown to be inhibited by IFN expression (22).
DHBV gene expression is required for the exclusion of DHBV-ClaI.
The need for gene expression by DHBV for the exclusion of DHBV-ClaI was examined by determining the effect of treating DHBV with UV radiation prior to infection of PDHs. UV irradiation results in cross-linking of the viral DNA, which inhibits transcription. PDHs were mock infected, infected with wild-type DHBV, or infected with UV-treated DHBV. The cells then were incubated with DHBV-ClaI at 3, 7, or 9 days later (Fig. 5). Intracellular virus was harvested 1 week after the last DHBV-ClaI infection (day 16) and analyzed. As expected, when cells were initially infected with wild-type DHBV, DHBV-ClaI was almost completely excluded by day 9 (Fig. 5A, lanes 19 to 21). However, there was evidence of DHBV-ClaI infection in cells that were initially infected with irradiated DHBV (Fig. 5A, lanes 22 to 24) or mock infected (lanes 25 to 27). The small amount of the PCR product corresponding to the size of wild-type DHBV in cells that were either mock infected or infected with UV-treated DHBV is likely due to incomplete digestion of the PCR product with the ClaI enzyme. Figure 5B shows a Western blot revealing the expression of two envelope proteins from mock-infected cells, cells infected with UV-treated DHBV, or cells infected with wild-type DHBV at various times postinfection. The expression of both surface antigens could be detected only in cells infected with wild-type DHBV. Neither of these antigens could be detected in cells infected with UV-treated DHBV, indicating that this virus was incapable of gene expression. A small amount of each surface antigen could be detected 3 days postinfection but not at subsequent time points and therefore likely represents the virus inoculum. Therefore, the ability of DHBV to exclude DHBV-ClaI is dependent on viral protein expression.
Identification of the DHBV protein responsible for superinfection exclusion.
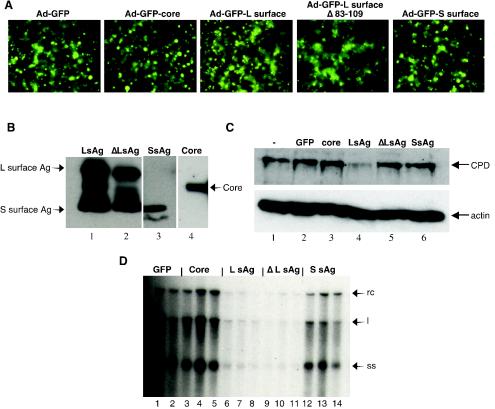
The results of the previous experiment suggested that gene expression is required for the exclusion of a second infection, in this case, DHBV-ClaI infection. However, they do not indicate which viral protein is involved. To determine which protein was responsible for the observed superinfection exclusion, individual DHBV proteins were tested for their ability to exclude DHBV infection. In addition, a DHBV L surface antigen with a deletion of the CPD binding domain was also analyzed for its ability to exclude DHBV infection. Recombinant adenoviruses were used to express DHBV proteins, since the transfection efficiency of PDHs is inefficient (approximately 1 to 5%). Recombinant adenoviruses Ad-core, Ad-LsAg, Ad-LsAgΔ83-109, Ad-SsAg, and Ad-GFP were used to infect PDHs at 2 days postplating. At 4 days after adenovirus infection, the cells were analyzed by fluorescence microscopy to determine the percentages of cells infected by the recombinant adenoviruses (Fig. 6A). As shown in Fig. 6A, the efficiencies of the adenovirus infections (measured as the percentages of GFP-expressing cells) were similar and ranged from 60 to 80% between experiments. In addition, on day 4, some cells were harvested to confirm the expression of the DHBV antigens and to examine the expression of CPD (Fig. 6B and C, respectively). Figure 6B shows that the appropriate proteins were expressed, and Fig. 6C shows that only Ad-LsAg decreased the expression of CPD. Also on day 4, the adenovirus-infected PDHs were infected with DHBV. One week later, intracellular viral DNA was harvested and analyzed by Southern blotting (Fig. 6D). Evidence of DHBV infection, as indicated by the presence of viral replicative intermediates, was seen in cells infected with Ad-GFP, Ad-core, and Ad-SsAg. Conversely, the level of DHBV replication was significantly reduced in cells infected with either Ad-LsAg or Ad-LsAgΔ83-109. These results indicate that the L surface antigen alone is capable of inhibiting DHBV infection of PDHs and that the region of this antigen which interacts with CPD (amino acids 83 to 109) is not necessary for exclusion.
FIG. 6.
Superinfection exclusion is mediated by the L surface antigen. (A) Efficiency of infection of PDHs by recombinant adenoviruses. Cells were infected with recombinant adenoviruses at an MOI of 50 at 2 days postplating. The percentages of infected cells were monitored by examining the expression of the GFP marker by fluorescence microscopy. Results shown were obtained 4 days after adenovirus infection. Magnification, ×40. (B) Western blot of viral antigens (Ag) expressed by recombinant adenoviruses Ad-LsAg, Ad-LsAgΔ83-109, Ad-SsAg, and Ad-core. Cells were harvested 4 days after adenovirus infection. The proteins were separated by SDS-PAGE and analyzed by Western blotting with antibodies specific for either DHBV L surface antigen (lanes 1 and 2), DHBV S surface antigen (lane 3), or DHBV core (lane 4). (C) Expression of CPD and actin in PDHs infected with recombinant adenoviruses. Uninfected PDHs (lane 1) or PDHs infected with adenoviruses expressing either GFP alone (lane 2) or GFP plus either core (lane 3), L surface antigen (lane 4), L surface antigen Δ83-109 (lane 5), or S surface antigen (lane 6) were harvested 4 days after adenovirus infection. The proteins were separated by SDS-PAGE and analyzed by Western blotting with antibodies specific for CPD (upper panel). The same blot was stripped and reprobed with antibodies specific for actin (lower panel). (D) Exclusion of DHBV by hepatocytes expressing L surface antigen. Cells were first infected with adenoviruses expressing either GFP alone (lanes 1 and 2) or GFP plus the following DHBV antigen: core (lanes 3 to 5), L surface antigen (lanes 6 to 8), L surface antigen Δ83-109 (lanes 9 to 11), or S surface antigen (lanes 12 to 14). Four days later, they were challenged with DHBV. Southern blot analysis of intracellular virus 1 week after DHBV infection is shown. The blot was probed with a 32P-labeled DHBV sequence. rc, relaxed circular, l, double-stranded linear, ss, single stranded.
Occasionally, the expression of the S surface antigen inhibited DHBV replication relative to the results obtained with the control adenovirus, Ad-GFP (Fig. 6D, lanes 12 to 14). However, the extent of the inhibition was substantially smaller than that seen with the L surface antigen and L surface antigen Δ83-109. In addition, unlike the inhibition seen with both forms of the L surface antigen, the slight inhibition of DHBV replication seen with the S surface antigen was not consistent between experiments.
Down-regulation of CPD expression does not correlate with exclusion of DHBV-ClaI.
The L surface antigen was the only DHBV protein that inhibited DHBV infection; thus, it is involved in the mechanism of superinfection exclusion. This protein was previously shown to down-regulate a DHBV receptor, CPD, at between 5 and 9 days after infection (2). This time is similar to the time at which DHBV-ClaI is excluded from hepatocytes previously infected with DHBV. Furthermore, receptor down-regulation by viral envelope proteins is a mechanism of superinfection exclusion used by a number of viruses, most notably, HIV, and it is possible that DHBV also mediates exclusion in this way. The previous experiment suggested that this is not the case, because a form of the L surface antigen which does not down-regulate CPD, L surface antigen Δ83-109, was still capable of inhibiting DHBV infection.
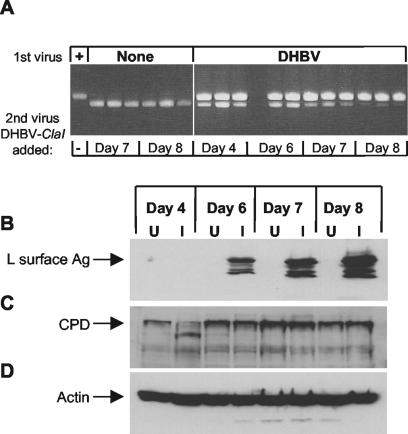
To further confirm that receptor down-regulation was not the mechanism of exclusion, the correlation between DHBV-ClaI exclusion and the decrease in CPD expression was examined. PDHs were first infected with wild-type DHBV and then, 4, 6, 7, or 8 days later, infected with DHBV-ClaI or harvested for Western blot analysis. To confirm that the cells were still susceptible to infection, PDHs that had not been infected with DHBV were infected with DHBV-ClaI at these time points. One week after the last DHBV-ClaI infection, intracellular virus was harvested and analyzed by PCR (Fig. 7A). In this experiment, partial exclusion of DHBV-ClaI was seen when DHBV-ClaI was introduced 6 days after the initial DHBV infection, and exclusion of DHBV-ClaI was almost complete by 7 or 8 days after DHBV infection (Fig. 7A). The L surface antigen was first detected at 6 days after DHBV infection (Fig. 7B); this time correlated with the time at which DHBV-ClaI exclusion first became evident. However, CPD expression remained approximately the same over the course of the experiment (Fig. 7C). A decrease in CPD levels was observed at 4 days postinfection; such a decrease was not observed in other experiments and so was presumed to be an artifact of the particular experiment shown. To confirm that equal amounts of protein were loaded in the lanes, this blot was stripped and reprobed with an antibody specific for actin (Fig. 7D).
FIG. 7.
Superinfection exclusion of DHBV-ClaI does not correlate with a decrease in CPD expression. (A) Exclusion of DHBV-ClaI in DHBV-infected PDHs. PDHs were first infected with DHBV and then infected with DHBV-ClaI 4, 6, 7, and 8 days later. To ensure that the cells could still be infected, uninfected cells were also infected with DHBV-ClaI on days 7 and 8. Intracellular virus was harvested 1 week later and analyzed for the presence of DHBV-ClaI by PCR. Each lane represents viral DNA from one well of a six-well culture dish. (B, C, and D) Western blot analysis of DHBV-infected PDHs. DHBV-infected (I) or uninfected (U) PDHs were harvested 4, 6, 7, and 8 days after DHBV infection. The proteins were separated by SDS-PAGE and analyzed by Western blotting with antibodies specific for the L surface antigen (Ag) (B), CPD (C), or actin (D).
Rhodamine-labeled DHBV binds to both uninfected and DHBV-infected PDHs.
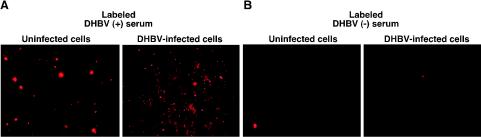
The cellular site at which superinfecting virus is blocked may provide clues about the mechanism of exclusion by the L surface antigen. It is possible that the L surface antigen interferes with the expression of an as-yet-unknown coreceptor. Alternatively, it is possible that even though the total cellular expression of CPD is not affected at the time of exclusion, surface levels of CPD have decreased. If either of these possibilities is the case, then the ability of DHBV-infected cells to bind to labeled virus should be lower than that of uninfected cells. Either uninfected or congenitally DHBV infected PDHs were incubated with rhodamine-labeled virus or labeled control serum under conditions which have been shown to be permissive for DHBV binding (5 h at 25°C) (25). After extensive washing with PBS, the cells were examined by fluorescence microscopy to detect any rhodamine-labeled virus that bound to cells. The labeled virus was able to bind to both uninfected and DHBV-infected hepatocytes (Fig. 8A). The same result was seen when labeled virus was incubated with the cells at 4°C (data not shown). Areas of intense, punctuate fluorescence were superimposed on diffuse fluorescence over the entire cell. The intense staining may have been the result of virus aggregation on the cellular membrane. No fluorescence was observed when cells were incubated with labeled control serum (from an uninfected duckling) (Fig. 8B), indicating that the fluorescence observed was due to labeled DHBV and not labeled serum proteins. Labeled DHBV did not bind to PDHs when it was incubated in the presence of unlabeled subviral particles, nor did it bind to Huh-7 and 239A cells, which are not susceptible to DHBV infection (data not shown). The binding was also decreased in the presence of L surface antigen-specific antibodies (data not shown). Although it was not possible to quantitate the amount of virus bound, it was clear that the virus was still capable of substantial binding to DHBV-infected hepatocytes. Therefore, it is unlikely that the block in DHBV infection of DHBV-infected hepatocytes occurs at the level of receptor binding.
FIG. 8.
Binding of rhodamine-labeled DHBV to DHBV-infected and uninfected PDHs. DHBV particles were partially purified from DHBV-positive serum and then labeled with the fluorescent dye rhodamine as described in Materials and Methods. Serum from an uninfected animal was similarly treated and labeled to serve as the control serum. Congenitally DHBV-infected (right panels) and uninfected (left panels) PDHs were incubated with either rhodamine-labeled DHBV-positive serum (A) or rhodamine-labeled control serum (B) for 5 h at room temperature. The cells were washed five times with PBS and analyzed by fluorescence microscopy. In order to obtain the correct focus on cells labeled with control serum (B), we had to search for a field with at least one fluorescent cell.
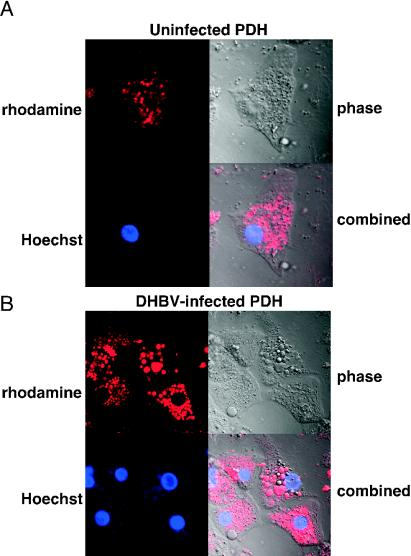
Confocal microscopy analysis demonstrates that DHBV is internalized in both uninfected and DHBV-infected PDHs.
Since the block in DHBV infection does not appear to be at the level of virus binding, we investigated the possibility that entry of the virus by endocytosis is inhibited in DHBV-infected hepatocytes. Confocal microscopy was used to determine whether rhodamine-labeled DHBV was able to enter hepatocytes. Rhodamine-labeled DHBV was incubated with DHBV-infected and uninfected PDHs at 37°C overnight. These conditions were previously shown to allow DHBV entry into permissive cells. The cells were washed extensively with PBS, and the nuclei were stained with Hoechst 33342 dye. The virus was able to enter both uninfected (Fig. 9A) and DHBV-infected (Fig. 9B) hepatocytes. The labeled virus (red) and the nucleus (blue) were both in the same focal plane, indicating that the virus was within the cell and not simply bound to the cell surface. Incubation of uninfected and DHBV-infected hepatocytes with labeled control serum did not result in any binding, as indicated by fluorescence microscopy; therefore, these cells were not included in the confocal microscopy analysis.
FIG. 9.
Confocal microscopy analysis of uninfected (A) and congenitally DHBV-infected (B) PDHs incubated with rhodamine-labeled DHBV. Cells were incubated with rhodamine-labeled DHBV overnight at 37°C and then washed extensively with PBS. Nuclei were stained with Hoechst 33342 prior to confocal microscopy analysis.
DISCUSSION
The results of these studies show that an existing DHBV infection can prevent subsequent infection by a second hepadnavirus. Neither DHBV-ClaI nor DHBV-M512V were able to efficiently establish infections in congenitally infected animals. The amount of virus used to challenge congenitally infected animals, approximately 109 and 1010 VGE of DHBV-M512V and DHBV-ClaI, respectively, was 10,000-fold higher than the amount of virus required to establish an infection in ducks. Thus, a preexisting DHBV infection imposes a profound block to superinfection.
Simultaneous inoculation of DHBV and DHBV-ClaI resulted in the establishment of a dual infection in animals. Analysis of viral DNA present in individual hepatocytes indicates that the majority of cells in these ducks are infected with both viruses. This presence of dual infections within cells makes it unlikely that the mechanism of exclusion involves the first infecting virus rapidly inducing a change in the cell to exclude the second virus. Further evidence for a slow mechanism of exclusion comes from the staggered coinfection experiments in which exclusion of the second virus became apparent only 5 days after the first infection. The high MOI (100 to 200 VGE/cell) used to infect the PDHs likely results in infection of all susceptible cells by the initial inoculum. Therefore, the delay in exclusion likely reflects the time required for sufficient expression of the viral antigen in the cell. A slow onset of exclusion is also consistent with the dependence of exclusion on the expression of the L surface antigen, as evidenced by the correlation of DHBV-ClaI exclusion with the expression of the L surface antigen (Fig. 7).
Exclusion does not depend on on-going viral replication. Inhibition of DHBV replication in congenitally infected ducklings with lamivudine did not, in 12 of 14 animals, prevent exclusion of a second virus. This is despite the fact that the second virus, DHBV-M512V, contains a mutation in the polymerase which renders it resistant to lamivudine and so should have a replicative advantage in the lamivudine-treated animals. Lamivudine inhibits viral replication but does not inhibit viral gene expression from the stable pool of nuclear cccDNA. The presence of DHBV-M512V in the two remaining animals could be related to the number of DHBV-infected hepatocytes at the time they were infected with DHBV-M512V. Congenitally infected animals vary in their level of viremia and a small percentage is able to clear the infection (unpublished data). This may explain why DHBV-M512V was able to establish an infection in two ducklings.
The exclusion mechanism is specific for DHBV as the unrelated viruses HSV 1, adenovirus, and VSV were not excluded from DHBV-infected hepatocytes. Thus, exclusion is unlikely to be mediated by antiviral cytokines such as IFNs. Previous studies have indicated that the small number of liver-resident macrophages (Kupffer cells) that are present in primary duck hepatocyte cultures can be artificially stimulated by endotoxin to produce IFN (15). The level of IFN produced is sufficient to inhibit DHBV replication. However, the highly IFN-sensitive virus VSV was still capable of infecting and killing DHBV-infected hepatocytes. Therefore, the initial DHBV infection is unlikely to be producing sufficient IFN to mediate the exclusion of DHBV-ClaI.
Exclusion of DHBV requires the expression of viral gene products. UV-treated DHBV, which did not express viral proteins, as shown by Western blotting, was incapable of excluding DHBV-ClaI infection of cultured hepatocytes. This is not surprising as the majority of viral interference mechanisms involve at least some viral gene expression. Since UV-treated virus can bind to the cells (data not shown), viral interference is not likely mediated by the transient occupancy of cellular receptors by the initial virus, a mechanism of exclusion seen with retroviruses, including Rous sarcoma virus and avian leukosis virus (29, 30).
Exclusion of DHBV infection is dependent on the expression of the L surface antigen. Recombinant adenoviruses were used to express the core, the L surface antigen, or the S surface antigen in primary hepatocytes to determine which viral protein mediates exclusion. The expression of GFP alone or core protein in hepatocytes did not inhibit DHBV infection relative to the results obtained with the control adenovirus, Ad-GFP. However, the expression of the L surface antigen and, to a lesser extent, the S surface antigen, in hepatocytes did result in decreased levels of DHBV replicative intermediates, a possible result of viral exclusion.
An alternative interpretation is that the decreased level of DHBV replication in cells expressing the envelope proteins is due to cytopathic effects caused by the overexpression of these proteins. However, while infection of primary hepatocytes with adenovirus does appear to have a limited cytopathic effect, this cytopathic effect was comparable with each of the viral proteins and appeared to be related more to the MOI used for infection (the higher the MOI, the greater the cytotoxicity). Western blot analysis of actin levels at the time of DHBV infection of the adenovirus-infected hepatocytes showed no significant differences in cell numbers in cultures expressing the various proteins at the time of DHBV infection.
Recently, it was shown that one of the putative receptors for DHBV, CPD, is down-regulated in DHBV-infected hepatocytes (2). Breiner et al. (2) showed that the L surface antigen binds to CPD in the ER, causing premature degradation of the receptor. Exclusion of DHBV could be explained if the level of receptor down-regulation in an infected cell is sufficient to prevent entry of potential superinfecting virus. Receptor down-regulation as a mechanism of superinfection exclusion is known to occur in a number of viral infections. For example, the HIV receptor, CD4, is down-regulated at the translational and posttranslational levels by the envelope, Vpu and Nef proteins of the virus (1). Surprisingly, the L surface antigen-mediated down-regulation of the receptor CPD, first demonstrated by Breiner et al. (2) and repeated in the present study, does not appear to be involved in exclusion. This conclusion is based on a number of observations. First, the time of DHBV-ClaI exclusion in PDHs did not correlate with a decrease in CPD expression. DHBV-ClaI was excluded 5 to 7 days after DHBV infection. At this time, Western blot analysis indicated that there was no significant decrease in total cellular CPD levels. However, it remains possible that cell surface levels of CPD are reduced at this time. The use of cell fractionation to examine specifically the plasma membrane levels of CPD might give an indication of cell surface CPD levels. However, CPD is localized primarily to the Golgi apparatus and only transiently cycles to the cell surface, and its detection on the cell surface of even uninfected hepatocytes is difficult (4).
Second, the expression of the L surface antigen containing a deletion of the CPD binding domain was still capable of excluding DHBV infection to levels comparable to those seen with the wild-type L surface antigen. The L surface antigen mediates the down-regulation of CPD by interacting with CPD in the ER, leading to premature degradation of CPD (2). It was speculated that deletion of the pre-S domain involved in the L surface antigen-CPD interaction would eliminate the intracellular interaction of these two proteins and prevent CPD down-regulation. Western blot analysis showed that the expression of L surface antigen Δ83-109 did not result in the down-regulation of CPD. Despite this, L surface antigen Δ 83-109 still inhibited DHBV infection.
Third, DHBV-infected hepatocytes were still capable of binding to rhodamine-labeled DHBV. Conversely, rhodamine-labeled control serum did not result in any significant binding, indicating that the observed binding was specific for DHBV. As well, binding was not observed when the rhodamine-labeled DHBV was incubated with cells known to be nonpermissive for DHBV infection. The binding of labeled virus was completely inhibited when cells were first incubated with an excess amount of subviral particles (data not shown). Binding was also partially inhibited when the labeled virus was preincubated with a monoclonal antibody specific for the pre-S region of the L surface antigen (data not shown).
Labeled virus was also capable of entering DHBV-infected hepatocytes, apparently by endocytosis, as indicated by the localization of fluorescence in endosome-like vesicles. It is the surface of the virus which becomes labeled with rhodamine, most likely through attachment of the rhodamine to the L and S surface antigens present in the viral envelope. Fusion of the viral and endosome membranes would disrupt the endosome membrane, allowing the release of the nucleocapsid into the cytoplasm. The fluorescence-labeled viral envelope would presumably remain associated with the disrupted endosome membrane. The fluorescence signal appears to be within endosomes in both uninfected and DHBV-infected hepatocytes.
In summary, superinfection exclusion in DHBV infection is not IFN mediated. Exclusion requires viral gene expression but not viral replication. The L surface antigen is capable of independently mediating exclusion. However, the block in superinfection occurs after attachment and entry of the virus into hepatocytes. The results of this study are consistent with a model of exclusion involving the L surface antigen and the establishment of the cccDNA pool. Establishment of the cccDNA pool occurs early in infection and is negatively regulated by the L surface antigen (32, 33, 37). Early in infection, when L surface antigen levels are low, nucleocapsids containing newly synthesized relaxed circular DNA are directed to the nucleus, where the relaxed circular DNA is converted to cccDNA. As the cccDNA pool increases, the amount of the L surface antigen also increases and the nucleocapsids are enveloped and exported from the cell as infectious virus. DHBV-infected hepatocytes likely contain sufficient levels of the L surface antigen to effectively block the amplification of the cccDNA of the “superinfecting” virus. Without establishing a pool of its specific cccDNA, the second virus would not produce detectable extracellular virus. This model is consistent with the results shown in this study. DHBV-ClaI was unable to establish an infection in either DHBV-infected cells or animals, yet rhodamine-labeled virus was able to bind to and enter DHBV-infected hepatocytes. This model also explains why a mutant L surface antigen was capable of inhibiting DHBV infection of PDHs independent of its ability to bind to CPD.
The existence of superinfection exclusion would explain the observation that the development of lamivudine resistance is more rapid and occurs at higher rates in liver transplant patients than in patients with chronic HBV infections and treated with the drug. Previous studies have shown that enrichment of wild-type DHBV over a replication-defective variant is rapid during the initial phase of infection, when DHBV is spreading within the liver. Once the majority of hepatocytes become infected, however, this enrichment of wild-type DHBV is much slower and appears to be dependent on an increase in liver mass (40, 41). This pattern is consistent with superinfection exclusion. Similarly, the spread of any variant arising in a single cell, such as lamivudine-resistant HBV, would be limited by the slow production of new, uninfected hepatocytes. In a patient undergoing a liver transplant, the uninfected hepatocytes of the new liver would be susceptible to infection by any lamivudine-resistant HBV in the viral population. Conversely, the liver of a chronically infected individual undergoing lamivudine therapy still has a low level of wild-type viral replication and a persistent pool of wild-type cccDNA, making it more difficult for the mutant to spread through the liver.
Superinfection exclusion also has implications for proposed antiviral therapy with HBV as a gene therapy vector. Infection of congenitally infected duck hepatocytes with a recombinant DHBV expressing GFP is significantly less efficient than infection of naive hepatocytes (24). In this study, more than 90% of uninfected hepatocytes were infected with the recombinant virus, compared with 1 to 4% of congenitally infected hepatocytes. Although these cells did show superinfection, it was extremely inefficient compared with infection of uninfected hepatocytes. The success of gene therapy for chronic HBV infection may therefore be limited by the ability of recombinant HBV to enter an infected cell and express the therapeutic gene.
Acknowledgments
We thank Heinz Schaller and Jesse Summers for providing anti-CPD and anti-DHBV core antisera. We also thank Jyy Huang and Gerald LaChance for expert care of the animals and Scott Slemko and Renata Jasinska for assistance with confocal and fluorescence microscopy.
K.-A.W. was supported by an Alberta Heritage Foundation for Medical Research studentship. M.A.J. was supported by a Canadian Institutes of Health Research postdoctoral fellowship. Funding for this work was provided by Glaxo Wellcome Canada and the Canadian Institutes of Health Research.
REFERENCES
- 1.Bour, S., R. Geleziunas, and M. Wainberg. 1995. The human immunodeficiency virus type 1 (HIV-1) CD4 receptor and its central role in promotion of HIV-1 infection. Microbiol. Rev. 59:63-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breiner, K., S. Urban, B. Glass, and H. Schaller. 2001. Envelope protein-mediated down-regulation of hepatitis B virus receptor in infected hepatocytes. J. Virol. 75:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiner, K. M., and H. Schaller. 2000. Cellular receptor traffic is essential for productive duck hepatitis B virus infection. J. Virol. 74:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breiner, K. M., S. Urban, and H. Schaller. 1998. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 72:8098-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christen, L., J. Seto, and E. Niles. 1990. Superinfection exclusion of vaccinia virus in virus-infected cell cultures. Virology 174:35-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckhardt, S., D. Milich, and A. McLachlan. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 65:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eng, F. J., E. G. Novikova, K. Kuroki, D. Ganem, and L. D. Fricker. 1998. gp180, a protein that binds duck hepatitis B virus particles, has metallocarboxypeptidase D-like enzymatic activity. J. Biol. Chem. 273:8382-8388. [DOI] [PubMed] [Google Scholar]
- 8.Fischer, K., K. Gutfreund, and D. Tyrrell. 2001. Lamivudine resistance in hepatitis B: mechanisms and clinical implications. Drug Resist. Update 2:118-128. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, K., and D. Tyrrell. 1996. Generation of duck hepatitis B virus polymerase mutants through site-directed mutagenesis which demonstrate resistance to lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine] in vitro. Antimicrob. Agents Chemother. 40:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, 4th ed. Lippincott/Williams & Wilkins, Philadelphia, Pa.
- 11.Gutfreund, K., M. Williams, R. George, V. Bain, M. Ma, E. Yoshida, J. Villeneuve, K. Fischer, and D. Tyrrell. 2000. Genotypic succession of mutations of the hepatitis B virus polymerase associated with lamivudine resistance. J. Hepatol. 33:469-475. [DOI] [PubMed] [Google Scholar]
- 12.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 14.Karpf, A., E. Lenches, E. Strauss, J. Strauss, and D. Brown. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klocker, U., U. Schultz, H. Schaller, and U. Protzer. 2000. Endotoxin stimulates liver macrophages to release mediators that inhibit an early step in hepadnavirus replication. J. Virol. 74:5525-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai, C., R. Chien, N. Leung, T. Chang, R. Guan, D. Tai, K. Ng, P. Wu, J. Dent, J. Barber, L. Stephenson, D. Gray, et al. 1998. A one-year trial of lamivudine for chronic hepatitis B. N. Engl. J. Med. 339:61-68. [DOI] [PubMed] [Google Scholar]
- 17.LeGuern, M., and J. Levy. 1992. Human immunodeficiency virus (HIV) type 1 can superinfect HIV-2-infected cells: pseudotype virions produced with expanded cellular host range. Proc. Natl. Acad. Sci. USA 89:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenhoff, R., C. Luscombe, and J. Summers. 1998. Competition in vivo between a cytopathic variant and a wild-type duck hepatitis B virus. Virology 251:85-95. [DOI] [PubMed] [Google Scholar]
- 19.Lenhoff, R., and J. Summers. 1994. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J. Virol. 68:4565-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mabit, H., K. M. Breiner, A. Knaust, B. Zachmann-Brand, and H. Schaller. 2001. Signals for bidirectional nucleocytoplasmic transport in the duck hepatitis B virus capsid protein. J. Virol. 75:1968-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandart, E., A. Kay, and F. Galibert. 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J. Virol. 49:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nery, J., D. Weppler, M. Rodriguiz, P. Ruiz, E. Schiff, and A. Tzakis. 1998. Efficacy of lamivudine in controlling hepatitis B virus recurrence after liver transplantation. Transplantation 65:1615-1621. [DOI] [PubMed] [Google Scholar]
- 24.Protzer, U., M. Nassal, P. Chiang, M. Kirschfink, and H. Schaller. 1999. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc. Natl. Acad. Sci. USA 96:10818-10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao, M., C. A. Scougall, A. Duszynski, and C. J. Burrell. 1999. Kinetics of early molecular events in duck hepatitis B virus replication in primary duck hepatocytes. J. Gen. Virol. 80:2127-2135. [DOI] [PubMed] [Google Scholar]
- 26.Schultz, U., and F. V. Chisari. 1999. Recombinant duck interferon gamma inhibits duck hepatitis B virus replication in primary hepatocytes. J. Virol. 73:3162-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz, U., J. Summers, P. Staeheli, and F. V. Chisari. 1999. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J. Virol. 73:5459-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seeger, C., and W. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 9:265-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steck, F. T., and H. Rubin. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology 29:628-641. [DOI] [PubMed] [Google Scholar]
- 30.Steck, F. T., and H. Rubin. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642-653. [DOI] [PubMed] [Google Scholar]
- 31.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 32.Summers, J., P. M. Smith, and A. L. Horwich. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summers, J., P. M. Smith, M. J. Huang, and M. S. Yu. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, S., B. Lee, W. Luo, D. Tovell, M. Robins, and D. Tyrrell. 1988. Inhibition of duck hepatitis B virus replication by purine 2′,3′-dideoxynucleosides. Biochem. Biophys. Res. Commun. 156:1144-1151. [DOI] [PubMed] [Google Scholar]
- 35.Tong, S., J. Li, and J. R. Wands. 1999. Carboxypeptidase D is an avian hepatitis B virus receptor. J. Virol. 73:8696-8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai, Y. 1999. Cost-effective one-step PCR amplification of cystic fibrosis Δ508 fragment in a single cell for preimplantation genetic diagnosis. Prenat. Diagn. 19:1048-1051. [PubMed] [Google Scholar]
- 37.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 38.Whitaker-Dowling, P., J. Youngner, C. Widnell, and D. Wilcox. 1983. Superinfection exclusion by vesicular stomatitis virus. Virology 131:137-143. [DOI] [PubMed] [Google Scholar]
- 39.Yeh, C., Y. Liaw, and J. Ou. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 64:6141-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y., and J. Summers. 1999. Enrichment of a precore-minus mutant of duck hepatitis B virus in experimental mixed infections. J. Virol. 73:3616-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., and J. Summers. 2000. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J. Virol. 74:5257-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]