Abstract
Background
Cadherin13 (CDH13) is a glycosylphosphatidylinositol-anchored cell adhesion molecule that plays a crucial role in morphogenesis and the maintenance of neuronal circuitry. CDH13 has been implicated in the susceptibility to a variety of psychiatric diseases. A recent genome-wide association study using Danish samples showed, for the first time, the involvement of a single nucleotide polymorphism (SNP) of CDH13 (intronic SNP rs8057927) in schizophrenia. Here, we investigated the association between other SNPs of CDH13 and schizophrenia and tried to replicate the association for the SNP of rs8057927, in the Japanese population.
Methods
Using TaqMan® SNP genotyping assays, five tag SNPs (rs12925602, rs7193788, rs736719, rs6565051, and rs7204454) in the promoter region of CDH13 were examined for their association with schizophrenia in two independent samples. The first sample comprised 665 patients and 760 controls, and the second sample comprised 677 patients and 667 controls. One tag SNP for rs8057927 was also examined for the association with schizophrenia in the first sample set.
Results
A GACAG haplotype of the five SNPs in the promoter region of CDH13 was significantly associated with schizophrenia in the first sample set (P=0.016 and corrected P=0.098). A combined analysis of the GACAG haplotype with the second sample set enhanced the significance (P=0.0026 and corrected P=0.021). We found no association between rs8057927 and schizophrenia in the first sample set.
Conclusion
Our results suggest that CDH13 may contribute to the genetic risk of schizophrenia. Further replication on the association of CDH13 with schizophrenia and functional studies are required to confirm the current findings.
Keywords: CDH13, promoter region, haplotype, SNP
Introduction
Schizophrenia is a severe mental disorder that ranks among the world’s top ten causes of long-term disability, with a worldwide prevalence of approximately 1%. Although the causes of schizophrenia are still largely unknown, previous studies have suggested that the inheritability of schizophrenia is high and that there is a small but significant environmental effect associated with the susceptibility to schizophrenia.1,2
Recent genome-wide association study (GWASs) has shown that common variants of single nucleotide polymorphisms (SNPs) with relatively weak effects may be associated with schizophrenia.3 Meanwhile, it is well established that macroscopic abnormalities, such as volume reductions of the prefrontal cortex, hippocampus, and generalized brain, are associated with schizophrenia.4,5 In addition, significant alterations in neuron size, morphology, and synaptic connectivity have been reported.6–8 These past studies suggest that neural development and mature brain function-related genes may also be schizophrenia- associated genes.
Cadherins (CDHs) belong to a superfamily of cell adhesion molecules that regulate morphogenesis by mediating cell adhesion. In the nervous system, CDHs play crucial roles in neural tube regionalization, neuronal migration, gray matter differentiation, neural circuit formation, spine morphology, and synapse formation and remodeling.9,10 The finding that the gene locus of the CDH superfamily overlaps with potential regions underlying schizophrenia susceptibility implicate an association between CDHs and schizophrenia.11,12 For example, protocadherin12 (PCDH12) and CDH18 are candidate genes that have been indicated to confer an increased risk for schizophrenia.7,12
CDH13, also known as H-cadherin or T-cadherin, belongs to the CDH superfamily. In humans, CDH13 is located on chromosome 16q23 and contains 1,169.8 kbp. Although the classical extracellular CDH structure is conserved, CDH13 lacks transmembrane and cytoplasmic domains and is anchored to the cellular membrane through glycosylphosphatidylinositol.13 CDH13 has been implicated in the susceptibility to a variety of psychiatric diseases. A GWAS of attention deficit hyperactivity disorder (ADHD) identified CDH13 as one of the genes that is most highly associated with ADHD,14 and a meta-analysis of ADHD linkage scans indicated the only genome-wide significant region overlapped with CDH13.15 GWASs have also indicated the involvement of CDH13 in depression,16 autism,17,18 alcohol dependence,19 nicotine dependence,20 and methamphetamine dependence.21 Recently, a GWAS of Danish samples indicated that rs8057927 in the intron of CDH13 is associated with schizophrenia.22 Although it was the first report to show an involvement of CDH13 in schizophrenia, rs8057927 in the intron of CDH13 is not a variant of coding region or promoter region. Therefore, the functional significance of rs8057927 in the intron of CDH13 remains unclear. In addition, there is a possibility that other SNPs in the coding region and/or promoter region of CDH13 are associated with schizophrenia.
Our present study was designed to investigate the association between coding or regulatory SNPs of CDH13 and schizophrenia, and to replicate the association for the SNP rs8057927, in the Japanese population. Here, we focused on five tag SNPs from the linkage disequilibrium (LD) block in the promoter region of CDH13 because we found neither cis-acting SNPs nor nonsynonymous SNPs after consulting the databases: mRNA by SNP Browser (http://www.sph.umich.edu/csg/liang/asthma/)23 and Japanese SNP (JSNP) DATABASE (http://snp.ims.u-tokyo.ac.jp).24
Materials and methods
Subjects
The present study was approved by the Ethical Committee for Genetic Studies of Kobe University Graduate School of Medicine and the Ethics Committee of Genetics at Niigata University School of Medicine. Informed consent was obtained from all of the participants. All of the participants were of Japanese descent and were recruited in the Kobe city area (the first set) or the Niigata area (the second set) of Japan.
The first set of participants consisted of 665 unrelated schizophrenia patients, including 344 males (with mean age ± standard deviation [SD] of 53.3±14.0 years) and 321 females (53.5±15.2 years), and 760 unrelated healthy volunteers (359 males [53.1±18.9 years]; 401 females [54.9±18.3 years]). There were no significant differences in the sex (χ2=1.277, P=0.258) and age (t=0.792, df=1381, P=0.429) distributions between the schizophrenia and the control groups. The second set consisted of 677 unrelated schizophrenia patients (363 males [39.5±13.3 years]; 314 females [39.7±14.3 years]) and 667 unrelated healthy volunteers (341 males [36.7±9.5 years]; 326 females [40.0±11.8 years]). There were no significant differences in the sex (χ2=0.838, P=0.360) and age (t=1.897, df=1,336, P=0.058) distributions between the schizophrenia and the control groups.
The psychiatric assessment of each participant was conducted as previously described.25,26 In brief, the patients were diagnosed by at least two psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition DSM-IV27 criteria for schizophrenia, based on unstructured interviews and reviews of their medical records at each hospital. None of the patients had a history of substance abuse (excluding nicotine dependence) or organic mental disorders. All of the control subjects were interviewed and screened for psychiatric disorders, based on an unstructured interview by a psychiatrist. None of the control subjects had any present, past, or family (up to first-degree relatives) histories of psychiatric disorders or substance abuse (excluding nicotine dependence).
SNP selection and genotyping
We first identified one LD block in the promoter region of CDH13 from the HapMap database (release#27, www.hapmap.org) (population: Japanese Tokyo, minor allele frequencies [MAFs] of more than 0.05), using the Haploview software program version 4.2 (http://www.broad.mit.edu/mpg/haploview/).28 We then selected five tagging SNPs (rs12925602, rs7193788, rs736719, rs6565051, and rs7204454) from the LD block, with the criterion of an r2 threshold greater than 0.8 in “pair-wise tagging only” mode, using the “Tagger” program in the Haploview software, and we used these SNPs in the following association analysis.
For genotype determination, peripheral blood was drawn from all of the participants, and the leukocyte DNA was extracted. We used TaqMan® assays (Applied Biosystems®; Life Technologies Corp, Carlsbad, CA, USA) for genotyping all of the SNPs. We selected predesigned TaqMan SNP genotyping assays from the Life Technologies database for all five SNPs that were examined. The genotyping was performed according to the protocol recommended by the manufacturer.
Although we also tried to investigate the intronic SNP rs8057927 previously reported for its involvement in schizophrenia in the Danish population,22 TaqMan assays for genotyping rs8057927 were not available. Therefore, we chose rs8049308 as the substitute for rs8057927 because rs8049308 is a tag SNP for rs8057927 (these two SNPs have strong LD to each other [D’=1.0, r2=0.946]) (Figure S1).
Statistics
We used the Haploview software to determine the Hardy–Weinberg equilibrium (HWE), LD, allelic/haplotype frequencies, and genetic association, between the schizophrenia and control groups. The allele-based association was tested using the χ2 test. If necessary, permutation tests based on 10,000 replications were performed to calculate the corrected P-values of the allelic or haplotypic analyses for multiple testing by the Haploview software. The genotype-based association was evaluated using the Cochran–Armitage trend test. The haplotype-based association was examined using the χ2 test and the Fisher’s exact test, using R version 2.15.0 (The R Foundation for Statistical Computing, Vienna, Austria). The power analysis was performed using the Power and Sample Size Calculations Version3.1.2 program with an α of 0.05.29 Statistical significance was defined at P<0.05.
Results
rs12925602, rs7193788, rs736719, rs6565051, and rs7204454
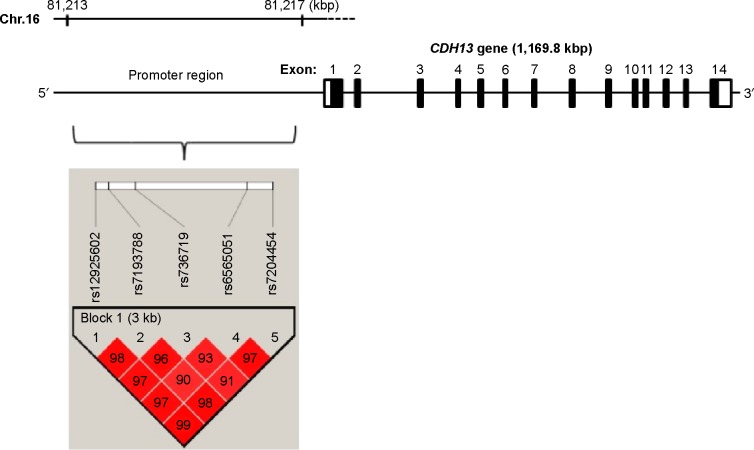
The distributions of all of the SNPs did not deviate from the HWE in each set. Using the solid spine method, five selected SNPs (rs12925602, rs7193788, rs736719, rs6565051, and rs7204454) in LD with each other formed one haplotype block (D’=0.90–0.99) (Figure 1). The allelic frequencies of the tag SNPs in the promoter region of CDH13 are shown in Table 1. Neither the genotype distribution nor the allelic frequency of these five SNPs was significantly associated with schizophrenia in either set. Even when the data of the first and second set were combined, no significant difference was found.
Figure 1.
Cadherin13 (CDH13) tag single nucleotide polymorphisms (SNPs) and the genetic structure of CDH13. The genetic structure of CDH13 is shown at the top. The gene consists of fourteen exons spanning 1,169.8 kbp. Linkage disequilibrium (D’ values) of five SNPs studied here are shown.
Table 1.
Association between CDH13 SNPs with schizophrenia
| Sample | SNP ID positiona | Phen | Genotype distribution
|
Minor allele
|
P-value
|
Power | OR (95% CI) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | Mm | mm | MAF | Allele | HWE | Genotypeb | Allelec | |||||
| rs12925602, rs7193788, rs736719, rs6565051, and rs7204454 | ||||||||||||
| First set SCZ, n=665 CON, n=760 |
rs12925602 | SCZ | 418 | 198 | 29 | 0.198 | A | 0.428 | 0.708 | 0.696 | 0.059 | 0.97 (0.80–1.16) |
| 81213402 | CON | 471 | 255 | 26 | 0.204 | 0.281 | (0.991) | |||||
| rs7193788 | SCZ | 203 | 304 | 139 | 0.449 | G | 0.269 | 0.327 | 0.369 | 0.099 | 1.08 (0.93–1.25) | |
| 81213661 | CON | 234 | 384 | 132 | 0.432 | 0.241 | (0.815) | |||||
| rs736719 | SCZ | 515 | 121 | 5 | 0.102 | T | 0.643 | 0.130 | 0.138 | 0.188 | 0.83 (0.66–1.06) | |
| 81214146 | CON | 575 | 161 | 9 | 0.12 | 0.698 | (0.436) | |||||
| rs6565051 | SCZ | 275 | 275 | 96 | 0.363 | G | 0.062 | 0.905 | 0.946 | 0.050 | 0.99 (0.85–1.16) | |
| 81216229 | CON | 310 | 337 | 105 | 0.364 | 0.479 | (1.000) | |||||
| rs7204454 | SCZ | 268 | 284 | 93 | 0.364 | C | 0.271 | 0.473 | 0.454 | 0.085 | 1.06 (0.91–1.24) | |
| 81216695 | CON | 315 | 342 | 92 | 0.350 | 0.982 | (0.892) | |||||
| Second set SCZ, n=677 CON, n=667 |
rs12925602 | SCZ | 433 | 215 | 27 | 0.199 | A | 1.000 | 0.3505 | 0.3476 | 0.100 | 1.10 (0.91–1.33) |
| 81213402 | CON | 444 | 196 | 25 | 0.185 | 0.629 | (0.786) | |||||
| rs7193788 | SCZ | 220 | 329 | 128 | 0.432 | G | 0.845 | 0.2825 | 0.6671 | 0.060 | 1.09 (0.94–1.27) | |
| 81213661 | CON | 244 | 316 | 123 | 0.424 | 0.575 | (0.985) | |||||
| rs736719 | SCZ | 522 | 140 | 15 | 0.126 | T | 0.180 | 0.1495 | 0.1455 | 0.176 | 1.19 (0.94–1.51) | |
| 81214146 | CON | 528 | 131 | 6 | 0.108 | 0.669 | (0.445) | |||||
| rs6565051 | SCZ | 265 | 308 | 101 | 0.378 | G | 0.497 | 0.3395 | 0.3389 | 0.104 | 0.93 (0.79–1.08) | |
| 81216229 | CON | 237 | 324 | 100 | 0.396 | 0.600 | (0.774) | |||||
| rs7204454 | SCZ | 289 | 296 | 83 | 0.346 | C | 0.639 | 0.7327 | 0.7267 | 0.056 | 0.97 (0.83–1.14) | |
| 81216695 | CON | 288 | 279 | 93 | 0.352 | 0.068 | (0.993) | |||||
| Combined SCZ, n=1,342 CON, n=1,427 |
rs12925602 | SCZ | 851 | 413 | 56 | 0.199 | A | 0.555 | 0.729 | 0.738 | 0.058 | 1.02 (0.90–1.17) |
| 81213402 | CON | 915 | 451 | 51 | 0.195 | 0.690 | (0.994) | |||||
| rs7193788 | SCZ | 423 | 633 | 267 | 0.440 | G | 0.334 | 0.161 | 0.366 | 0.098 | 1.08 (0.97–1.20) | |
| 81213661 | CON | 478 | 700 | 255 | 0.428 | 0.670 | (0.811) | |||||
| rs736719 | SCZ | 1,037 | 261 | 20 | 0.114 | T | 0.511 | 0.999 | 0.992 | 0.050 | 1.00 (0.85–1.18) | |
| 81214146 | CON | 1,103 | 292 | 15 | 0.114 | 0.462 | (1.000) | |||||
| rs6565051 | SCZ | 540 | 583 | 197 | 0.371 | G | 0.067 | 0.503 | 0.520 | 0.072 | 0.96 (0.86–1.07) | |
| 81216229 | CON | 547 | 661 | 205 | 0.379 | 0.909 | (0.931) | |||||
| rs7204454 | SCZ | 557 | 580 | 176 | 0.355 | C | 0.243 | 0.806 | 0.788 | 0.056 | 1.01 (0.91–1.13) | |
| 81216695 | CON | 603 | 621 | 185 | 0.351 | 0.245 | (0.997) | |||||
| rs8049308 (as the substitute for rs8057927) | ||||||||||||
| First set SCZ, n=665 CON, n=760 |
rs8049308 | SCZ | 309 | 267 | 69 | 0.314 | C | 0.357 | 0.634 | 0.630 | 0.066 | 1.04 (0.89–1.22) |
| 81252503 | CON | 363 | 313 | 72 | 0.305 | 0.753 | (0.658) | |||||
Notes:
SNP ID number and positions are available at http://hapmap.ncbi.nlm.nih.gov/.
Genotypic P-values were tested with the Cochran-Armitage test for trend.
Allelic P-values were tested with χ2; corrections for multiple comparisons are in parentheses (for 10,000 permutations).
Abbreviations: CDH13, cadherin13; CI, confidence interval; CON, control; HWE, Hardy–Weinberg equilibrium; M, major allele; m, minor allele; MAF, minor allele frequency; OR, odds ratio; Phen, phenotype; SCZ, schizophrenia; SNP, single nucleotide polymorphism; SNP ID, single nucleotide polymorphism identification.
Detailed haplotype frequencies between the schizophrenia and control groups are shown in Table 2. Each haplotype analysis of the LD block revealed a nominal significant distribution of the GACAG haplotype between the schizophrenia and control groups in the first set (P=0.016). Although no significant difference was found in the second set, the distributions of each haplotype between the schizophrenia and control groups were similar to those in the first sample. When the data of the first and second set were combined, the significance was enhanced for the GACAG haplotype (P=0.0026). The GACAG haplotype was also significantly associated with schizophrenia even after correction for multiple testing (corrected P=0.021). The frequency of the GACAG haplotype in the schizophrenia group (0.006) was lower than that in the control group (0.014).
Table 2.
Association between haplotypes in the promoter region of CDH13 and schizophrenia
| Sample | Haplotype | Haplotype frequency
|
χ2 | P-value* | Global P-values | OR (95% CI) | |
|---|---|---|---|---|---|---|---|
| Schizophrenia | Control | ||||||
| rs12925602–rs7204454 | |||||||
| First set SCZ, n=665 CON, n=760 |
GACGG | 0.343 | 0.336 | 0.162 | 0.688 (0.999) |
χ2=9.87, df=6 P-value =0.130 (P-value =0.125 by Fisher’s exact test) |
1.03 (0.88–1.21) |
| GGCAC | 0.261 | 0.232 | 3.072 | 0.080 (0.492) | 1.17 (0.98–1.39) | ||
| AACAG | 0.191 | 0.201 | 0.441 | 0.507 (0.997) | 0.94 (0.78–1.13) | ||
| GGTAC | 0.097 | 0.107 | 0.863 | 0.353 (0.979) | 0.89 (0.70–1.14) | ||
| GGCAG | 0.071 | 0.066 | 0.262 | 0.609 (0.999) | 1.08 (0.81–1.45) | ||
| GGCGG | 0.011 | 0.011 | 0.019 | 0.890 (1.000) | 0.95 (0.47–1.94) | ||
| GACAG | 0.009 | 0.020 | 5.842 | 0.016 (0.098)** | 0.44 (0.21–0.87)** | ||
| Second set SCZ, n=677 CON, n=667 |
GACGG | 0.362 | 0.384 | 1.389 | 0.239 (0.881) |
χ2=7.26, df=6 P-value =0.298 (P-value =0.303 by Fisher’s exact test) |
0.91 (0.78–1.06) |
| GGCAC | 0.224 | 0.242 | 1.208 | 0.272 (0.917) | 0.90 (0.76–1.08) | ||
| AACAG | 0.200 | 0.185 | 0.911 | 0.340 (0.967) | 1.10 (0.91–1.33) | ||
| GGTAC | 0.120 | 0.107 | 1.113 | 0.292 (0.938) | 1.14 (0.90–1.44) | ||
| GGCAG | 0.070 | 0.061 | 0.773 | 0.380 (0.975) | 1.15 (0.85–1.56) | ||
| GGCGG | 0.014 | 0.012 | 0.341 | 0.559 (0.996) | 1.22 (0.62–2.40) | ||
| GACAG | 0.003 | 0.008 | 2.612 | 0.106 (0.575) | 0.40 (0.13–1.26) | ||
| Combined SCZ, n=1,342 CON, n=1,427 |
GACGG | 0.352 | 0.359 | 0.229 | 0.632 (1.000) |
χ2=9.90, df=6 P-value =0.129 (P-value =0.122 by Fisher’s exact test) |
0.97 (0.87–1.09) |
| GGCAC | 0.242 | 0.237 | 0.176 | 0.675 (1.000) | 1.03 (0.91–1.16) | ||
| AACAG | 0.196 | 0.194 | 0.033 | 0.855 (1.000) | 1.01 (0.89–1.16) | ||
| GGTAC | 0.109 | 0.107 | 0.033 | 0.8559 (1.000) | 1.02 (0.86–1.21) | ||
| GGCAG | 0.071 | 0.064 | 0.953 | 0.3289 (0.995) | 1.11 (0.90–1.37) | ||
| GGCGG | 0.012 | 0.012 | 0.040 | 0.8418 (1.000) | 1.05 (0.65–1.71) | ||
| GACAG | 0.006 | 0.014 | 9.100 | 0.0026 (0.021)** | 0.41 (0.23–0.75)** | ||
Notes:
This column shows the nominal P-values and the corrected P-values for multiple testing (for 10,000 permutations).
Significant differences between the schizophrenia and control groups.
Abbreviations: CDH13, cadherin13; CI, confidence interval; CON, control; OR, odds ratio; SCZ, schizophrenia.
rs8049308 (as the substitute for rs8057927)
The allelic frequency of rs8049308 in the first set is shown in Table 1. The distributions of this SNP did not differ from the HWE in the first set. Neither the genotype distribution nor the allelic frequency of rs8049308 was significantly associated with schizophrenia.
Discussion
Here we showed that SNPs in the promoter region of CDH13 are associated with schizophrenia in the Japanese population. Although CDH13 has been implicated in the susceptibility to a variety of psychiatric diseases,14–21 there has been no report regarding the association between CDH13 and schizophrenia except for a recent GWAS of a Danish population sample.22 In addition, this recent GWAS found the association between schizophrenia and an intron of CDH13 but not the promoter region. Therefore, our present study was the first to investigate the association of the promoter region of CDH13 with schizophrenia in the Japanese population.
In the human adult brain, CDH13 expression is detected in the prefrontal cortex, hippocampus, hypothalamus, amygdala, and substantia nigra (http://www.gtexportal.org/),30 which overlap with regions linked to a variety of psychiatric diseases including schizophrenia.13,31 CDH13 might have a role as an axonal pathfinder during neurodevelopment and play a role in the maintenance of inhibitory and excitatory synapses after maturation of neuronal circuits.32 In addition, altered excitation/inhibition balance caused by the dysfunction or loss of inhibitory interneurons has been associated with the pathophysiology of schizophrenia.33,34 These past studies suggest the involvement of CDH13 in the pathophysiology of schizophrenia. Therefore, the attention to CDH13 in this manuscript may be reasonable, and further studies are needed to confirm the role of CDH13 in the pathophysiology of schizophrenia.
Our results showed significant differences in the distribution of the GACAG haplotype in the promoter region of CDH13 between schizophrenia patients and healthy controls. Based on the frequency of the haplotype, the GACAG haplotype may have a protective role. None of the SNPs in the promoter region of CDH13 evaluated in this study revealed a statistically significant association of the CDH13 locus with schizophrenia. One reason is that the sample size was too small to detect an association of CDH13 SNPs with schizophrenia. Based on the observed allele frequencies of rs12925602, rs7193788, rs736719, rs6565051, and rs7204454, the current combined samples provide powers of 0.058, 0.098, 0.050, 0.072, and 0.056, respectively, to detect nominally significant results. A recent mega analysis by the Psychiatric Genomics Consortium did not identify any association between CDH13 SNPs and schizophrenia.35 Although their analysis included 492 schizophrenia and 427 control Japanese samples, most of their samples were from European populations. Genetic association of CDH13 SNPs with schizophrenia may be variable in different ethnic populations. Therefore, further studies with larger samples in the Japanese and other Asian populations are needed.
As shown in Table S1, the genotype and allele frequencies of the SNPs (rs12925602, rs7193788, rs736719, rs6565051, and rs7204454) are different among populations. The distributions of haplotypes of the five SNPs (rs12925602–rs7204454) are also different (Table S2). The frequency of the GACAG haplotype is rare among Asian and Caucasian populations, while the frequency of this haplotype in Africans is 0.024–0.126.28 Therefore, replication studies, especially in other Asian populations and African populations, are required to confirm the findings of our present study.
Although we also conducted a case-control study for the intronic SNP rs8049308 as the substitute for rs8057927, which previously indicated an association with schizophrenia in the Danish samples,22 neither the genotype distribution nor the allelic frequency of rs8049308 was significantly associated with schizophrenia in the first set. As shown in Table S1, the genotype and allele frequencies of rs8057927 and rs8049308 in the Caucasian populations are significantly lower compared with the Asian populations. These differences may explain why the result identified in the Danish samples was not replicated in our Japanese samples.
A limitation in the present study should be considered. The number of subjects in the association study was small and may not have been large enough to detect a significant difference because the genetic impact of CDH13 on schizophrenia may be mild. Therefore, further investigations with larger sample sizes are needed to confirm the present results.
The results reported here raise the question: do nucleotide substitutions in the CDH13 promoter actually affect the transcriptional activity of the CDH13 promoter? Our computational analysis using the TFBIND (http://tfbind.hgc.jp/)36 revealed that most of the SNPs we studied here were located in the putative transcription factor binding sites (Table S3). This suggests that nucleotide substitution in the CDH13 promoter region may affect the transcriptional activity of this promoter region by affecting the ability of this promoter region to bind to transcription factors. To test this hypothesis, transcriptional assays, such as a luciferase assay, are required in future studies.
Conclusion
The present study suggests that haplotype variants in the promoter region of CDH13 may affect the susceptibility to schizophrenia. To confirm this result, further replication studies using larger sample sizes and different populations and functional studies are required.
Supplementary materials
The intronic SNPs (rs8057927 and rs8049308) have strong LD to each other (D’=1.0, r2=0.946). rs8049308 is a tag SNP for rs8057927 with the criteria of r2 threshold greater than 0.8 in ‘pair-wise tagging only’ mode using the ‘Tagger’ program in the Haploview software.
Abbreviations: CDH13, cadherin13; SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium.
Table S1.
Genotype frequencies and allele frequencies of CDH13 SNPs in different ethnic populations
| SNP | Population | Genotype frequencies
|
Allele frequencies
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Freq | Count | Genotype | Freq | Count | Genotype | Freq | Count | Total | Allele | Freq | Count | Allele | Freq | Count | Total | ||
| rs12925602 | JPT | G/G | 0.708 | 80 | A/G | 0.265 | 30 | A/A | 0.027 | 3 | 113 | G | 0.841 | 190 | A | 0.159 | 36 | 226 |
| CHB | G/G | 0.708 | 97 | A/G | 0.255 | 35 | A/A | 0.036 | 5 | 137 | G | 0.836 | 229 | A | 0.164 | 45 | 274 | |
| CHD | G/G | 0.642 | 70 | A/G | 0.339 | 37 | A/A | 0.018 | 2 | 109 | G | 0.812 | 177 | A | 0.188 | 41 | 218 | |
| GIH | G/G | 0.802 | 81 | A/G | 0.149 | 15 | A/A | 0.050 | 5 | 101 | G | 0.876 | 177 | A | 0.124 | 25 | 202 | |
| CEU | G/G | 0.850 | 96 | A/G | 0.142 | 16 | A/A | 0.009 | 1 | 113 | G | 0.920 | 208 | A | 0.080 | 18 | 226 | |
| TSI | G/G | 0.745 | 76 | A/G | 0.255 | 26 | A/A | 0.000 | 0 | 102 | G | 0.873 | 178 | A | 0.127 | 26 | 204 | |
| ASW | G/G | 0.750 | 42 | A/G | 0.250 | 14 | A/A | 0.000 | 0 | 56 | G | 0.875 | 98 | A | 0.125 | 14 | 112 | |
| LWK | G/G | 0.782 | 86 | A/G | 0.191 | 21 | A/A | 0.027 | 3 | 110 | G | 0.877 | 193 | A | 0.123 | 27 | 220 | |
| MKK | G/G | 0.833 | 130 | A/G | 0.160 | 25 | A/A | 0.006 | 1 | 156 | G | 0.913 | 285 | A | 0.087 | 27 | 312 | |
| YRI | G/G | 0.789 | 116 | A/G | 0.204 | 30 | A/A | 0.007 | 1 | 147 | G | 0.891 | 262 | A | 0.109 | 32 | 294 | |
| MEX | G/G | 0.741 | 43 | A/G | 0.224 | 13 | A/A | 0.034 | 2 | 58 | G | 0.853 | 99 | A | 0.147 | 17 | 116 | |
| rs7193788 | JPT | A/A | 0.265 | 30 | A/G | 0.504 | 57 | G/G | 0.230 | 26 | 113 | A | 0.518 | 117 | G | 0.482 | 109 | 226 |
| CHB | A/A | 0.285 | 39 | A/G | 0.445 | 61 | G/G | 0.270 | 37 | 137 | A | 0.507 | 139 | G | 0.493 | 135 | 274 | |
| CHD | A/A | 0.275 | 30 | A/G | 0.560 | 61 | G/G | 0.165 | 18 | 109 | A | 0.555 | 121 | G | 0.445 | 97 | 218 | |
| GIH | A/A | 0.584 | 59 | A/G | 0.366 | 37 | G/G | 0.050 | 5 | 101 | A | 0.767 | 155 | G | 0.233 | 47 | 202 | |
| CEU | A/A | 0.690 | 78 | A/G | 0.274 | 31 | G/G | 0.035 | 4 | 113 | A | 0.827 | 187 | G | 0.173 | 39 | 226 | |
| TSI | A/A | 0.784 | 80 | A/G | 0.186 | 19 | G/G | 0.029 | 3 | 102 | A | 0.877 | 179 | G | 0.123 | 25 | 204 | |
| ASW | A/A | 0.719 | 41 | A/G | 0.246 | 14 | G/G | 0.035 | 2 | 57 | A | 0.842 | 96 | G | 0.158 | 18 | 114 | |
| LWK | A/A | 0.691 | 76 | A/G | 0.273 | 30 | G/G | 0.036 | 4 | 110 | A | 0.827 | 182 | G | 0.173 | 38 | 220 | |
| MKK | A/A | 0.679 | 106 | A/G | 0.282 | 44 | G/G | 0.038 | 6 | 156 | A | 0.821 | 256 | G | 0.179 | 56 | 312 | |
| YRI | A/A | 0.796 | 117 | A/G | 0.190 | 28 | G/G | 0.014 | 2 | 147 | A | 0.891 | 262 | G | 0.109 | 32 | 294 | |
| MEX | A/A | 0.741 | 43 | A/G | 0.241 | 14 | G/G | 0.017 | 1 | 58 | A | 0.862 | 100 | G | 0.138 | 16 | 116 | |
| rs736719 | JPT | C/C | 0.779 | 88 | C/T | 0.186 | 21 | T/T | 0.035 | 4 | 113 | C | 0.872 | 197 | T | 0.128 | 37 | 226 |
| CHB | C/C | 0.679 | 93 | C/T | 0.277 | 38 | T/T | 0.044 | 6 | 133 | C | 0.818 | 224 | T | 0.182 | 50 | 274 | |
| CHD | C/C | 0.688 | 75 | C/T | 0.303 | 33 | T/T | 0.009 | 1 | 109 | C | 0.839 | 183 | T | 0.161 | 35 | 218 | |
| GIH | C/C | 0.762 | 77 | C/T | 0.228 | 23 | T/T | 0.010 | 1 | 101 | C | 0.876 | 177 | T | 0.124 | 25 | 202 | |
| CEU | C/C | 0.699 | 79 | C/T | 0.265 | 30 | T/T | 0.035 | 4 | 113 | C | 0.832 | 188 | T | 0.168 | 38 | 226 | |
| TSI | C/C | 0.784 | 80 | C/T | 0.186 | 19 | T/T | 0.029 | 3 | 102 | C | 0.877 | 179 | T | 0.123 | 25 | 204 | |
| ASW | C/C | 0.737 | 42 | C/T | 0.228 | 13 | T/T | 0.035 | 2 | 57 | C | 0.851 | 97 | T | 0.149 | 17 | 114 | |
| LWK | C/C | 0.700 | 77 | C/T | 0.264 | 29 | T/T | 0.036 | 4 | 110 | C | 0.832 | 183 | T | 0.168 | 37 | 220 | |
| MKK | C/C | 0.679 | 106 | C/T | 0.288 | 45 | T/T | 0.032 | 5 | 156 | C | 0.824 | 257 | T | 0.176 | 55 | 312 | |
| YRI | C/C | 0.796 | 117 | C/T | 0.190 | 28 | T/T | 0.014 | 2 | 147 | C | 0.891 | 262 | T | 0.109 | 32 | 294 | |
| MEX | C/C | 0.776 | 45 | C/T | 0.207 | 12 | T/T | 0.017 | 1 | 58 | C | 0.879 | 102 | T | 0.121 | 14 | 116 | |
| rs6565051 | JPT | G/G | 0.133 | 15 | A/G | 0.469 | 53 | A/A | 0.398 | 45 | 113 | G | 0.367 | 83 | A | 0.633 | 143 | 226 |
| CHB | G/G | 0.146 | 20 | A/G | 0.416 | 57 | A/A | 0.438 | 60 | 137 | G | 0.354 | 97 | A | 0.646 | 177 | 274 | |
| CHD | G/G | 0.148 | 16 | A/G | 0.463 | 50 | A/A | 0.389 | 42 | 108 | G | 0.380 | 82 | A | 0.620 | 134 | 216 | |
| GIH | G/G | 0.079 | 8 | A/G | 0.356 | 36 | A/A | 0.564 | 57 | 101 | G | 0.257 | 83 | A | 0.743 | 150 | 202 | |
| CEU | G/G | 0.071 | 8 | A/G | 0.354 | 40 | A/A | 0.575 | 65 | 113 | G | 0.248 | 56 | A | 0.752 | 170 | 226 | |
| TSI | G/G | 0.108 | 11 | A/G | 0.461 | 47 | A/A | 0.431 | 44 | 102 | G | 0.338 | 69 | A | 0.662 | 135 | 204 | |
| ASW | G/G | 0.088 | 5 | A/G | 0.421 | 24 | A/A | 0.491 | 28 | 57 | G | 0.298 | 34 | A | 0.702 | 80 | 114 | |
| LWK | G/G | 0.073 | 8 | A/G | 0.355 | 39 | A/A | 0.573 | 63 | 110 | G | 0.250 | 55 | A | 0.750 | 165 | 220 | |
| MKK | G/G | 0.052 | 8 | A/G | 0.426 | 66 | A/A | 0.523 | 81 | 155 | G | 0.265 | 82 | A | 0.735 | 228 | 310 | |
| YRI | G/G | 0.095 | 14 | A/G | 0.442 | 65 | A/A | 0.463 | 68 | 147 | G | 0.316 | 93 | A | 0.684 | 201 | 294 | |
| MEX | G/G | 0.140 | 8 | A/G | 0.509 | 29 | A/A | 0.351 | 20 | 57 | G | 0.395 | 45 | A | 0.605 | 69 | 114 | |
| rs7204454 | JPT | G/G | 0.319 | 36 | C/G | 0.540 | 61 | C/C | 0.142 | 16 | 113 | G | 0.588 | 133 | C | 0.412 | 93 | 226 |
| CHB | G/G | 0.382 | 52 | C/G | 0.441 | 60 | C/C | 0.176 | 24 | 136 | G | 0.603 | 164 | C | 0.397 | 108 | 272 | |
| CHD | G/G | 0.394 | 43 | C/G | 0.486 | 53 | C/C | 0.119 | 13 | 109 | G | 0.638 | 139 | C | 0.362 | 79 | 218 | |
| GIH | G/G | 0.158 | 16 | C/G | 0.406 | 41 | C/C | 0.436 | 44 | 101 | G | 0.361 | 73 | C | 0.639 | 129 | 202 | |
| CEU | G/G | 0.100 | 11 | C/G | 0.436 | 48 | C/C | 0.464 | 51 | 110 | G | 0.318 | 70 | C | 0.682 | 150 | 220 | |
| TSI | G/G | 0.147 | 15 | C/G | 0.598 | 61 | C/C | 0.255 | 26 | 102 | G | 0.446 | 91 | C | 0.554 | 113 | 204 | |
| ASW | G/G | 0.263 | 15 | C/G | 0.439 | 25 | C/C | 0.298 | 17 | 57 | G | 0.482 | 55 | C | 0.518 | 59 | 114 | |
| LWK | G/G | 0.164 | 18 | C/G | 0.536 | 59 | C/C | 0.300 | 33 | 110 | G | 0.432 | 95 | C | 0.568 | 125 | 220 | |
| MKK | G/G | 0.141 | 22 | C/G | 0.449 | 70 | C/C | 0.410 | 64 | 156 | G | 0.365 | 114 | C | 0.635 | 198 | 312 | |
| YRI | G/G | 0.284 | 40 | C/G | 0.504 | 71 | C/C | 0.213 | 30 | 141 | G | 0.535 | 151 | C | 0.465 | 131 | 282 | |
| MEX | G/G | 0.310 | 18 | C/G | 0.500 | 29 | C/C | 0.190 | 11 | 58 | G | 0.560 | 65 | C | 0.440 | 51 | 116 | |
| rs8057927 | JPT | T/T | 0.478 | 54 | C/T | 0.425 | 48 | C/C | 0.097 | 11 | 113 | T | 0.690 | 156 | C | 0.310 | 70 | 226 |
| CHB | T/T | 0.478 | 65 | C/T | 0.412 | 56 | C/C | 0.110 | 15 | 136 | T | 0.684 | 186 | C | 0.316 | 86 | 272 | |
| CHD | T/T | 0.514 | 56 | C/T | 0.394 | 43 | C/C | 0.092 | 10 | 109 | T | 0.711 | 155 | C | 0.289 | 63 | 218 | |
| GIH | T/T | 0.901 | 91 | C/T | 0.089 | 9 | C/C | 0.010 | 1 | 101 | T | 0.946 | 191 | C | 0.054 | 11 | 202 | |
| CEU | T/T | 0.876 | 99 | C/T | 0.124 | 14 | C/C | 0 | 0 | 113 | T | 0.938 | 212 | C | 0.062 | 14 | 226 | |
| TSI | T/T | 0.853 | 87 | C/T | 0.147 | 15 | C/C | 0 | 0 | 102 | T | 0.926 | 189 | C | 0.074 | 15 | 204 | |
| ASW | T/T | 0.632 | 36 | C/T | 0.351 | 20 | C/C | 0.018 | 1 | 57 | T | 0.807 | 92 | C | 0.193 | 22 | 114 | |
| LWK | T/T | 0.620 | 67 | C/T | 0.324 | 35 | C/C | 0.056 | 6 | 108 | T | 0.782 | 169 | C | 0.218 | 47 | 216 | |
| MKK | T/T | 0.692 | 108 | C/T | 0.250 | 39 | C/C | 0.058 | 9 | 156 | T | 0.817 | 255 | C | 0.183 | 57 | 312 | |
| YRI | T/T | 0.623 | 91 | C/T | 0.336 | 49 | C/C | 0.041 | 6 | 146 | T | 0.791 | 231 | C | 0.209 | 61 | 292 | |
| MEX | T/T | 0.807 | 46 | C/T | 0.175 | 10 | C/C | 0.018 | 1 | 57 | T | 0.895 | 102 | C | 0.105 | 12 | 114 | |
| rs8049308 | JPT | T/T | 0.455 | 20 | C/T | 0.500 | 22 | C/C | 0.045 | 2 | 44 | T | 0.705 | 62 | C | 0.295 | 26 | 88 |
| CHB | T/T | 0.364 | 16 | C/T | 0.477 | 21 | C/C | 0.159 | 7 | 44 | T | 0.602 | 53 | C | 0.398 | 35 | 88 | |
| CEU | T/T | 0.650 | 39 | C/T | 0.333 | 20 | C/C | 0.017 | 1 | 60 | T | 0.817 | 98 | C | 0.183 | 22 | 120 | |
| YRI | T/T | 0.583 | 35 | C/T | 0.383 | 23 | C/C | 0.033 | 2 | 60 | T | 0.775 | 93 | C | 0.225 | 27 | 120 | |
Note: Genotype frequencies and allele frequencies data were determined by the HapMap database (HapMap data release 28, Phase 2+3, August 10), on NCBI B36 assembly, dbSNP b126 (http://hapmap.ncbi.nlm.nih.gov/).
Abbreviations: ASW, African ancestry in southwest USA; CDH13, cadherin13; CEU, residents of UT, USA with Northern and Western European ancestry, from the Centre d’Etude du Polymorphisme Humain collection; CHB, Han Chinese in Beijing, People’s Republic of China; CHD, Chinese in metropolitan Denver, CO, USA; freq, frequency; GIH, Gujarati Indians in Houston, TX, USA; JPT, Japanese in Tokyo, Japan; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, CA, USA; MKK, Maasai in Kinyawa, Kenya; SNP, single nucleotide polymorphism; TSI, Tuscan in Italy; YRI, Yoruba in Ibadan, Nigeria.
Table S2.
Haplotype frequencies of CDH13 SNPs (rs12925602–rs7204454) in different ethnic populations1
| JPT | CHB | CHD | GIH | CEU | TSI | ASW | LWK | MKK | YRI | MEX | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GACGG | 0.355 | 0.351 | 0.347 | 0.233 | 0.244 | 0.335 | 0.286 | 0.289 | 0.231 | 0.287 | 0.394 |
| CGCAC | 0.285 | 0.220 | 0.206 | 0.085 | |||||||
| AACAG | 0.151 | 0.161 | 0.194 | 0.119 | 0.081 | 0.114 | 0.127 | 0.106 | 0.091 | 0.130 | 0.154 |
| GGTAC | 0.128 | 0.179 | 0.135 | 0.142 | 0.162 | 0.114 | 0.103 | 0.167 | 0.154 | 0.087 | 0.106 |
| GGCAG | 0.070 | 0.077 | 0.076 | ||||||||
| GGCGG | 0.012 | 0.006 | 0.018 | 0.006 | |||||||
| GACAG | 0.006 | 0.071 | 0.067 | 0.024 | 0.126 | 0.010 | |||||
| AACGG | 0.006 | 0.017 | |||||||||
| GGCGC | 0.012 | ||||||||||
| AGCAG | 0.006 | ||||||||||
| AACAC | 0.004 | ||||||||||
| GACGC | 0.017 | 0.006 | 0.008 | 0.004 | |||||||
| GGCAC | 0.009 | 0.008 | 0.003 | 0.019 | |||||||
| AGTAC | 0.008 | ||||||||||
| GGTGG | 0.014 | ||||||||||
| GGTGC | 0.006 | 0.014 | 0.022 | 0.010 | |||||||
| GACAC | 0.006 | 0.392 | 0.500 | 0.415 | 0.389 | 0.367 | 0.469 | 0.343 | 0.308 |
Note: Haplotype frequencies data were determined by the Haploview software program (version 4.2; Broad Institute, Cambridge, MA, USA) (http://www.broad.mit.edu/mpg/haploview/).
Abbreviations: ASW, African ancestry in southwest USA; CDH13, cadherin13; CEU, residents of UT, USA with Northern and Western European ancestry, from the Centre d’Etude du Polymorphisme Humain collection; CHB, Han Chinese in Beijing, People’s Republic of China; CHD, Chinese in metropolitan Denver, CO, USA; GIH, Gujarati Indians in Houston, TX, USA; JPT, Japanese in Tokyo, Japan; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, CA, USA; MKK, Maasai in Kinyawa, Kenya; SNPs, single nucleotide polymorphisms; TSI, Tuscan in Italy; YRI, Yoruba in Ibadan, Nigeria.
Table S3.
Putative transcription factor binding site in each SNP on the promoter region of CDH13
| SNP ID | Allele | Sequence | Predicted TF | Binding site | Function |
|---|---|---|---|---|---|
| rs12925602 | A | TCTGCCTACATC[A] AGGAAATTCAGA |
c-Ets- | ATCAAGGAAATT | Regulates numerous genes and involved in stem cell development, cell senescence and death, and tumorigenesis |
| GATA-1 | CATCAAGGA | Regulates the switch of fetal hemoglobin to adult hemoglobin for erythroid development | |||
| CdxA | CA TCAAG | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes | |||
| G | TCTGCCTACATC[G] AGGAAATTCAGA |
c-Ets- | ATCGAGGAAATT | Regulates numerous genes and involved in stem cell development, cell senescence and death, and tumorigenesis | |
| GATA-1 | CATCGAGGA | Regulates the switch of fetal hemoglobin to adult hemoglobin for erythroid development | |||
| rs7193788 | A | GCACGCAGCAGT[A] AAAATACAGAAA |
CdxA | TAAAAATAAAAATA | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes |
| AhR/Ar | GAGCACGCAGCAGTAA | A ligand-activated transcription factor involved in the regulation of biological responses to planar aromatic hydrocarbons | |||
| Sox5 | GTAAAAATAC | A transcription factor involved in the regulation of embryonic development and in the determination of cell fate | |||
| G | GCACGCAGCAGT[G] AAAATACAGAAA |
AhR/Ar | ACGCAGCAGTGAAAA | A ligand-activated transcription factor involved in the regulation of biological responses to planar aromatic hydrocarbons | |
| rs736719 | C | CAGGAAGAAACA[C] GAAGCAGTGTTT |
SRY | AAACACG | A transcription factor and a member of the HMG-box family of DNA binding proteins, which may directly generate some male-specific properties of the brain |
| T | CAGGAAGAAACA[T] GAAGCAGTGTTT |
SRY | AAACATG | A transcription factor and a member of the HMG-box family of DNA binding proteins, which may directly generate some male specific properties of the brain | |
| HNF-3b | GAAGAAACA TGA | A transcription factor and a member of the forkhead class of DNA-binding proteins | |||
| rs6565051 | A | ACCTTCCCTGGA[A] TGGAGAAAAGTC |
C/EBPb | GAATG GAGAAAAGT | A transcription factor that can bind as a homodimer to certain DNA regulatory regions and can also form heterodimers with other C/EBP |
| G | ACCTTCCCTGGA[G] TGGAGAAAAGTC |
C/EBPb | GAGTG GAGAAAAGT | A transcription factor that can bind as a homodimer to certain DNA regulatory regions and can also form heterodimers with other C/EBP | |
| MZF1 | AGTG GAGA | A member of the SCAN domain family transcription factors that form dimers through their highly conserved SCAN motifs | |||
| rs7204454 | C | GTGAGTTCAGTA[C] AATTTGTGTTTT |
CdxA | TACAATT | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes |
| G | GTGAGTTCAGTA[G] AATTTGTGTTTT |
CdxA | TAGAATT | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes |
Abbreviations: CDH13, cadherin13; SNP, single nucleotide polymorphism; SNP ID, single nucleotide polymorphism identification; TF, transcription factor.
Reference
- 1.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Smoking Research Foundation. We thank Y Nagashima and N Yamazaki for technical assistance.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Mueser KT, McGurk SR. Schizophrenia. Lancet. 2004;363(9426):2063–2072. doi: 10.1016/S0140-6736(04)16458-1. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13(8):537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarley RW, Wible CG, Frumin M, et al. MRI anatomy of schizophrenia. Biol Psychiatry. 1999;45(9):1099–1119. doi: 10.1016/s0006-3223(99)00018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49(1–2):1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122(Pt4):593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- 7.Gregório SP, Sallet PC, Do KA, Lin E, Gattaz WF, Dias-Neto E. Polymorphisms in genes involved in neurodevelopment may be associated with altered brain morphology in schizophrenia: preliminary evidence. Psychiatry Res. 2009;165(1–2):1–9. doi: 10.1016/j.psychres.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Stephan KE, Friston KJ, Frith CD. Dysconnection in schizophrenia: from abnormal synaptic plasticity to failures of self-monitoring. Schizophr Bull. 2009;35(3):509–527. doi: 10.1093/schbul/sbn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8(1):11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 10.Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol Rev. 2012;92(2):597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- 11.Pedrosa E, Stefanescu R, Margolis B, et al. Analysis of protocadherin alpha gene enhancer polymorphism in bipolar disorder and schizophrenia. Schizophr Res. 2008;102(1–3):210–219. doi: 10.1016/j.schres.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Rivero O, Sich S, Popp S, Schmitt A, Franke B, Lesch KP. Impact of the ADHD-susceptibility gene CDH13 on development and function of brain networks. Eur Neuropsychopharmacol. 2013;23(6):492–507. doi: 10.1016/j.euroneuro.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Neale BM, Lasky-Su J, Anney R, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou K, Dempfle A, Arcos-Burgos M, et al. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terracciano A, Tanaka T, Sutin AR, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68(9):811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders SJ, Ercan-Sencicek AG, Hus V, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treutlein J, Cichon S, Ridinger M, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66(77):773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhl GR, Drgon T, Johnson C, et al. Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement. Pharmacogenomics. 2010;11(3):357–367. doi: 10.2217/pgs.09.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhl GR, Drgon T, Liu QR, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008;65(3):345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- 22.Børglum AD, Demontis D, Grove J, et al. GROUP investigators 10 Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry. 2014;19(3):325–333. doi: 10.1038/mp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 24.Hirakawa M, Tanaka T, Hashimoto, et al. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 2002;30:158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe Y, Muratake T, Kaneko N, Nunokawa A, Someya T. No association between the brain-derived neurotrophic factor gene and schizophrenia in a Japanese population. Schizophr Res. 2006;84(1):29–35. doi: 10.1016/j.schres.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Okazaki S, Watanabe Y, Hishimoto A, et al. Association analysis of putative cis-acting polymorphisms of interleukin-19 gene with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014;50:151–156. doi: 10.1016/j.pnpbp.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 27.American Psychiatric Association . The Diagnostic and Statistical Manual of Mental Disorders, Fourth edition, text revision (DSM-IV TR) American Psychiatric Association Press; 2000. [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Dupont WD, Plummer WD. Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19(6):589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 30.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benes FM. Amygdalocortical circuitry in schizophrenia: from circuits to molecules. Neuropsychopharmacology. 2010;35(1):239–257. doi: 10.1038/npp.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paradis S, Harrar DB, Lin Y, et al. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53(2):217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477(7363):171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao W, Watanabe T, Cho S, et al. Shank1 regulates excitatory synaptic transmission in mouse hippocampal parvalbumin-expressing inhibitory interneurons. Eur J Neurosci. 2015;41(8):1025–1035. doi: 10.1111/ejn.12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15(7–8):622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The intronic SNPs (rs8057927 and rs8049308) have strong LD to each other (D’=1.0, r2=0.946). rs8049308 is a tag SNP for rs8057927 with the criteria of r2 threshold greater than 0.8 in ‘pair-wise tagging only’ mode using the ‘Tagger’ program in the Haploview software.
Abbreviations: CDH13, cadherin13; SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium.
Table S1.
Genotype frequencies and allele frequencies of CDH13 SNPs in different ethnic populations
| SNP | Population | Genotype frequencies
|
Allele frequencies
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Freq | Count | Genotype | Freq | Count | Genotype | Freq | Count | Total | Allele | Freq | Count | Allele | Freq | Count | Total | ||
| rs12925602 | JPT | G/G | 0.708 | 80 | A/G | 0.265 | 30 | A/A | 0.027 | 3 | 113 | G | 0.841 | 190 | A | 0.159 | 36 | 226 |
| CHB | G/G | 0.708 | 97 | A/G | 0.255 | 35 | A/A | 0.036 | 5 | 137 | G | 0.836 | 229 | A | 0.164 | 45 | 274 | |
| CHD | G/G | 0.642 | 70 | A/G | 0.339 | 37 | A/A | 0.018 | 2 | 109 | G | 0.812 | 177 | A | 0.188 | 41 | 218 | |
| GIH | G/G | 0.802 | 81 | A/G | 0.149 | 15 | A/A | 0.050 | 5 | 101 | G | 0.876 | 177 | A | 0.124 | 25 | 202 | |
| CEU | G/G | 0.850 | 96 | A/G | 0.142 | 16 | A/A | 0.009 | 1 | 113 | G | 0.920 | 208 | A | 0.080 | 18 | 226 | |
| TSI | G/G | 0.745 | 76 | A/G | 0.255 | 26 | A/A | 0.000 | 0 | 102 | G | 0.873 | 178 | A | 0.127 | 26 | 204 | |
| ASW | G/G | 0.750 | 42 | A/G | 0.250 | 14 | A/A | 0.000 | 0 | 56 | G | 0.875 | 98 | A | 0.125 | 14 | 112 | |
| LWK | G/G | 0.782 | 86 | A/G | 0.191 | 21 | A/A | 0.027 | 3 | 110 | G | 0.877 | 193 | A | 0.123 | 27 | 220 | |
| MKK | G/G | 0.833 | 130 | A/G | 0.160 | 25 | A/A | 0.006 | 1 | 156 | G | 0.913 | 285 | A | 0.087 | 27 | 312 | |
| YRI | G/G | 0.789 | 116 | A/G | 0.204 | 30 | A/A | 0.007 | 1 | 147 | G | 0.891 | 262 | A | 0.109 | 32 | 294 | |
| MEX | G/G | 0.741 | 43 | A/G | 0.224 | 13 | A/A | 0.034 | 2 | 58 | G | 0.853 | 99 | A | 0.147 | 17 | 116 | |
| rs7193788 | JPT | A/A | 0.265 | 30 | A/G | 0.504 | 57 | G/G | 0.230 | 26 | 113 | A | 0.518 | 117 | G | 0.482 | 109 | 226 |
| CHB | A/A | 0.285 | 39 | A/G | 0.445 | 61 | G/G | 0.270 | 37 | 137 | A | 0.507 | 139 | G | 0.493 | 135 | 274 | |
| CHD | A/A | 0.275 | 30 | A/G | 0.560 | 61 | G/G | 0.165 | 18 | 109 | A | 0.555 | 121 | G | 0.445 | 97 | 218 | |
| GIH | A/A | 0.584 | 59 | A/G | 0.366 | 37 | G/G | 0.050 | 5 | 101 | A | 0.767 | 155 | G | 0.233 | 47 | 202 | |
| CEU | A/A | 0.690 | 78 | A/G | 0.274 | 31 | G/G | 0.035 | 4 | 113 | A | 0.827 | 187 | G | 0.173 | 39 | 226 | |
| TSI | A/A | 0.784 | 80 | A/G | 0.186 | 19 | G/G | 0.029 | 3 | 102 | A | 0.877 | 179 | G | 0.123 | 25 | 204 | |
| ASW | A/A | 0.719 | 41 | A/G | 0.246 | 14 | G/G | 0.035 | 2 | 57 | A | 0.842 | 96 | G | 0.158 | 18 | 114 | |
| LWK | A/A | 0.691 | 76 | A/G | 0.273 | 30 | G/G | 0.036 | 4 | 110 | A | 0.827 | 182 | G | 0.173 | 38 | 220 | |
| MKK | A/A | 0.679 | 106 | A/G | 0.282 | 44 | G/G | 0.038 | 6 | 156 | A | 0.821 | 256 | G | 0.179 | 56 | 312 | |
| YRI | A/A | 0.796 | 117 | A/G | 0.190 | 28 | G/G | 0.014 | 2 | 147 | A | 0.891 | 262 | G | 0.109 | 32 | 294 | |
| MEX | A/A | 0.741 | 43 | A/G | 0.241 | 14 | G/G | 0.017 | 1 | 58 | A | 0.862 | 100 | G | 0.138 | 16 | 116 | |
| rs736719 | JPT | C/C | 0.779 | 88 | C/T | 0.186 | 21 | T/T | 0.035 | 4 | 113 | C | 0.872 | 197 | T | 0.128 | 37 | 226 |
| CHB | C/C | 0.679 | 93 | C/T | 0.277 | 38 | T/T | 0.044 | 6 | 133 | C | 0.818 | 224 | T | 0.182 | 50 | 274 | |
| CHD | C/C | 0.688 | 75 | C/T | 0.303 | 33 | T/T | 0.009 | 1 | 109 | C | 0.839 | 183 | T | 0.161 | 35 | 218 | |
| GIH | C/C | 0.762 | 77 | C/T | 0.228 | 23 | T/T | 0.010 | 1 | 101 | C | 0.876 | 177 | T | 0.124 | 25 | 202 | |
| CEU | C/C | 0.699 | 79 | C/T | 0.265 | 30 | T/T | 0.035 | 4 | 113 | C | 0.832 | 188 | T | 0.168 | 38 | 226 | |
| TSI | C/C | 0.784 | 80 | C/T | 0.186 | 19 | T/T | 0.029 | 3 | 102 | C | 0.877 | 179 | T | 0.123 | 25 | 204 | |
| ASW | C/C | 0.737 | 42 | C/T | 0.228 | 13 | T/T | 0.035 | 2 | 57 | C | 0.851 | 97 | T | 0.149 | 17 | 114 | |
| LWK | C/C | 0.700 | 77 | C/T | 0.264 | 29 | T/T | 0.036 | 4 | 110 | C | 0.832 | 183 | T | 0.168 | 37 | 220 | |
| MKK | C/C | 0.679 | 106 | C/T | 0.288 | 45 | T/T | 0.032 | 5 | 156 | C | 0.824 | 257 | T | 0.176 | 55 | 312 | |
| YRI | C/C | 0.796 | 117 | C/T | 0.190 | 28 | T/T | 0.014 | 2 | 147 | C | 0.891 | 262 | T | 0.109 | 32 | 294 | |
| MEX | C/C | 0.776 | 45 | C/T | 0.207 | 12 | T/T | 0.017 | 1 | 58 | C | 0.879 | 102 | T | 0.121 | 14 | 116 | |
| rs6565051 | JPT | G/G | 0.133 | 15 | A/G | 0.469 | 53 | A/A | 0.398 | 45 | 113 | G | 0.367 | 83 | A | 0.633 | 143 | 226 |
| CHB | G/G | 0.146 | 20 | A/G | 0.416 | 57 | A/A | 0.438 | 60 | 137 | G | 0.354 | 97 | A | 0.646 | 177 | 274 | |
| CHD | G/G | 0.148 | 16 | A/G | 0.463 | 50 | A/A | 0.389 | 42 | 108 | G | 0.380 | 82 | A | 0.620 | 134 | 216 | |
| GIH | G/G | 0.079 | 8 | A/G | 0.356 | 36 | A/A | 0.564 | 57 | 101 | G | 0.257 | 83 | A | 0.743 | 150 | 202 | |
| CEU | G/G | 0.071 | 8 | A/G | 0.354 | 40 | A/A | 0.575 | 65 | 113 | G | 0.248 | 56 | A | 0.752 | 170 | 226 | |
| TSI | G/G | 0.108 | 11 | A/G | 0.461 | 47 | A/A | 0.431 | 44 | 102 | G | 0.338 | 69 | A | 0.662 | 135 | 204 | |
| ASW | G/G | 0.088 | 5 | A/G | 0.421 | 24 | A/A | 0.491 | 28 | 57 | G | 0.298 | 34 | A | 0.702 | 80 | 114 | |
| LWK | G/G | 0.073 | 8 | A/G | 0.355 | 39 | A/A | 0.573 | 63 | 110 | G | 0.250 | 55 | A | 0.750 | 165 | 220 | |
| MKK | G/G | 0.052 | 8 | A/G | 0.426 | 66 | A/A | 0.523 | 81 | 155 | G | 0.265 | 82 | A | 0.735 | 228 | 310 | |
| YRI | G/G | 0.095 | 14 | A/G | 0.442 | 65 | A/A | 0.463 | 68 | 147 | G | 0.316 | 93 | A | 0.684 | 201 | 294 | |
| MEX | G/G | 0.140 | 8 | A/G | 0.509 | 29 | A/A | 0.351 | 20 | 57 | G | 0.395 | 45 | A | 0.605 | 69 | 114 | |
| rs7204454 | JPT | G/G | 0.319 | 36 | C/G | 0.540 | 61 | C/C | 0.142 | 16 | 113 | G | 0.588 | 133 | C | 0.412 | 93 | 226 |
| CHB | G/G | 0.382 | 52 | C/G | 0.441 | 60 | C/C | 0.176 | 24 | 136 | G | 0.603 | 164 | C | 0.397 | 108 | 272 | |
| CHD | G/G | 0.394 | 43 | C/G | 0.486 | 53 | C/C | 0.119 | 13 | 109 | G | 0.638 | 139 | C | 0.362 | 79 | 218 | |
| GIH | G/G | 0.158 | 16 | C/G | 0.406 | 41 | C/C | 0.436 | 44 | 101 | G | 0.361 | 73 | C | 0.639 | 129 | 202 | |
| CEU | G/G | 0.100 | 11 | C/G | 0.436 | 48 | C/C | 0.464 | 51 | 110 | G | 0.318 | 70 | C | 0.682 | 150 | 220 | |
| TSI | G/G | 0.147 | 15 | C/G | 0.598 | 61 | C/C | 0.255 | 26 | 102 | G | 0.446 | 91 | C | 0.554 | 113 | 204 | |
| ASW | G/G | 0.263 | 15 | C/G | 0.439 | 25 | C/C | 0.298 | 17 | 57 | G | 0.482 | 55 | C | 0.518 | 59 | 114 | |
| LWK | G/G | 0.164 | 18 | C/G | 0.536 | 59 | C/C | 0.300 | 33 | 110 | G | 0.432 | 95 | C | 0.568 | 125 | 220 | |
| MKK | G/G | 0.141 | 22 | C/G | 0.449 | 70 | C/C | 0.410 | 64 | 156 | G | 0.365 | 114 | C | 0.635 | 198 | 312 | |
| YRI | G/G | 0.284 | 40 | C/G | 0.504 | 71 | C/C | 0.213 | 30 | 141 | G | 0.535 | 151 | C | 0.465 | 131 | 282 | |
| MEX | G/G | 0.310 | 18 | C/G | 0.500 | 29 | C/C | 0.190 | 11 | 58 | G | 0.560 | 65 | C | 0.440 | 51 | 116 | |
| rs8057927 | JPT | T/T | 0.478 | 54 | C/T | 0.425 | 48 | C/C | 0.097 | 11 | 113 | T | 0.690 | 156 | C | 0.310 | 70 | 226 |
| CHB | T/T | 0.478 | 65 | C/T | 0.412 | 56 | C/C | 0.110 | 15 | 136 | T | 0.684 | 186 | C | 0.316 | 86 | 272 | |
| CHD | T/T | 0.514 | 56 | C/T | 0.394 | 43 | C/C | 0.092 | 10 | 109 | T | 0.711 | 155 | C | 0.289 | 63 | 218 | |
| GIH | T/T | 0.901 | 91 | C/T | 0.089 | 9 | C/C | 0.010 | 1 | 101 | T | 0.946 | 191 | C | 0.054 | 11 | 202 | |
| CEU | T/T | 0.876 | 99 | C/T | 0.124 | 14 | C/C | 0 | 0 | 113 | T | 0.938 | 212 | C | 0.062 | 14 | 226 | |
| TSI | T/T | 0.853 | 87 | C/T | 0.147 | 15 | C/C | 0 | 0 | 102 | T | 0.926 | 189 | C | 0.074 | 15 | 204 | |
| ASW | T/T | 0.632 | 36 | C/T | 0.351 | 20 | C/C | 0.018 | 1 | 57 | T | 0.807 | 92 | C | 0.193 | 22 | 114 | |
| LWK | T/T | 0.620 | 67 | C/T | 0.324 | 35 | C/C | 0.056 | 6 | 108 | T | 0.782 | 169 | C | 0.218 | 47 | 216 | |
| MKK | T/T | 0.692 | 108 | C/T | 0.250 | 39 | C/C | 0.058 | 9 | 156 | T | 0.817 | 255 | C | 0.183 | 57 | 312 | |
| YRI | T/T | 0.623 | 91 | C/T | 0.336 | 49 | C/C | 0.041 | 6 | 146 | T | 0.791 | 231 | C | 0.209 | 61 | 292 | |
| MEX | T/T | 0.807 | 46 | C/T | 0.175 | 10 | C/C | 0.018 | 1 | 57 | T | 0.895 | 102 | C | 0.105 | 12 | 114 | |
| rs8049308 | JPT | T/T | 0.455 | 20 | C/T | 0.500 | 22 | C/C | 0.045 | 2 | 44 | T | 0.705 | 62 | C | 0.295 | 26 | 88 |
| CHB | T/T | 0.364 | 16 | C/T | 0.477 | 21 | C/C | 0.159 | 7 | 44 | T | 0.602 | 53 | C | 0.398 | 35 | 88 | |
| CEU | T/T | 0.650 | 39 | C/T | 0.333 | 20 | C/C | 0.017 | 1 | 60 | T | 0.817 | 98 | C | 0.183 | 22 | 120 | |
| YRI | T/T | 0.583 | 35 | C/T | 0.383 | 23 | C/C | 0.033 | 2 | 60 | T | 0.775 | 93 | C | 0.225 | 27 | 120 | |
Note: Genotype frequencies and allele frequencies data were determined by the HapMap database (HapMap data release 28, Phase 2+3, August 10), on NCBI B36 assembly, dbSNP b126 (http://hapmap.ncbi.nlm.nih.gov/).
Abbreviations: ASW, African ancestry in southwest USA; CDH13, cadherin13; CEU, residents of UT, USA with Northern and Western European ancestry, from the Centre d’Etude du Polymorphisme Humain collection; CHB, Han Chinese in Beijing, People’s Republic of China; CHD, Chinese in metropolitan Denver, CO, USA; freq, frequency; GIH, Gujarati Indians in Houston, TX, USA; JPT, Japanese in Tokyo, Japan; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, CA, USA; MKK, Maasai in Kinyawa, Kenya; SNP, single nucleotide polymorphism; TSI, Tuscan in Italy; YRI, Yoruba in Ibadan, Nigeria.
Table S2.
Haplotype frequencies of CDH13 SNPs (rs12925602–rs7204454) in different ethnic populations1
| JPT | CHB | CHD | GIH | CEU | TSI | ASW | LWK | MKK | YRI | MEX | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GACGG | 0.355 | 0.351 | 0.347 | 0.233 | 0.244 | 0.335 | 0.286 | 0.289 | 0.231 | 0.287 | 0.394 |
| CGCAC | 0.285 | 0.220 | 0.206 | 0.085 | |||||||
| AACAG | 0.151 | 0.161 | 0.194 | 0.119 | 0.081 | 0.114 | 0.127 | 0.106 | 0.091 | 0.130 | 0.154 |
| GGTAC | 0.128 | 0.179 | 0.135 | 0.142 | 0.162 | 0.114 | 0.103 | 0.167 | 0.154 | 0.087 | 0.106 |
| GGCAG | 0.070 | 0.077 | 0.076 | ||||||||
| GGCGG | 0.012 | 0.006 | 0.018 | 0.006 | |||||||
| GACAG | 0.006 | 0.071 | 0.067 | 0.024 | 0.126 | 0.010 | |||||
| AACGG | 0.006 | 0.017 | |||||||||
| GGCGC | 0.012 | ||||||||||
| AGCAG | 0.006 | ||||||||||
| AACAC | 0.004 | ||||||||||
| GACGC | 0.017 | 0.006 | 0.008 | 0.004 | |||||||
| GGCAC | 0.009 | 0.008 | 0.003 | 0.019 | |||||||
| AGTAC | 0.008 | ||||||||||
| GGTGG | 0.014 | ||||||||||
| GGTGC | 0.006 | 0.014 | 0.022 | 0.010 | |||||||
| GACAC | 0.006 | 0.392 | 0.500 | 0.415 | 0.389 | 0.367 | 0.469 | 0.343 | 0.308 |
Note: Haplotype frequencies data were determined by the Haploview software program (version 4.2; Broad Institute, Cambridge, MA, USA) (http://www.broad.mit.edu/mpg/haploview/).
Abbreviations: ASW, African ancestry in southwest USA; CDH13, cadherin13; CEU, residents of UT, USA with Northern and Western European ancestry, from the Centre d’Etude du Polymorphisme Humain collection; CHB, Han Chinese in Beijing, People’s Republic of China; CHD, Chinese in metropolitan Denver, CO, USA; GIH, Gujarati Indians in Houston, TX, USA; JPT, Japanese in Tokyo, Japan; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, CA, USA; MKK, Maasai in Kinyawa, Kenya; SNPs, single nucleotide polymorphisms; TSI, Tuscan in Italy; YRI, Yoruba in Ibadan, Nigeria.
Table S3.
Putative transcription factor binding site in each SNP on the promoter region of CDH13
| SNP ID | Allele | Sequence | Predicted TF | Binding site | Function |
|---|---|---|---|---|---|
| rs12925602 | A | TCTGCCTACATC[A] AGGAAATTCAGA |
c-Ets- | ATCAAGGAAATT | Regulates numerous genes and involved in stem cell development, cell senescence and death, and tumorigenesis |
| GATA-1 | CATCAAGGA | Regulates the switch of fetal hemoglobin to adult hemoglobin for erythroid development | |||
| CdxA | CA TCAAG | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes | |||
| G | TCTGCCTACATC[G] AGGAAATTCAGA |
c-Ets- | ATCGAGGAAATT | Regulates numerous genes and involved in stem cell development, cell senescence and death, and tumorigenesis | |
| GATA-1 | CATCGAGGA | Regulates the switch of fetal hemoglobin to adult hemoglobin for erythroid development | |||
| rs7193788 | A | GCACGCAGCAGT[A] AAAATACAGAAA |
CdxA | TAAAAATAAAAATA | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes |
| AhR/Ar | GAGCACGCAGCAGTAA | A ligand-activated transcription factor involved in the regulation of biological responses to planar aromatic hydrocarbons | |||
| Sox5 | GTAAAAATAC | A transcription factor involved in the regulation of embryonic development and in the determination of cell fate | |||
| G | GCACGCAGCAGT[G] AAAATACAGAAA |
AhR/Ar | ACGCAGCAGTGAAAA | A ligand-activated transcription factor involved in the regulation of biological responses to planar aromatic hydrocarbons | |
| rs736719 | C | CAGGAAGAAACA[C] GAAGCAGTGTTT |
SRY | AAACACG | A transcription factor and a member of the HMG-box family of DNA binding proteins, which may directly generate some male-specific properties of the brain |
| T | CAGGAAGAAACA[T] GAAGCAGTGTTT |
SRY | AAACATG | A transcription factor and a member of the HMG-box family of DNA binding proteins, which may directly generate some male specific properties of the brain | |
| HNF-3b | GAAGAAACA TGA | A transcription factor and a member of the forkhead class of DNA-binding proteins | |||
| rs6565051 | A | ACCTTCCCTGGA[A] TGGAGAAAAGTC |
C/EBPb | GAATG GAGAAAAGT | A transcription factor that can bind as a homodimer to certain DNA regulatory regions and can also form heterodimers with other C/EBP |
| G | ACCTTCCCTGGA[G] TGGAGAAAAGTC |
C/EBPb | GAGTG GAGAAAAGT | A transcription factor that can bind as a homodimer to certain DNA regulatory regions and can also form heterodimers with other C/EBP | |
| MZF1 | AGTG GAGA | A member of the SCAN domain family transcription factors that form dimers through their highly conserved SCAN motifs | |||
| rs7204454 | C | GTGAGTTCAGTA[C] AATTTGTGTTTT |
CdxA | TACAATT | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes |
| G | GTGAGTTCAGTA[G] AATTTGTGTTTT |
CdxA | TAGAATT | A transcription factor that binds to DNA to regulate the expression of genes, in particular the Hox genes |
Abbreviations: CDH13, cadherin13; SNP, single nucleotide polymorphism; SNP ID, single nucleotide polymorphism identification; TF, transcription factor.