Abstract
The hepatitis C virus (HCV) core protein represents the first 191 amino acids of the viral precursor polyprotein and is cotranslationally inserted into the membrane of the endoplasmic reticulum (ER). Processing at position 179 by a recently identified intramembrane signal peptide peptidase leads to the generation and potential cytosolic release of a 179-amino-acid matured form of the core protein. Using confocal microscopy, we observed that a fraction of the mature core protein colocalized with mitochondrial markers in core-expressing HeLa cells and in Huh-7 cells containing the full-length HCV replicon. Subcellular fractionation confirmed this observation and showed that the core protein associates with purified mitochondrial fractions devoid of ER contaminants. The core protein also fractionated with mitochondrion-associated membranes, a site of physical contact between the ER and mitochondria. Using immunoelectron microscopy and in vitro mitochondrial import assays, we showed that the core protein is located on the mitochondrial outer membrane. A stretch of 10 amino acids within the hydrophobic C terminus of the processed core protein conferred mitochondrial localization when it was fused to green fluorescent protein. The location of the core protein in the outer mitochondrial membrane suggests that it could modulate apoptosis or lipid transfer, both of which are associated with this subcellular compartment, during HCV infection.
Hepatitis C virus (HCV), the major causative agent of non-A, non-B hepatitis, is estimated to infect 3% of the world's population (11). Viral infection persists in ∼80% of infected individuals, causing chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (1, 13). Analyses of viral kinetics showed that HCV particles are continuously produced and cleared during chronic infection (46, 54). The lack of an efficient infectious cell culture model has prevented a comprehensive understanding of the viral life cycle. Important progress was made with the development of autonomously replicating HCV RNAs which produce nonstructural viral proteins upon transfection into hepatoma cells (7, 27). Although these subgenomic replicons were recently extended to the full-length HCV genome encoding all viral proteins, no viral particle formation was observed (52).
The HCV genome, a single plus-stranded RNA molecule of 9,600 nucleotides, encodes a single precursor polyprotein of about 3,000 amino acids (for reviews, see references 6 and 58). The precursor consists of N-terminal structural and C-terminal nonstructural components. During and after translation, the precursor protein undergoes an ordered series of proteolytic cleavages by viral and host proteases that generate the individual viral proteins. The functional properties of these proteins have been deduced by analogy with related RNA viruses and by cDNA expression experiments in mammalian cells (for a review, see reference 65).
There are at least three viral proteins that have structural properties, including the core protein and the envelope glycoproteins E1 and E2. The core protein is believed to form the viral nucleocapsid, binds to RNA in vitro (60), and engages in homotypic interactions that are necessary for particle formation (23, 33). In addition, overexpression of the core protein influences many cellular functions, including cell signaling, apoptosis, carcinogenesis, and lipid metabolism (for reviews, see references 16, 35, and 56). The core protein has been reported to interact with various cellular proteins, including p53 (28), tumor necrosis factor receptor 1 (74), the lymphotoxin β receptor (32), the transcription factors LZIP (21) and Stat3 (72), the heterogeneous nuclear ribonucleoprotein K (20), a putative RNA helicase (73), and the human DEAD-box protein DDX3 (49). The core protein also associates directly with triglycerides in lipid storage droplets (4) and inhibits microsomal triglyceride transfer protein activity (51). These properties have been linked to the accumulation of intracellular lipids during chronic HCV infection, a condition called steatosis. Steatosis and hepatocellular carcinoma are also prominent symptoms in mice that are transgenic for the core protein (41, 43). Therefore, the core protein may play an important role in HCV pathogenesis.
The core protein represents the first 191 amino acids of the viral precursor polyprotein and is separated from the subsequent E1 protein by a highly hydrophobic signal sequence (60). During translation of the viral RNA, this signal sequence mediates an association of the precursor polyprotein with the membrane of the endoplasmic reticulum (ER) and directs portions of the subsequent E1 and E2 proteins into the ER lumen, while most of the core protein remains at the cytosolic side. The core protein is released from the rest of the polyprotein through signal peptidase cleavage at the luminal side of the ER. It was previously thought to receive further processing at the carboxylic side of amino acid 173 (25, 60). Processing of the core protein is now believed to occur between amino acids 179 and 180 by a recently identified intramembrane-cleaving presenilin-type signal peptide peptidase (SPP) (24, 36, 70). Cleavage by SPP is essential for the maturation of the core protein and loosens its attachment to the ER membrane. The mature core protein has been found at the ER membrane (26), at the surface of lipid droplets (4), and in the cell nucleus (26, 34, 41, 55).
In liver cell lines transfected with the core protein and in core protein-transgenic mice, the core protein was also detected on rough ER membranes surrounding the mitochondria (39) and in direct association with mitochondria (41, 48). This association has been linked to the mitochondrial injury and enhanced oxidative stress observed in core protein-transgenic mice (42, 48). However, the form of core protein associated with mitochondria and the submitochondrial localization of the core protein have not been determined. We report here that a fraction of the mature core protein localizes to the outer surfaces of mitochondria. The core protein associates with the mitochondrial outer membrane via a 10-amino-acid domain in the C terminus of the processed protein. Since the mitochondrial outer membrane is central to the regulation of host cell apoptosis and lipid metabolism (for reviews, see references 30 and 67), we propose that the presence of the core protein at the surfaces of mitochondria might directly influence these important cellular functions.
MATERIALS AND METHODS
Plasmids.
The full-length 191-amino-acid core protein coding sequence (genotype 1b) was cloned into plasmid pTet-Splice (Life Technologies) to generate Tet-Core or into pcDNA3.1+ (Invitrogen) to generate CMV-Core. In addition, the coding sequence for the 173-amino-acid core protein was cloned into pcDNA3.1+ and used for in vitro transcription and translation reactions. During PCR amplification, the sequence for the Flag octapeptide (DYKDDDDK) was added at the 5′ end of the core open reading frame.
The full-length untagged core protein amplified by PCR from the HCV Con1 consensus sequence (27) was cloned into a modified version of the lentiviral pHR′ vector (45). This vector is a minimal nonreplicative human immunodeficiency virus type 1 genome flanked by two long terminal repeats containing viral cis-acting sequences that are necessary for packaging and infection and a heterologous promoter (EF-1α) driving core protein expression. The plasmids, expressing residues 139 to 158 (C2), 139 to 148 (C2a), and 149 to 158 (C2b) of the core protein (genotype 1) fused to the amino terminus of enhanced green fluorescent protein (EGFP), were constructed by cloning of the PCR-amplified EGFP sequence from pEGFP-1 (BD Clontech Laboratories) into the SalI and NotI restriction sites of the vector pCMV/myc/cyto (Invitrogen) to generate pCMV/EGFP/myc/cyto. The sequence for C2 was generated by PCR and inserted between the PstI and SalI sites of pCMV/EGFP/myc/cyto. Double-stranded oligonucleotides were used to generate the C2a and C2b sequences. Their sequences were as follows: GCTCGTCGGCGCCCCTCTTGGAGGCGCTGCCG and TCGACGGCAGCGCCTCCAAGAGGGGCGCCGACGAGCTGCA for C2a and GAGGGCCCTGGCGCATGGCGTCCGGGTTCTGG and TCGACCAGAACCCGGACGCCATGCGCCAGGGCCCTCTGCA for C2b. After annealing, each double-stranded oligonucleotide was ligated between the PstI and SalI sites of pCMV/EGFP/myc/cyto. All constructs were confirmed by DNA sequencing. A plasmid expressing the tetracycline-controlled transactivator (pTet-tTA) was obtained from D. Schatz, Yale School of Medicine, New Haven, Conn. (64). The plasmids pDsRed2-Mito and pDsRed2-ER were purchased from BD Clontech Laboratories.
Cell lines and transfection-transduction methods.
HeLa, Huh-7, HEK293T, and HeLa Tet-Off cells stably expressing the tetracycline-controlled transactivator (tTA) were grown under standard cell culture conditions. HeLa Tet-Off cells (obtained from I. Hoffmann, Deutsches Krebsforschungszentrum, Heidelberg, Germany) also received 600 μg of G418 (Life Technologies)/ml in the medium. Replicon cell lines 21-5 and 20-1, containing the stably replicating HCV sfl genome (52), were cultured in the presence of 250 μg of G418/ml. For the generation of stable core protein-expressing Jurkat cells, Jurkat D4 cells (obtained from J. Sodroski, Dana-Farber Cancer Institute, Boston, Mass.) stably expressing the tTA were transfected by electroporation with Tet-Core and a hygromycin resistance gene derived from pREP7 (Invitrogen). Transfected cells were selected in the presence of hygromycin (200 μg/ml; Calbiochem), and single-cell clones were generated by limiting dilution. Core protein expression was tightly controlled, as verified by Western blot analysis in the absence and presence of doxycycline (2 μg/ml; Sigma). HeLa and HeLa Tet-Off cells were transiently transfected by a standard calcium phosphate method and Huh-7 cells were transfected by the use of FuGENE6 (Roche) according to the manufacturer's recommendations. Tetracycline-regulated core protein expression in Huh-7 cells was achieved by transient cotransfection of Tet-Core and pTet-tTA.
Lentiviral vectors pseudotyped with the glycoprotein of vesicular stomatitis virus were prepared by a previously described three-plasmid transfection method (75). Briefly, the vesicular stomatitis virus glycoprotein-encoding construct pMD.G and the human immunodeficiency virus-based packaging plasmid pCMVΔR8.91 (both provided by D. Trono, University of Geneva, Geneva, Switzerland) were cotransfected with the core protein-expressing lentiviral transfer vector into HEK293T cells by standard calcium phosphate transfection procedures. HeLa cells grown on coverslips were infected at a multiplicity of infection of ∼5.
Antibodies.
The following antibodies were obtained commercially: anti-core (clone C7-50; Affinity BioReagents), anti-mtHsp70 (clone JG1; Affinity Bioreagents), anti-manganese superoxide dismutase (MnSOD; Stressgen Biotechnologies), anti-calreticulin (Stressgen Biotechnologies), anti-Hsp90α (Stressgen Biotechnologies), anti-cytochrome c oxidase subunit IV (clone 20E8-C12; Molecular Probes), anti-cytochrome c (clone 7H8.2C12; Pharmingen), anti-Flag M2 (Sigma), and anti-Myc (Invitrogen).
Immunofluorescence and confocal microscopy.
HeLa Tet-Off cells grown on glass coverslips were incubated with 40 nM MitoTracker (CMXRos; Molecular Probes) 20 h after transfection. The cells were fixed in 3.7% paraformaldehyde for 15 min at 37°C, permeabilized with 0.2% Triton X-100, blocked with 1% bovine serum albumin, and immunostained with a monoclonal anti-core antibody (1:100) followed by incubation with a Cy2-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories). Coverslips were mounted on glass slides and analyzed by confocal laser scanning microscopy (Zeiss LSM 510 UV). For ER colocalization experiments, pDsRed-ER was cotransfected with the core protein-expressing plasmids into HeLa Tet-Off cells, which were subsequently processed for anti-core immunostaining as described above. Replicon-containing Huh-7 cells were grown on coverslips, fixed in 2.5% paraformaldehyde, and permeabilized with 0.5% Triton X-100 for 5 min at room temperature. The cells were blocked with 1% bovine serum albumin and costained with monoclonal anti-core and polyclonal anti-manganese superoxide dismutase (1:300) antibodies to stain the mitochondria or with polyclonal anti-calreticulin antibodies (1:300) to stain the ER, followed by incubation with Cy2-conjugated anti-mouse immunoglobulin G and Cy5-conjugated anti-rabbit immunoglobulin G secondary antibodies suitable for multilabeling experiments (both from Jackson ImmunoResearch Laboratories). Confocal images were acquired with a Leica TCS SP laser scanning confocal system. Huh-7 cells were seeded into two-well Lab-Tek II chamber slides (Nalgene Nunc International), cotransfected with the core-GFP fusion protein and DsRed-Mito expression constructs, and 20 h after transfection, were processed for confocal microscopy with a Zeiss Axioplan Z or Leica TCS SP system.
Electron microscopic immunolocalization.
Immunolocalization and processing for electron microscopy were performed as described previously (22). Briefly, lentivirally infected HeLa cells or Huh-7 replicon cells were grown on glass coverslips, fixed in 2.5% paraformaldehyde, and permeabilized with 0.05% Triton X-100 for 3 to 5 min. Anti-core antibodies (1:40) were added for 5 h, followed by a 6-h incubation with 5-nm-diameter-gold-coupled anti-mouse secondary antibodies (Amersham Life Sciences). The cells were further processed for electron microscopy by standard procedures and were analyzed with a Zeiss EM 910 microscope. In Huh-7 replicon cells, secondary antibodies coupled to 1.4-nm-diameter gold particles (Nanogold; Biotrend) were used. After binding for 4 h, the samples were washed and fixed in 2.5% glutaraldehyde (in sodium cacodylate buffer, pH 7.2) for 15 min, and the gold particles were enlarged with an HQ-Silver enhancement kit (Biotrend).
Mitochondrial import assays.
Mammalian mitochondria were isolated from HEK293T cells as described previously (50). Radiolabeled core protein 173 was generated from CMV-Core173 by in vitro transcription and translation reactions (Promega) in the presence of [35S]methionine. Import assays were performed as described previously (63). For each import reaction, 50 μg of freshly isolated mammalian mitochondria was mixed with the radiolabeled protein and incubated at 30°C for 20 min. To maintain the coupling of isolated mitochondria, we added 2 mM ATP and 10 mM sodium succinate. When indicated, the mitochondrial transmembrane potential was disrupted by the preincubation of mitochondria with 8 μM antimycin, 20 μM oligomycin, and 1 μM valinomycin (all Sigma) before the addition of the radiolabeled protein. Mitochondrial import was stopped by the addition of valinomycin (1 μM) and a transfer to 0°C. When indicated, the samples were treated with proteinase K (50 μg/ml) for 10 min on ice. The protease treatment was stopped by the addition of 2 mM phenylmethylsulfonyl fluoride. Mitochondria were reisolated by centrifugation at 10,000 × g for 5 min at 4°C, washed in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM morpholinepropanesulfonic acid [MOPS]-KOH, pH 7.2), and recentrifuged as described above. Mitochondrial pellets were resuspended in sodium dodecyl sulfate (SDS) sample buffer and subjected to SDS-polyacrylamide gel electrophoresis. Dried gels were exposed to Biomax MR film (Kodak).
Differential centrifugation.
The details of the differential centrifugation experiments were described previously (63, 71). Briefly, core protein-expressing Jurkat cells (7 × 107) were resuspended in ice-cold buffer A (250 mM sucrose, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 20 mM HEPES-KOH, pH 7.5) containing protease inhibitors (Roche), incubated on ice for 10 min, and subjected to Dounce homogenization (Wheaton). After the removal of the nuclei and unbroken cells by two rounds of centrifugation at 800 × g for 5 min, the heavy-membrane (HM) fraction was prepared by centrifugation (10,000 × g for 10 min). The resulting supernatant was centrifuged at 100,000 × g for 1 h to produce the light-membrane (LM) fraction (pellet). The HM and LM fractions were normalized for cell equivalents, and the distribution of the core protein and marker proteins between these fractions was assessed by immunoblotting.
Subcellular fractionation and density gradient centrifugation.
Crude mitochondrial fractions obtained by centrifugation (5,000 × g for 10 min) of the postnuclear supernatant of the core protein-expressing Jurkat cell homogenate (109 cells) were washed with HES buffer (250 mM sucrose, 1 mM EDTA, 10 mM HEPES-KOH, pH 7.5) and layered on top of a linear 20 to 50% (wt/wt) sucrose gradient prepared in HES buffer. After centrifugation at 52,000 × g for 90 min in an SW41 rotor (Beckman Coulter), a dense band corresponding to purified mitochondria was harvested. Mitochondrion-associated membranes (MAMs) were isolated from the gradient as a diffuse white band located above the mitochondrial fraction. Both fractions were slowly diluted to 0.25 M sucrose. The mitochondrial fraction was centrifuged (30,000 × g for 30 min), and the mitochondrial pellet was washed in HES buffer followed by reisolation at 6,300 × g for 10 min. The MAM fraction was pelleted at 100,000 × g for 30 min at 4°C (62). To prepare cytosolic and microsomal fractions, we further centrifuged (10,000 × g for 10 min) the supernatant of the initial 5,000 × g centrifugation step to remove residual mitochondria. The resulting supernatant was centrifuged at 100,000 × g for 1 h to separate the microsomes (pellet fraction) from the cytosol (supernatant fraction). All fractions were solubilized in TXIP-1 buffer (63) containing protease inhibitors and were frozen at −70°C. The total protein content of each fraction was determined by using the Bio-Rad DC protein assay, and equal amounts were analyzed by immunoblotting.
Alkaline extraction experiments.
Crude mitochondrial fractions isolated from the cell homogenate and LM fractions, which were enriched in microsomes, were prepared as described above and resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2). Equal total protein amounts (250 μg) from both fractions were transferred to TLS-55 rotor centrifuge tubes. SEM buffer was added to a final volume of 2 ml, and the membranes were pelleted for 20 min at 100,000 × g. The supernatants were removed and the pellets were directly resuspended in freshly prepared ice-cold 0.1 M sodium carbonate (pH 11.5) at a final concentration of 500 μg of protein per ml of sodium carbonate. After incubation on ice for 15 min, the extracts were layered onto sucrose carbonate cushions (0.1 M sodium carbonate, 250 mM sucrose) and centrifuged at 130,000 × g for 15 min as described previously (31). The pellet was directly solubilized in SDS sample buffer. The proteins in the supernatant were first concentrated by trichloroacetate precipitation and then solubilized in SDS sample buffer before Western blot analysis.
Protease accessibility assays.
Crude mitochondrial fractions obtained from core protein-expressing Jurkat cells or Huh-7 cells transfected with GFP fusion proteins were resuspended in SM buffer (250 mM sucrose, 10 mM MOPS-KOH, pH 7.2), and equal amounts of mitochondria were divided into aliquots and analyzed as described previously (40). Briefly, mitochondria were sedimented (10,000 × g for 5 min), the supernatants were discarded, and the pellets were resuspended in SM buffer or SMTX-1 buffer (SM buffer containing 1% [wt/vol] Triton X-100). When indicated, proteinase K was added to a final concentration of 150 μg/ml. The samples were incubated on ice for 15 min and then proteolysis was stopped by the addition of 2 mM phenylmethylsulfonyl fluoride. The proteins were precipitated by the addition of trichloroacetic acid to a final concentration of 15%. The precipitated proteins were collected by centrifugation, washed with acetone, recentrifuged, and analyzed by immunoblotting with an anti-FLAG or anti-Myc (1:4,000) antibody or with antibodies against mitochondrial marker proteins.
RESULTS
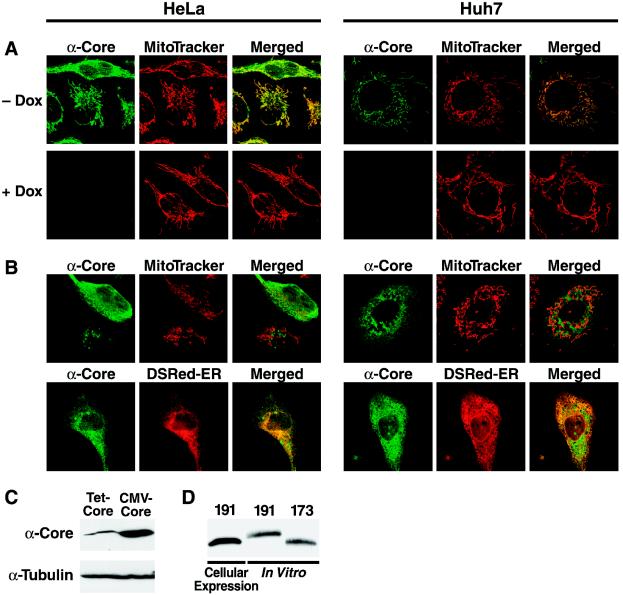
When we transiently expressed the full-length core protein (191 amino acids) in HeLa cells by using a tetracycline-regulated construct, we noted a rod-like distribution of the core protein-specific signal in the majority of transfected cells. This distribution is characteristic for proteins that localize to the mitochondria. Costaining with the fixable mitochondrion-selective marker MitoTracker confirmed the mitochondrial localization of the core protein in these cells (Fig. 1A, left panel). Mitochondrial localization of the core protein was predominantly observed when it was expressed from a construct that was responsive to the tTA expressed in the HeLa cells used for this study (HeLa Tet-Off cells). No core protein was detected in cells grown in the presence of doxycycline (2 μg/ml), confirming the specificity of the core protein signal. The same result was observed for Huh-7 hepatoma cells that were transiently cotransfected with the tetracycline-controlled core protein expression vector and the tTA transactivator (Fig. 1A, right panel).
FIG. 1.
A fraction of the HCV core protein colocalizes with a mitochondrial marker in transfected cells. Confocal microscopic images of HeLa Tet-Off and Huh-7 cells expressing the core protein from a tetracycline-controlled (A) or a CMV-driven expression vector (B) are shown. Living cells were incubated with MitoTracker Red or were cotransfected with a construct expressing the ER-targeted Discosoma red fluorescent protein (DsRed-ER). Immunostaining for the core protein was performed with anti-core protein and Cy2-labeled anti-mouse antibodies 20 h after transfection. (C) Western blot analysis of core protein expressed from the tetracycline-controlled or CMV-driven vector. The blot was stripped and reprobed for tubulin as a loading control. (D) Comparison of core protein expressed from the CMV-driven vector with in vitro-synthesized truncated core protein (173 amino acids) and unprocessed core protein (191 amino acids) generated in the absence of microsomal membranes.
In a minority of cells, we also observed a reticular or granular staining pattern for the core protein that was typical for protein localization to the ER or to lipid droplets. This distribution was observed in the vast majority of HeLa Tet-Off or Huh-7 cells when core protein expression was driven by the constitutive immediate-early promoter of cytomegalovirus (CMV) (Fig. 1B). Cotransfection with a construct expressing an ER-resident Discosoma red fluorescent protein (DsRed-ER) confirmed the localization of the core protein to the ER (Fig. 1B). Interestingly, the core protein expression in cells transfected with the CMV-driven plasmid was stronger than that in cells transfected with the tetracycline-controlled vector, as determined by Western blot analysis (Fig. 1C). With both transfection systems, the core protein was expressed as a full-length protein (191 amino acids) and was recovered as a single band that was similar in size to that of the in vitro-synthesized processed core protein (173 amino acids) and smaller than an in vitro-synthesized full-length core protein that had been generated in the absence of microsomal membranes (Fig. 1D). This indicated proper processing of the core protein at the ER membrane after cellular expression. These data confirm the subcellular localization of the HCV core protein to mitochondria. They further suggest that the predominant subcellular localization of the core protein might depend on the level of protein expression in cells.
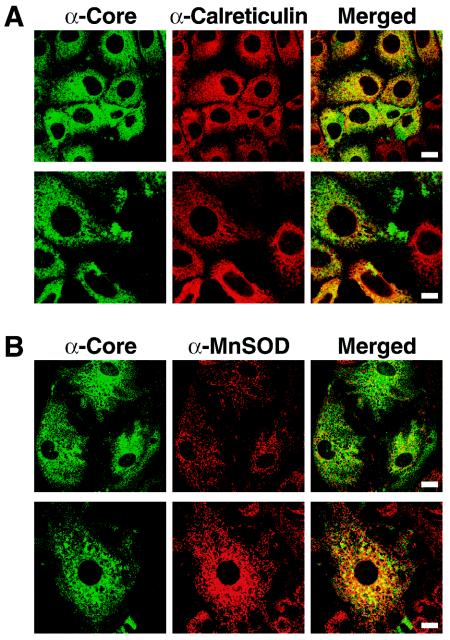
To further study the mitochondrial localization of the core protein, we examined Huh-7 hepatoma cells that were stably transfected with a full-length HCV replicon. These cells autonomously replicate the complete HCV RNA genome at high levels and express all viral proteins (52). Core protein expression was confirmed by Western blot analysis (data not shown) and by confocal microscopy (Fig. 2). We observed a strong, exclusively cytoplasmic, core protein-specific signal that overlapped in part with the signal for the ER marker calreticulin (Fig. 2A) and to a slightly lesser extent with the mitochondrion-resident manganese superoxide dismutase (MnSOD; Fig. 2B). This is consistent with the model that during HCV replication a fraction of core protein localizes to mitochondria.
FIG. 2.
Partial localization of core protein to mitochondria in Huh-7 cells expressing a full-length HCV replicon. Huh-7 cells (clone 21-5) containing a full-length HCV replicon were costained for the core protein and calreticulin (A) or the core protein and MnSOD (B). Bars: 20 μm in upper panel and 10 μm in lower panel (A) or 20 μm (B).
HCV core protein cofractionates with mitochondria.
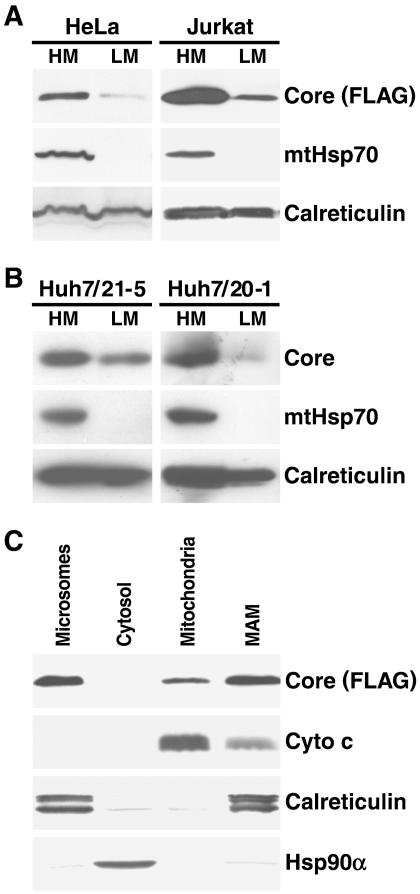
Next, we performed cellular fractionation experiments. Crude HM and LM fractions were prepared by differential centrifugation from cell homogenates prepared from core protein-expressing HeLa Tet-Off and Jurkat Tet-Off cells. HeLa cells were transiently transfected while Jurkat cells were stably transfected with the tetracycline-controlled core protein expression plasmid. Cell equivalents from HM and LM fractions were analyzed by Western blot analysis with antibodies specific for the core protein and for organelle marker proteins (Fig. 3A).
FIG. 3.
Core protein cofractionates with mitochondria. (A) Western blot analysis of HM and LM fractions isolated from core protein-expressing HeLa and Jurkat Tet-Off cells by differential centrifugation. Cell equivalents from each fraction were loaded, and the distributions of FLAG-tagged core protein, mitochondrial heat shock protein 70 (mtHsp70), and the ER-resident calreticulin protein were analyzed. (B) Western blot analysis of HM and LM fractions of two Huh-7 cell lines (21-5 and 20-1) expressing the full-length replicon with core protein- and organelle-specific antibodies. (C) Jurkat homogenates were further separated by sucrose density gradient centrifugation to obtain purified mitochondria and MAMs. Microsomes and cytosolic fractions were prepared by differential centrifugation of the same cell homogenates. Equal protein amounts (30 μg) from each fraction were analyzed by Western blotting with antibodies specific for FLAG-tagged core protein, cytochrome c (Cyto c), calreticulin, and heat shock protein 90α (Hsp90α).
In both cell types, the core protein was strongly enriched in the HM fraction, which contained all cellular mitochondria, as shown by the exclusive recovery of the mitochondrial matrix protein mtHsp70 (Fig. 3A). The core protein was also found in the LM, or microsomal, fraction (Fig. 3A). Calreticulin was also recovered from the HM fraction, raising the concern that the core protein could cofractionate with ER contaminants rather than with the mitochondria in this fraction. However, while calreticulin was present in both fractions in similar quantities, the core protein was enriched in the HM compared to the LM fraction, indicating that its cofractionation with HMs cannot be explained solely by an ER association. The same was observed for Huh-7 cells expressing the full-length HCV replicon. Differential centrifugation was performed with two stably transfected cell lines (21-5 and 20-1). In both clones, the core protein was enriched in the mitochondrial HM fraction compared to the microsomal LM fraction (Fig. 3B). Again, the enrichment of the core protein in the HM fraction was higher than that observed with calreticulin, indicating that the core protein in HCV replicon cells associates with mitochondria.
To better separate the mitochondria from the ER, we further fractionated crude mitochondrial fractions from core protein-expressing Jurkat cells on a linear 20 to 50% sucrose density gradient. Centrifugation yielded a dense band corresponding to purified mitochondria and a diffuse white band located above the mitochondrial band, previously described as the MAM fraction (59, 66). MAMs represent a defined subcellular compartment of physical contact sites on mitochondrial and ER membranes (29, 57). Equal amounts of protein from purified mitochondria, MAMs, and crude microsomal and cytosolic fractions were analyzed by Western blot analysis. The core protein was present in all membrane fractions, including purified mitochondria, and was relatively enriched in MAMs and microsomes (Fig. 3C). No core protein was detected in the soluble cytosolic fraction, indicating that the core protein does not exist in substantial amounts as a free protein in the cytosol. The purity of the different fractions was confirmed by immunoblotting with antibodies directed against established organelle marker proteins. The mitochondrial fraction was devoid of calreticulin, confirming that a complete separation between the mitochondrial and ER fractions had occurred. Calreticulin cofractionated with MAMs and microsomes, as expected. These data demonstrate that in core protein-expressing cells, a fraction of the core protein localizes to mitochondria and to contact sites between the ER and mitochondria.
Localization of HCV core protein to the mitochondrial outer membrane.
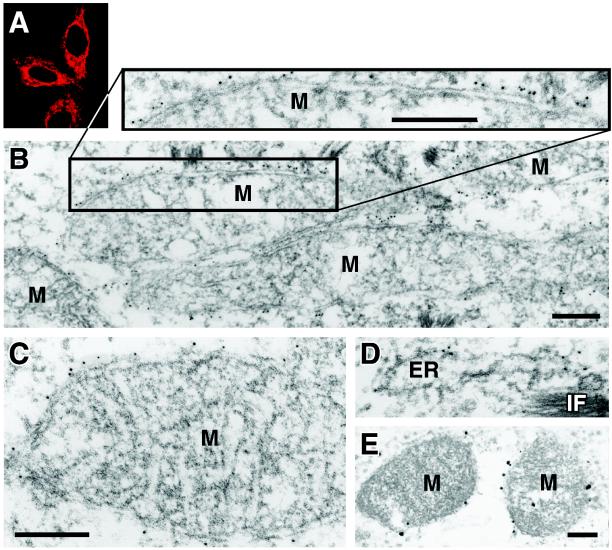
Mitochondria are complex organelles bounded by an inner and an outer membrane. Mitochondrial proteins can be localized to either the mitochondrial matrix, the inner membrane, the intermembrane space, or the outer membrane. Two different strategies were employed to examine the submitochondrial localization of the core protein. First, we performed immunoelectron microscopy of HeLa cells transduced with an HCV core protein-expressing lentiviral vector. Upon infection, core protein expression was driven by the constitutive elongation factor 1α promoter and was detected by immunofluorescence microscopy in rod-shaped structures in the cytoplasm of transduced cells (Fig. 4A). In the immunoelectron microscopic analysis, the mitochondria were strongly labeled with core protein-specific antibodies (Fig. 4B and C). Core protein-specific gold particles (5-nm diameter) were found in close proximity and at a uniform distance from the mitochondrial outer membrane, consistent with the model that a fraction of the core protein directly interacts with the outer surface of this membrane (Fig. 4B, enlarged detail).
FIG. 4.
Core protein localizes to the outer surfaces of mitochondria. The images shown are confocal (A) and immunoelectron (B to D) microscopic images of the core protein in HeLa cells transduced with a full-length core protein-expressing lentiviral vector and an immunoelectron microscopic image (E) of the core protein in Huh-7 cells containing a full-length HCV replicon. Core protein-specific gold particles (5-nm diameter) were predominantly found at the outer surfaces of mitochondria (M). For panel E, nanogold particles (1.4-nm diameter) were used and enhanced with a silver enhancement step. Bars: 0.2 μm. IF, intermediate filaments.
We counted the gold particles in 20 micrographs and found that 70% of the immunolabeling was associated with the outer mitochondrial membrane. Thirteen percent of the particles were associated with cisternae of the ER (Fig. 4D), 6% were associated with vesicular structures (with diameters of about 100 to 150 nm), and 11% were bound to elements in the cytoplasm that could not be identified. No immune reaction was observed in uninfected control cells or when experiments were performed with the gold-labeled secondary antibody alone (data not shown). We also examined full-length replicon-expressing Huh-7 cells by immunoelectron microscopy (Fig. 4E). Although cytoplasmic membrane structures were less preserved in this analysis because of an additional silver enhancement step, we found 30% of the immunolabeling to be directly bound to the outer mitochondrial membrane. We further noted that 100% of the mitochondria that we analyzed were decorated with gold labels at their outer surfaces, indicating that in full-length replicon-expressing cells, a fraction of the core protein also localizes to the outer mitochondrial membrane.
HCV core protein is not transported across the mitochondrial outer membrane in vitro and in vivo.
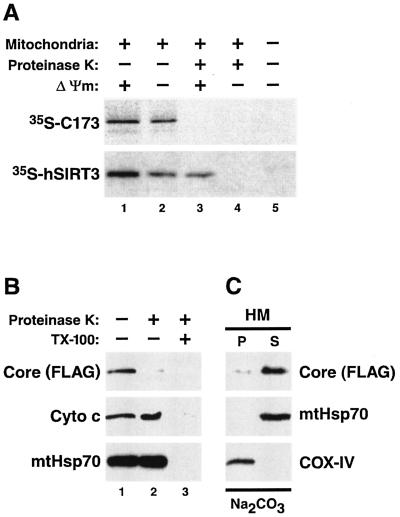
To examine whether the core protein could be actively transported across the mitochondrial membranes or if it stays bound to the outer mitochondrial surface, we performed in vitro mitochondrial import assays. Isolated mitochondria were incubated with an in vitro-synthesized radiolabeled core protein (173 amino acids) under stringent binding conditions in the presence of 80 mM KCl. After centrifugation and several washes, the recovered core protein in mitochondrial pellets was analyzed by autoradiography (Fig. 5A). Core 173 sedimented in the presence (Fig. 5A, lane 1), but not in the absence (Fig. 5A, lane 5), of mitochondria, excluding the possibility that protein aggregation was the cause of pellet recovery of the core protein. Binding of the core protein to mitochondria was not affected by a treatment with inhibitors of the mitochondrial transmembrane potential (Fig. 5A, lane 2), indicating that the core protein is not transported across the inner mitochondrial membrane. Under the same conditions, the signal for human SIRT3, a known mitochondrial matrix protein, was reduced, indicating that SIRT3 is actively imported across the inner mitochondrial membrane as previously described (Fig. 5A, lane 2) (63). When binding reactions were treated with proteinase K, the core protein-specific signal disappeared, whereas a significant fraction of SIRT3 was protected from protease digestion (Fig. 5A, compare the upper and lower panels of lane 3). Inhibitors of the mitochondrial transmembrane potential abolished the mitochondrial import of SIRT3 and rendered the protein accessible to proteinase K treatment, demonstrating the efficiency of the inhibitors used in this experiment (Fig. 5A, lane 4). These results collectively indicate that the core protein is not imported into mitochondria in vitro.
FIG. 5.
Nonintegral attachment of core protein to the mitochondrial outer membrane. (A) In vitro import assays using isolated mitochondria and in vitro-synthesized radiolabeled core protein (173 amino acids) or human SIRT3 (hSIRT3) were analyzed by autoradiography. The core protein was not imported into mitochondria and was accessible to the proteolytic activity of proteinase K (top, lane 3). SIRT3 was partially protected from digestion by proteinase K because of active mitochondrial import (bottom, lane 3). Accordingly, inhibitors of the mitochondrial membrane potential (Δψm) reduced the signal for SIRT3 (bottom, lane 2) but had no effect on the core protein (top, lane 2). SIRT3 was completely digested by proteinase K when active mitochondrial import was inhibited (bottom, lane 4). (B) Proteinase K digestion of mitochondria isolated from core protein-expressing Jurkat Tet-Off cells in the presence or absence of detergent (TX-100). A Western blot analysis showed that while the FLAG-tagged core protein was readily digested by proteinase K in the absence of TX-100 (lane 2), proteins located in the intermembrane space, such as cytochrome c (Cyto c), and mtHsp70, which is localized in the mitochondrial matrix, were protected. (C) Sodium carbonate extracts of HM fractions isolated from core protein-expressing Jurkat Tet-Off cells were analyzed for the distribution of the FLAG-tagged core protein, mtHsp70, which in the matrix is peripherally attached to the inner mitochondrial membrane, and COX-IV, a polytopic inner mitochondrial membrane protein.
To further characterize the core protein association with mitochondria in vivo, we performed proteinase K accessibility assays with mitochondrial fractions isolated from core protein-expressing Jurkat cells. Proteinase K treatment led to the nearly complete disappearance of the core protein-specific signal in a Western blot analysis, confirming that the core protein in cells localizes predominantly to the outer mitochondrial surface, where it is accessible for protease digestion (Fig. 5B, lane 2). To examine the integrity of the mitochondrial membranes during proteinase K treatment, we probed the blots with an antibody against cytochrome c, which is located in the mitochondrial intermembrane space, or against the mitochondrial matrix protein mtHsp70. The stability of both markers was not affected by the addition of proteinase K, indicating that the mitochondrial membranes remained intact (Fig. 5B, lane 2). Permeabilization of the membranes with a detergent rendered both marker proteins susceptible to protease digestion, proving the effectiveness of the proteinase K preparation used in this experiment (Fig. 5B, lane 3). These results demonstrate that the core protein also localizes to the outer surface of mitochondria in vivo.
Peripheral attachment of HCV core protein to the mitochondrial outer membrane.
To determine if the core protein inserts into mitochondrial outer membranes as an integral protein, we performed alkaline extraction experiments. Equal amounts of crude microsomal or mitochondrial fractions isolated from core protein-expressing Jurkat cells were treated with sodium carbonate, followed by centrifugation over a sucrose carbonate cushion (15, 31). The pellet containing the integral membrane proteins was directly solubilized in gel loading buffer, while the supernatant containing soluble and peripheral membrane proteins was concentrated by trichloracetate precipitation. Both samples were analyzed by Western blot analysis. For microsomal fractions, the alkaline treatment led to the complete extraction of the core protein into the supernatant, as described previously (36; data not shown). The same result was obtained for mitochondrial fractions. The majority of the core protein was recovered in the supernatant, indicating peripheral binding of the core protein to the mitochondrial outer surface (Fig. 5C). However, a small amount of core protein was also found in the pellet, suggesting that a small fraction of the core protein may become integrally inserted. To exclude an incomplete separation of the integral and peripheral proteins in this experiment, we analyzed the distribution of established mitochondrial proteins. As expected, cytochrome c oxidase IV (COX-IV), a polytopic membrane-spanning protein, was exclusively found in the pellet, whereas mtHsp70, which is peripherally attached to the inner mitochondrial membrane within the mitochondrial matrix, was completely extracted into the supernatant (Fig. 5C).
Mitochondrial targeting signal in HCV core protein.
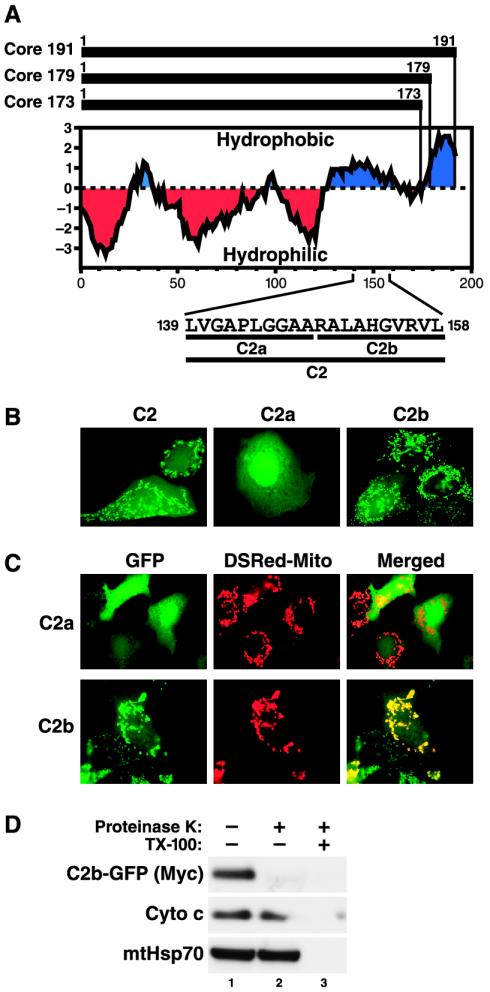
Unlike mitochondrial matrix-targeted proteins, which mostly contain cleavable presequences, the signals that target proteins to the mitochondrial outer membrane are contained within the mature protein sequence (for reviews, see references 37 and 47). Frequently, these signals are contained within a hydrophobic C terminus. The mature core protein also contains a hydrophobic C terminus (Fig. 6A). Therefore, we fused stretches of amino acids within this region to the amino terminus of GFP and examined the subcellular localization of the resulting fusion proteins in Huh-7 cells by confocal microscopy. We identified a domain of 20 amino acids (C2; amino acids 139 to 158) that conferred a rod-like cytoplasmic localization to GFP, consistent with targeting to the mitochondria (Fig. 6B). A further division of this domain into N-terminal (C2a) and C-terminal (C2b) halves demonstrated that the C-terminal 10-amino-acid motif (RALAHGVRVL) was sufficient to target GFP to these rod-like structures (Fig. 6B). These 10 amino acids are highly conserved among different viral isolates, with the central 7 amino acids (ALAHGVR) present in the core proteins of all HCV genotypes (9). Cotransfection with a construct expressing the mitochondrion-resident form of DsRed (DsRed-Mito) confirmed the localization of C2b-GFP, but not C2a-GFP, to the mitochondria (Fig. 6C). To further examine the submitochondrial localization of C2b-GFP, we conducted proteinase K accessibility assays with mitochondrial fractions isolated from Huh-7 cells expressing the C2b-GFP fusion protein. C2b-GFP, but not cytochrome c or mtHsp70, was readily digested by proteinase K in the absence of Triton X-100, indicating that the fusion protein, like the full-length core protein, localized to the mitochondrial outer membrane (Fig. 6D, lane 2). The digestion of cytochrome c and mtHsp70 in the presence of detergent confirmed the efficiency of the proteinase K treatment (Fig. 6D, lane 3).
FIG. 6.
A 10-amino-acid motif functions as a mitochondrial targeting signal in the core protein. (A) Kyte-Doolittle hydrophobicity plot of full-length (Core 191) and processed (Core 179 and Core 173) core proteins. The amino acid sequences and positions of domain C2 and the subdivisions C2a and C2b within the full-length core protein are indicated at the bottom. (B) Confocal microscopy of Huh-7 cells transfected with fusion proteins of domains C2, C2a, or C2b to GFP. (C) Confocal microscopy of Huh-7 cells cotransfected with the indicated GFP fusion proteins and a construct expressing the mitochondrion-targeted Discosoma red fluorescent protein (DsRed-Mito). (D) Proteinase K digestion of crude mitochondrial fractions isolated from Huh-7 cells transfected with C2b-GFP fusion protein in the absence or presence of detergent (TX-100). A Western blot analysis was performed with antibodies against the Myc epitope to detect the Myc-tagged fusion protein, against cytochrome c (Cyto c), and against mtHsp70.
DISCUSSION
Many RNA viruses establish specific associations with intracellular membranes, and all HCV proteins, with the exception of NS3, contain predicted or defined membrane association domains (reviewed in reference 14). The full-length core protein, residing at the cytosolic side of the ER membrane, contains a signal sequence at its C terminus that is cotranslationally inserted into the ER membrane (60). This membrane anchor is nearly entirely removed from the core protein after cleavage by SPP, thus potentially allowing the processed protein to leave the ER and travel to other organelles within the cell. Indeed, the core protein has been shown to be partially localized to the surfaces of lipid droplets (4, 18, 19, 36) and to mitochondria (41, 48).
We have examined in detail the relationship of the core protein with mitochondria. First, we observed by both confocal and electron microscopy that the fully processed core protein partially localized to mitochondria in transiently transfected cells (Fig. 1 and 4), as was previously shown (48). We further observed partial localization of the core protein to mitochondria in Huh-7 cells stably replicating the full-length HCV genome (Fig. 2). The association with mitochondria was confirmed by the finding that a significant portion of the core protein cofractionated with mitochondria isolated from transfected cells and cells expressing the full-length replicon (Fig. 3). Second, we demonstrated that the mitochondrion-associated core protein was highly susceptible to protease digestion in the absence of any detergent and also could be extracted by an alkaline treatment (Fig. 5), showing that this protein is a peripheral membrane protein on the outer surfaces of mitochondria. Third, we were able to identify a small, highly conserved region in the C terminus of the processed core protein that allowed the targeting of a heterologous protein to the outer surfaces of mitochondria (Fig. 6). Our data collectively demonstrate that a fraction of the processed core protein localizes to the surfaces of mitochondria via a targeting signal near its C terminus.
When we used sucrose density gradient centrifugation to carefully separate mitochondria from ER membranes, we found that the core protein was associated not only with mitochondria and the bulk ER, but also with so-called mitochondrion-associated ER membranes. In HeLa cells, regions of contact between mitochondria and the ER comprise 5 to 20% of the total mitochondrial network (57), forming a functionally specialized subregion of the ER that was initially described as fraction X and later renamed as MAMs (66). These membranes have a defined function in the direct transfer of lipids between the ER and mitochondria. They are enriched in enzymes involved in lipid synthesis, such as phosphatidylserine synthase (reviewed in reference 68), and are specifically associated with contact sites between the mitochondrial inner and outer membranes (2, 3). Recent evidence suggests that these membranes form transient membrane bridges that support protein movements between the ER and mitochondria. The dynamin-like protein DLP1 is believed to regulate dynamic fusion events between mitochondrial and ER membranes (53). A newly identified N-linked glycosylated protein (10) and the human CMV immediate-early regulator proteins gpUL37 and pUL37x1 (12) are also believed to traffic from the ER to mitochondria via these membrane bridges.
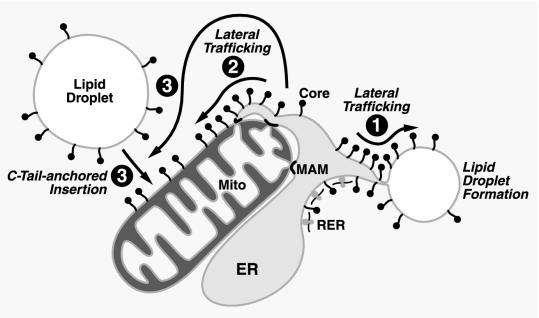
A model was previously proposed in which the core protein, after SPP cleavage, remains anchored to the outer leaflet of the ER membrane via residual amino acids of the original transmembrane domain (36). It was further proposed that the core protein travels along the ER membrane by lateral diffusion, reaching the surfaces of lipid droplets that form between the two leaflets of the ER membrane (Fig. 7, point 1). Our finding that the core protein cofractionates with microsomes and MAMs supports and extends this model. We propose that the processed core protein attached to the ER diffuses laterally, not only to lipid droplets, but also to specific subregions within the ER network, including the MAMs. From there, it can then diffuse onto the surfaces of mitochondria via adjacent or partially fused membranes of both compartments (Fig. 7, point 2). The potential saturation of these membrane bridges, together with the dynamics of lateral diffusion, might limit the core protein's access to the mitochondrial outer membrane and might explain why the majority of the core protein remains in the ER after strong protein overexpression (Fig. 1B).
FIG. 7.
Model of mitochondrial translocation of core protein. See the text for details.
Our model was supported by the fact that the core protein isolated from core protein-expressing cells was fully processed, indicating that it underwent SPP cleavage in the ER membrane. In addition, we noticed that a mutant of the core protein (A180V/S183L/C184V) which showed significantly reduced cleavage by SPP in vivo and in vitro (36) also showed reduced mitochondrial localization (data not shown). Since SPP is only known to act within the ER membrane, these data indicate that the localization of the core protein to mitochondria in cells requires prior insertion and cleavage at the ER. Hypothetically, the core protein could also be released from the ER membrane and/or lipid droplets after SPP cleavage and then become inserted into the mitochondrial outer membrane (Fig. 7, point 3). Many proteins at the mitochondrial outer membrane and the cytosolic side of the ER membrane are posttranslationally inserted through a characteristic C-terminal transmembrane anchor (reviewed in references 8 and 69). Such C-tail-anchored proteins include members of the family of translocases of the outer mitochondrial membrane necessary for mitochondrial import, several enzymes, metabolic cofactors, and many members of the Bcl-2 family. Recently, it was reported that the HCV RNA-dependent RNA polymerase NS5B inserts into the ER membrane via a 21-amino-acid C-tail anchor (61). However, tail-anchored proteins are generally integral membrane proteins, whereas we showed here that the majority of the mitochondrial core protein is peripherally attached (Fig. 5C). Alternatively, the core protein could bind another protein at the mitochondrial surface via its mitochondrial targeting motif. Indeed, the TOM20 translocase was found to support the association of a tail-anchored protein with the mitochondrial outer membrane (38). In addition, many Bcl-2-like proteins are known to assist other Bcl-2 family members to localize to mitochondria (reviewed in reference 17).
The specific association of the core protein with mitochondria strengthens the idea that the core protein could modify mitochondrial functions during an HCV infection. Several mitochondrial abnormalities have been observed in HCV-infected or core protein-expressing cells. They include mitochondria with an abnormal shape, with vacuoles and thinned and fragmented cristae, observed in Huh-7 cells containing the full-length HCV replicon (52) and in liver biopsies from individuals with chronic HCV infection (5). Disrupted mitochondrial membrane structures were also reported for hepatocytes isolated from core protein-transgenic mice (43). Core protein-induced mitochondrial injury was linked to enhanced lipid peroxidation and the presence of reactive oxygen species (48). Finally, the core protein has been shown to influence apoptosis (reviewed in references 16 and 56), a process in which mitochondria play a key role.
The role of the mitochondrial targeting of the core protein in the viral life cycle is unknown at present. Interestingly, a study of subgenomic HCV replicons found the NS3 and NS4A proteins to be preferentially localized to ER cisternae in close proximity to mitochondria, indicating that MAMs might be used for RNA replication (44). If so, the core protein present on mitochondria would be in a good position to encapsidate progeny positive-stranded RNAs released from the replication complex. Another, not mutually exclusive, possibility is that an alteration of mitochondrial function by the core protein may change the cellular physiology to favor viral replication. Further experiments are needed to test these hypotheses and to fully characterize the mode by which the core protein is targeted to the mitochondria.
Acknowledgments
We gratefully acknowledge I. Hassepass, A. Mion, M. Volkening, C. Kuntzen, E. Verdin, C. Hetzer-Egger, and M. Bissel for helpful discussions; B. Martoglio, G. Daum, J. C. Reed, T. Kaufmann, C. Borner, and W. Just for technical advice; John Carroll and Chris Goodfellow for graphics; and Stephen Ordway and Gary Howard for editorial assistance. We thank B. J. North and H. Spring for support with confocal microscopy, B. Li for initial domain mapping experiments, and A. Pedal, B. Hub, J. Eglinger, and L. Romanyuk for technical assistance.
This work was supported by European Union grant QLK2-CT-2002-01329 (R.B.), University of California AIDS Research Program grant ID02-NCIRE-040 (T.S.B.Y.), and University of California Liver Center Pilot Feasibility Project Award P30 DK26743-22 (M.O.).
Footnotes
This paper is dedicated to Harald zur Hausen on the occasion of his retirement as scientific director of the German Cancer Research Center and for his support, mentoring, and motivation.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 2.Ardail, D., F. Gasnier, F. Lerme, C. Simonot, P. Louisot, and O. Gateau-Roesch. 1993. Involvement of mitochondrial contact sites in the subcellular compartmentalization of phospholipid biosynthetic enzymes. J. Biol. Chem. 268:25985-25992. [PubMed] [Google Scholar]
- 3.Ardail, D., F. Lerme, and P. Louisot. 1991. Involvement of contact sites in phosphatidylserine import into liver mitochondria. J. Biol. Chem. 266:7978-7981. [PubMed] [Google Scholar]
- 4.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates with cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaro, G., G. Di Lorenzo, A. Asti, M. Ribersani, G. Belloni, B. Grisorio, G. Filice, and G. Barbarini. 1999. Hepatocellular mitochondrial alterations in patients with chronic hepatitis C: ultrastructural and biochemical findings. Am. J. Gastroenterol. 94:2198-2205. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 8.Borgese, N., S. Colombo, and E. Pedrazzini. 2003. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra, N. C., M. J. Spiro, and R. G. Spiro. 1998. Identification of a glycoprotein from rat liver mitochondrial inner membrane and demonstration of its origin in the endoplasmic reticulum. J. Biol. Chem. 273:19715-19721. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Colberg-Poley, A. M., M. B. Patel, D. P. Erezo, and J. E. Slater. 2000. Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J. Gen. Virol. 81:1779-1789. [DOI] [PubMed] [Google Scholar]
- 13.Di Bisceglie, A. M. 1998. Hepatitis C. Lancet 351:351-355. [DOI] [PubMed] [Google Scholar]
- 14.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 15.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannini, C., and C. Brechot. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl. 1):S27-S38. [DOI] [PubMed] [Google Scholar]
- 17.Harris, M. H., and C. B. Thompson. 2000. The role of the Bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell Death Differ. 7:1182-1191. [DOI] [PubMed] [Google Scholar]
- 18.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 19.Hope, R. G., D. J. Murphy, and J. McLauchlan. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261-4270. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh, T. Y., M. Matsumoto, H. C. Chou, R. Schneider, S. B. Hwang, A. S. Lee, and M. M. Lai. 1998. Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem. 273:17651-17659. [DOI] [PubMed] [Google Scholar]
- 21.Jin, D. Y., H. L. Wang, Y. Zhou, A. C. Chun, K. V. Kibler, Y. D. Hou, H. Kung, and K. T. Jeang. 2000. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 19:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kartenbeck, J., H. Stukenbrok, and A. Helenius. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel, M., M. Lorinczi, R. Rijnbrand, S. M. Lemon, and S. J. Watowich. 2001. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 75:2119-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemberg, M. K., and B. Martoglio. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735-744. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo, S. Y., F. Masiarz, S. B. Hwang, M. M. Lai, and J. H. Ou. 1995. Differential subcellular localization of hepatitis C virus core gene products. Virology 213:455-461. [DOI] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgnomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 28.Lu, W., S. Y. Lo, M. Chen, K. Wu, Y. K. Fung, and J. H. Ou. 1999. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology 264:134-141. [DOI] [PubMed] [Google Scholar]
- 29.Marsh, B. J., D. N. Mastronarde, K. F. Buttle, K. E. Howell, and J. R. McIntosh. 2001. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl. Acad. Sci. USA 98:2399-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinou, J. C., and D. R. Green. 2001. Breaking the mitochondrial barrier. Nat. Rev. Mol. Cell. Biol. 2:63-67. [DOI] [PubMed] [Google Scholar]
- 31.Martoglio, B., S. Hauser, and B. Dobberstein. 1998. Cotranslational translocation of proteins into microsomes derived from the rough endoplasmic reticulum of mammalian cells, p. 265-273. In J. E. Celis (ed.), Cell biology: a laboratory handbook, 2nd ed. Academic Press, San Diego, Calif.
- 32.Matsumoto, M., T. Y. Hsieh, N. Zhu, T. VanArsdale, S. B. Hwang, K. S. Jeng, A. E. Gorbalenya, S. Y. Lo, J. H. Ou, C. F. Ware, and M. M. Lai. 1997. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J. Virol. 71:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto, M., S. B. Hwang, K. S. Jeng, N. Zhu, and M. M. Lai. 1996. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218:43-51. [DOI] [PubMed] [Google Scholar]
- 34.Matsuura, Y., T. Harada, M. Makimura, M. Sato, H. Aizaki, T. Suzuki, and T. Miyamura. 1994. Characterization of HCV structural proteins expressed in various animal cells. Intervirology 37:114-118. [DOI] [PubMed] [Google Scholar]
- 35.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 36.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mihara, K. 2000. Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. Bioessays 22:364-371. [DOI] [PubMed] [Google Scholar]
- 38.Millar, D. G., and G. C. Shore. 1996. Signal anchor sequence insertion into the outer mitochondrial membrane. Comparison with porin and the matrix protein targeting pathway. J. Biol. Chem. 271:25823-25829. [DOI] [PubMed] [Google Scholar]
- 39.Moradpour, D., C. Englert, T. Wakita, and J. R. Wands. 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222:51-63. [DOI] [PubMed] [Google Scholar]
- 40.Morgan, K. S. 2003. Characterization of cellular proteins, p. 5.2.8-5.2.10. In J. S. Bonifacino (ed.), Current protocols in cell biology. John Wiley & Sons, Inc., New York, N.Y.
- 41.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 42.Moriya, K., K. Nakagawa, T. Santa, Y. Shintani, H. Fujie, H. Miyoshi, T. Tsutsumi, T. Miyazawa, K. Ishibashi, T. Horie, K. Imai, T. Todoroki, S. Kimura, and K. Koike. 2001. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 61:4365-4370. [PubMed] [Google Scholar]
- 43.Moriya, K., H. Yotsuyanagi, Y. Shintani, H. Fujie, K. Ishibashi, Y. Matsuura, T. Miyamura, and K. Koike. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78:1527-1531. [DOI] [PubMed] [Google Scholar]
- 44.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgnomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 45.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 46.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 47.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 48.Okuda, M., K. Li, M. R. Beard, L. A. Showalter, F. Scholle, S. M. Lemon, and S. A. Weinman. 2002. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 122:366-375. [DOI] [PubMed] [Google Scholar]
- 49.Owsianka, A. M., and A. H. Patel. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330-340. [DOI] [PubMed] [Google Scholar]
- 50.Pallotti, F., and G. Lenaz. 2001. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 65:1-35. [DOI] [PubMed] [Google Scholar]
- 51.Perlemuter, G., A. Sabile, P. Letteron, G. Vona, A. Topilco, Y. Chretien, K. Koike, D. Pessayre, J. Chapman, G. Barba, and C. Brechot. 2002. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 16:185-194. [DOI] [PubMed] [Google Scholar]
- 52.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pitts, K. R., Y. Yoon, E. W. Krueger, and M. A. McNiven. 1999. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell 10:4403-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramratnam, B., S. Bonhoeffer, J. Binley, A. Hurley, L. Zhang, J. E. Mittler, M. Markowitz, J. P. Moore, A. S. Perelson, and D. D. Ho. 1999. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 354:1782-1785. [DOI] [PubMed] [Google Scholar]
- 55.Ravaggi, A., G. Natoli, D. Primi, A. Albertini, M. Levrero, and E. Cariani. 1994. Intracellular localization of full-length and truncated hepatitis C virus core protein expressed in mammalian cells. J. Hepatol. 20:833-836. [DOI] [PubMed] [Google Scholar]
- 56.Ray, R. B., and R. Ray. 2001. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 202:149-156. [DOI] [PubMed] [Google Scholar]
- 57.Rizzuto, R., P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft, and T. Pozzan. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763-1766. [DOI] [PubMed] [Google Scholar]
- 58.Rosenberg, S. 2001. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313:451-464. [DOI] [PubMed] [Google Scholar]
- 59.Rusinol, A. E., Z. Cui, M. H. Chen, and J. E. Vance. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269:27494-27502. [PubMed] [Google Scholar]
- 60.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 62.Schumacher, M. M., J. Y. Choi, and D. R. Voelker. 2002. Phosphatidylserine transport to the mitochondria is regulated by ubiquitination. J. Biol. Chem. 277:51033-51042. [DOI] [PubMed] [Google Scholar]
- 63.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shockett, P., M. Difilippantonio, N. Hellman, and D. G. Schatz. 1995. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc. Natl. Acad. Sci. USA 92:6522-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 66.Vance, J. E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265:7248-7256. [PubMed] [Google Scholar]
- 67.van der Vusse, G. J., M. van Bilsen, J. F. Glatz, D. M. Hasselbaink, and J. J. Luiken. 2002. Critical steps in cellular fatty acid uptake and utilization. Mol. Cell. Biochem. 239:9-15. [DOI] [PubMed] [Google Scholar]
- 68.Voelker, D. R. 2000. Interorganelle transport of aminoglycerophospholipids. Biochim. Biophys. Acta 1486:97-107. [DOI] [PubMed] [Google Scholar]
- 69.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 70.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215-2218. [DOI] [PubMed] [Google Scholar]
- 71.Yang, J., X. Liu, K. Bhalla, C. N. Kim, A. M. Ibrado, J. Cai, T. I. Peng, D. P. Jones, and X. Wang. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275:1129-1132. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida, T., T. Hanada, T. Tokuhisa, K. Kosai, M. Sata, M. Kohara, and A. Yoshimura. 2002. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J. Exp. Med. 196:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.You, L. R., C. M. Chen, T. S. Yeh, T. Y. Tsai, R. T. Mai, C. H. Lin, and Y. H. Lee. 1999. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 73:2841-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]