Abstract
This study examined the effect of naturally occurring Epstein-Barr virus (EBV) latent membrane protein 1 (LMP-1) gene sequence variation on the LMP-1 half-life in epithelial cells. The LMP-1 half-life was not influenced by sequence variation in amino acids 250 to 307 or amino acids 343 to 352. The LMP-1 half-life was short when the amino acid encoded at position 129 was methionine, the initiation codon product of lytic LMP-1 (lyLMP-1). The mutation of amino acid 129 to isoleucine greatly increased the LMP-1 half-life. Expression of lyLMP-1 in trans down-regulated the LMP-1 half-life in a dose-dependent manner and restored a short-half-life phenotype to the mutated LMP-1 construct lacking the cis ability to express lyLMP-1. This observed dominant negative effect of lyLMP-1 expression on the LMP-1 half-life in epithelial cells in vitro may have implications for EBV epithelial oncogenesis in vivo.
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus that is associated with numerous malignancies, especially nasopharyngeal carcinoma (NPC). Oncogenic EBV latent membrane protein 1 (LMP-1) is a 63-kDa protein of 386 amino acids (strain B958) encoded by the BNLF1 gene (Fig. 1). LMP-1 inserts into the plasma membrane and functions as a ligand-independent, constitutively active growth factor receptor, similar to CD40 of the tumor necrosis factor receptor (TNFR) family. After oligomerization, LMP-1 binds TNFR-associated factors (TRAFs) and the TNFR-associated death domain protein (26, 38) to activate intracellular signaling through the NF-κB, cJun N-terminal kinase, and p38 mitogen-activated protein kinase pathways (10, 11, 42). LMP-1 also activates the JAK-STAT signaling pathway, possibly through interaction with Janus kinase 3 (JAK3) (19, 22). Cumulatively, these signals generate many effects on host cell growth, differentiation, apoptosis, and immune response.
FIG. 1.
LMP-1 structure and gene expression. (A) LMP-1 of EBV strain B958 consists of a short cytoplasmic amino-terminal domain (amino acids [aa] 1 to 25), six transmembrane domains (amino acids 26 to 186), and a long cytoplasmic carboxy-terminal domain (amino acids 187 to 386). The following functional domains important to LMP-1 protein-protein interactions and intracellular signal transduction are shown: TRAFs (PXQXT, amino acids 204 to 208), TNFR-associated death domain protein (TRADD; YYD, amino acids 384 to 386), and JAK3 (Box1a [PHPPLD], amino acids 275 to 280; Box1b [PHPPLD], amino acids 302 to 307; and Box2 [PPQLTEEVENK], amino acids 320 to 330). The following important sites of naturally occurring LMP-1 gene sequence variation are shown: amino acid 129, the repeat unit region (amino acids 250 to 307), and the 30-nucleotide domain (deletion or duplication; encoding amino acids 343 to 352). (B) LMP-1 gene sequences may be expressed from any of three different promoters. The ED-L1 and ED-L1E promoters are located upstream of the LMP-1 ORF. Transcripts from either of these two promoters (ED-L1, 2.8-kb message; ED-L1E, 3.7-kb message) express only the complete LMP-1. The ED-L1A promoter is located within the first intron of the LMP-1 gene. Transcripts from this promoter (ED-L1A, 2.5-kb message) express the truncated lyLMP-1 only when the amino acid encoded at position 129 is methionine and the lyLMP-1 ORF is created, as found for EBV strain B958.
The LMP-1 gene manifests remarkable natural sequence heterogeneity (9, 35, 48). Although a specific LMP-1 genotype-disease phenotype relationship has not yet been identified (48), NPC-derived LMP-1 is both structurally and functionally different from the LMP-1 derived from the laboratory EBV strain B958 in the following ways: (i) NPC LMP-1 activates NF-κB signaling and AP-1 transactivation better than B958 LMP-1 (5, 7, 15, 16, 27, 33, 34), (ii) NPC LMP-1 down-regulates cell immune markers, blocks cell apoptosis, and up-regulates the epidermal growth factor receptor (EGFR) better than B958 LMP-1 (7, 8, 15, 27, 33, 34, 52), and (iii) NPC LMP-1 transforms cells and forms tumors in mice more efficiently than B958 LMP-1 (8, 24, 28, 33, 52). Chimeric studies of LMP-1 have mapped some of these functional differences to the carboxy-terminal 30-nucleotide domain encoding amino acids 343 to 352 (28) and transmembrane domain amino acids 85 to 129 (5, 34).
The intracellular quantity of expressed LMP-1 and the cell type background together influence the strength of LMP-1 signaling and its ultimate phenotypic effect on the cell (4, 16, 17, 20, 24, 28, 49, 50). The intracellular half-life of LMP-1 is generally short (Table 1), and the kinetics of LMP-1 turnover resemble those of a cellular growth factor receptor (31). Rapid LMP-1 turnover negatively regulates constitutive LMP-1 signaling (31, 32). LMP-1s with longer half-lives generate proportionally more signal activity and exert a greater phenotypic effect on the cell (5, 31, 32). Regulation of the LMP-1 half-life has been alternatively mapped to carboxy-terminal domain amino acids 331 to 364 (32, 37) or to transmembrane domain amino acids 45 to 192 (5).
TABLE 1.
LMP-1 intracellular half-life: summary of the published literature
| LMP-1 strain | Cell line | LMP-1 half-life (h) | Reference |
|---|---|---|---|
| B958 | HEp2 (human epithelial) | 8 | 32 |
| BALB/3T3 (murine fibroblast) | 3.3 | 32 | |
| BALB/3T3 (murine fibroblast) | 2 | 31 | |
| BALB/3T3 (murine fibroblast) | 2-3.5 | 4 | |
| LCL (human B Lymphocyte) | 5 | 3 | |
| EA-B1 (human B lymphocyte) | 2 | 2 | |
| COS 7/5 (monkey kidney) | 2 | 2 | |
| 293 (human embryonic kidney) | 3.8 | 43 | |
| 293 (human embryonic kidney) | 2.9-3.9 | 5 | |
| CAO | 293 (human embryonic kidney) | 7-7.3 | 5 |
| Jijoye | Burkitt's lymphoma (human B lymphocyte) | 2 | 29 |
| Burkitt's lymphoma (human B lymphocyte) | 2 | 36 |
In the present study, variant recombinant LMP-1 expression constructs were created to test the hypothesis that naturally occurring LMP-1 sequence variation influences the intracellular half-life of LMP-1 in RHEK-1 human keratinocyte epithelial cells. The functional equivalent of an LMP-1 cDNA clone (clone B958WT) was created using a PCR-based splicing-by-overlap extension technique (23) to assemble the three LMP-1 gene exons from B958 lymphoblastoid cell line DNA and clone into the pSG5 expression vector (Stratagene). Naturally occurring LMP-1 sequence variants contained in previously cloned LMP-1 sequences (48) or in the Raji lymphoblastoid cell line were utilized as templates for specific sequence variations that were introduced into clone B958WT by the same splicing-by-overlap extension and cloning technique. In total, 13 recombinant LMP-1 expression clones in three different structural variant groups were created, each with a different specific sequence variation superimposed upon an otherwise identical B958 sequence background (Table 2). Each clone also expressed a FLAG epitope at the amino terminus of LMP-1. All clones were sequenced to verify the specific variant sequence structure created for each clone.
TABLE 2.
Structural characteristics of the LMP-1 clones and their associated intracellular LMP-1 half-lives in RHEK-1 epithelial cellsa
| LMP-1 clone by group | No. of repeat units (aa 250-307) | JAK3-Box1a 1/2 repeat unit (aa 275-280)b | JAK3-Box1b incomplete repeat (aa 302-307)b | 30-Nucleotide domain (aa 343-352)b | Amino acids deleted | Amino acid at position 129 | Protein half-life (h) (mean ± SD) |
|---|---|---|---|---|---|---|---|
| Group 1 | |||||||
| B958WT | 4.5 | + | + | + | None | Met | 5.0 ± 0.5 |
| A | 3 | − | + | + | 272-287 | Met | 5.8 ± 0.1 |
| B | 2.5 | + | + | + | 250-271 | Met | 6.7 ± 0.5 |
| C | 2 | − | + | + | 272-298 | Met | 6.4 ± 0.7 |
| D | 1 | − | + | + | 250-287 | Met | 7.8 ± 1.0 |
| E | 0 | − | + | + | 250-298 | Met | 6.0 ± 0.6 |
| F | 0 | − | − | + | 250-307 | Met | 5.6 ± 0.4 |
| Group 2 | |||||||
| B958WT | 4.5 | + | + | + | None | Met | 5.0 ± 0.5 |
| Gc | 4.5 | + | + | ++e | None | Met | 7.2 ± 1.0 |
| Hc | 4.5 | + | + | + | None | Met | 7.8 ± 0.8 |
| Id | 4.5 | + | + | − | 343-352 | Met | 6.3 ± 1.0 |
| Group 3 | |||||||
| B958WT | 4.5 | + | + | + | None | Met | 5.0 ± 0.5 |
| Jf | 4.5 | + | + | + | None | Ile | 19.3 ± 1.0 |
| K | 4.5 | + | + | + | None | Ile | 18.3 ± 1.8 |
| Lf | 4.5 | + | + | + | None | Met | 5.3 ± 0.9 |
aa, amino acids.
+, present; −, absent.
Amino acid sequence differs from that of clone B958WT at positions 349 (Asp versus Ala), 352 (His versus Arg), and 366 (Ser versus Thr).
Amino acid sequence differs from that of clone B958WT at position 366 (Ser versus Thr).
Amino acids 343 to 352 are duplicated in tandem.
Amino acid sequence differs from that of clone B958WT at positions 85, 106, and 122 (see Table 3).
LMP-1 repeat unit region sequences do not influence LMP-1 protein half-life.
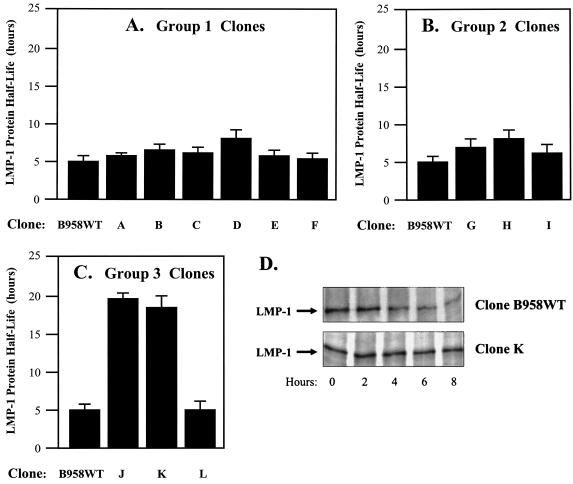
The carboxy-terminal domain repeat unit region (corresponding to amino acids 250 to 307) of the LMP-1 gene of the B958 strain comprises 4[1/2] repeat units consisting of three perfect copies of a 33-nucleotide repeat unit, another repeat unit with a one-half-repeat-unit insertion that encodes a JAK3 binding domain (JAK3-Box1a), and an incomplete repeat unit encoding another JAK3 binding domain (JAK3-Box1b) (Fig. 1). Naturally occurring LMP-1 gene variants comprise zero to seven repeat units, and intrastrain recombination in vivo can alter the number of repeat units present in a single LMP-1 strain (48). Seven group 1 LMP-1 clones were studied to determine the effect of variation in the numbers of repeat units and JAK3-Box1 sequences upon the LMP-1 half-life. The LMP-1s expressed from all seven group 1 clones had similar half-lives (Table 2 and Fig. 2). These results suggest that variation in the repeat unit region sequences is likely not involved in the regulation of LMP-1 turnover in RHEK-1 epithelial cells.
FIG. 2.
Effect of LMP-1 gene sequence variation on the intracellular half-life of LMP-1 expressed in RHEK-1 epithelial cells. LMP-1 expression plasmids (25 to 50 μg) were electroporated into 5 × 106 RHEK-1 cells. The transfected cells were maintained in Dulbecco's minimal essential medium (DMEM) with 5% fetal bovine serum for 48 h. The cells were incubated in methionine-cysteine-free DMEM for 1 h (starve), incubated in methionine-cysteine-free DMEM containing [35S]methionine-cysteine for 1 h (pulse), grown in complete DMEM (chase), and harvested at various times over the next 24 h. Protein from cell lysates was immunoprecipitated with anti-LMP-1 antibodies (S-12 [BD-Pharmingen] or CS1-4 [DAKO]) or anti-FLAG antibody (Sigma-Aldrich). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, radiolabeled LMP-1 was viewed by autoradiography and quantitated by spot densitometry using an AlphaImager system (Alpha-Innotech Corporation). The LMP-1 half-life was calculated by plotting protein quantity versus time and interpolating the time at which a 50% reduction in quantity occurred. The results are presented as bar graphs representing the mean LMP-1 half-life (+standard deviation) (in hours) of three independent experiments for each LMP-1 clone. (A) Group 1 clones (see Table 2) demonstrated LMP-1 half-lives that were not significantly different from each other (for clone B958WT versus clone D, the P value was >0.05 by the t test). (B) Group 2 clones (see Table 2) demonstrated LMP-1 half-lives that were not significantly different from each other (for clone B958WT versus clone H, the P value was >0.05 by the t test). (C) Group 3 clones (see Tables 2 and 3) demonstrated significant differences in the LMP-1 half-life (for clone B958WT versus clone K, the P value was <0.05 by the t test; for clone J versus clone L, the P value was <0.05 by the t test). (D) Representative autoradiographs demonstrating a short LMP-1 half-life for clone B958WT and a long LMP-1 half-life for clone K. In conclusion, the LMP-1 half-life is determined by the amino acid encoded at position 129 (methionine or isoleucine).
LMP-1 deletion-duplication sequences do not influence LMP-1 half-life.
The carboxy-terminal 30-nucleotide domain (corresponding to amino acids 343 to 352) is present in the LMP-1 gene of the B958 strain. However, these 30 nucleotides are frequently involved in intrastrain recombinant deletion or duplication in vivo (48). The presence or absence of the 10 amino acids encoded by this 30-nucleotide domain has been implicated in functional differences between LMP-1 strains, including the regulation of the LMP-1 half-life (28, 32, 37). Four group 2 LMP-1 clones were studied to determine the effects of both deletion and duplication of amino acids 343 to 352 upon the LMP-1 half-life. The LMP-1s expressed from all four group 2 clones had similar half-lives (Table 2 and Fig. 2). These results suggest that sequence variation in amino acids 343 to 352 (absence, presence, or duplication) is likely not involved in the regulation of LMP-1 turnover in RHEK-1 epithelial cells.
LMP-1 amino acid 129 and the lytic LMP-1 (lyLMP-1) open reading frame (ORF) influence LMP-1 half-life.
A region of the transmembrane domains of the LMP-1 gene of the NPC EBV strains CAO and C15 (encoding amino acids 85 to 129) has been implicated in functional differences from EBV strain B958, including the regulation of the LMP-1 half-life (5, 34). Four group 3 LMP-1 clones were studied to determine the effect of sequence variation in the transmembrane domains upon the LMP-1 half-life. Clone J differed from clone B958WT by only four amino acid changes (Table 2 and Table 3), the same four changes in amino acids 85 to 129 found in common between the NPC EBV strains CAO and C15. The half-life of clone J was significantly longer than that of clone B958WT (Table 2 and Fig. 2).
TABLE 3.
Amino acid differences within the transmembrane domains (amino acids 85 to 129) of the group 3 LMP-1 clones
| Position | Amino acid in clone:
|
|||
|---|---|---|---|---|
| B958WT | J | K | L | |
| 85 | Ile | Leu | Ile | Leu |
| 106 | Phe | Tyr | Phe | Tyr |
| 122 | Ile | Leu | Ile | Leu |
| 129 | Met | Ile | Ile | Met |
PCR-based site-directed mutagenesis (QuickChange kit; Stratagene) was performed to mutate amino acid 129 in clone B958WT from methionine to isoleucine to eliminate the lyLMP-1 ORF product (clone K) (Tables 2 and 3). Similarly, amino acid 129 was mutated in clone J from isoleucine to methionine to create the lyLMP-1 ORF product (clone L) (Tables 2 and 3). Remarkably, clone K (with Ile129) demonstrated a long LMP-1 half-life similar to that of clone J (with Ile129), and clone L (with Met129) demonstrated a short LMP-1 half-life similar to that of clone B958WT (with Met129) (Table 2 and Fig. 2). These results suggest that the presence or absence of the lyLMP-1 ORF (as defined by the amino acid at position 129) is a significant determinant of the LMP-1 half-life in RHEK-1 epithelial cells.
In vivo, the lyLMP-1 ORF is expressed from the ED-L1A promoter located in the first intron of the LMP-1 gene (Fig. 1) (25). Although the LMP-1-expressing clones used in the present study did not contain intron sequences, the LMP-1 ORF of EBV strain B958 contains six internal ATG codons (corresponding to amino acids 34, 51, 89, 129, 184, and 339) that could potentially serve as internal protein translation initiation sites in the transcripts expressed from these clones in vitro. Western blot analysis demonstrated a 45-kDa band consistent with lyLMP-1 expression from clone B958WT (with Met129; short half-life) but not clone K (with Ile129; long half-life) (Fig. 3). These results suggest that lyLMP-1 expression may influence the LMP-1 half-life.
FIG. 3.
Effect of lyLMP-1 expression on intracellular half-life of LMP-1 expressed in RHEK-1 epithelial cells. The indicated amounts of clones B958WT, K, and lyLMP-1 were transfected or cotransfected into RHEK-1 cells. (A) Western blot detection of expressed LMP-1 (63-kDa band) and lyLMP-1 (45-kDa band) was achieved by using anti-LMP-1 monoclonal antibody S-12 (BD-Pharmingen). Expression of lyLMP-1 was detected with clone lyLMP-1 (lane 1) and clone B958WT (lane 3), which contains the lyLMP-1 ORF product, but not with clone K (lane 2), which lacks the lyLMP-1 ORF. lyLMP-1 was expressed from 100 μg of clone lyLMP-1 in the clone K-cotransfected cells (lane 6) at concentrations approximately equimolar to those of native lyLMP-1 expressed from clone B958WT (lane 3). (B) The intracellular LMP-1 half-life was determined by pulse-chase labeling, immunoprecipitation, and autoradiography as described in the legend to Fig. 2. The results are presented as a bar graph representing the mean LMP-1 half-lives (+standard deviation) (in hours) for three independent experiments in the presence of the indicated quantities of the lyLMP-1 expression plasmid. These data demonstrate a significant dose-dependent reduction in the clone K LMP-1 half-life by clone lyLMP-1. (C) Representative autoradiographs demonstrating a long LMP-1 half-life for clone K cotransfected with 25 μg of clone lyLMP-1 and a short LMP-1 half-life for clone K cotransfected with 100 μg of clone lyLMP-1. In conclusion, lyLMP-1 in trans generates a short-half-life phenotype for LMP-1 when lyLMP-1 is expressed either natively from an lyLMP-1 ORF-containing LMP-1 gene or ectopically from expression plasmid clone lyLMP-1.
Expression of lyLMP-1 specifically down-regulates LMP-1 half-life.
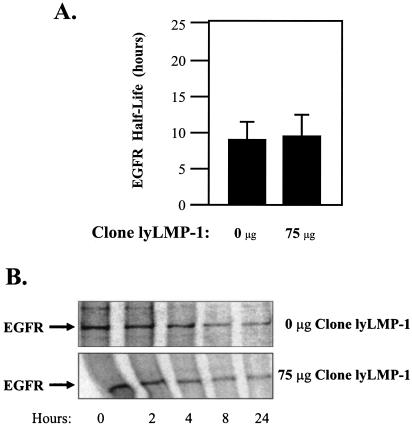
Clone lyLMP-1 was created by PCR amplification of only the lyLMP-1 ORF from clone B958WT, followed by restriction cloning of the product into pSG5. To test the hypothesis that lyLMP-1 down-regulates the LMP-1 half-life, RHEK-1 epithelial cells were cotransfected with clone K and clone lyLMP-1. Increasing quantities of clone lyLMP-1 significantly reduced the clone K LMP-1 half-life in a dose-dependent manner (Fig. 3). Expression of lyLMP-1 in trans from clone lyLMP-1 restored a short LMP-1 half-life phenotype to the mutant clone K that lacked the cis ability to express lyLMP-1. Transfection with clone lyLMP-1 alone had no effect on the half-life of either total cellular protein (data not shown) or the EGFR, a cell-encoded plasma membrane protein with turnover kinetics similar to those of LMP-1 (Fig. 4). These results suggest that expression of lyLMP-1 in trans specifically down-regulates the LMP-1 half-life in RHEK-1 epithelial cells.
FIG. 4.
Effect of lyLMP-1 expression on intracellular half-life of EGFR in RHEK-1 epithelial cells. The indicated amounts of clone lyLMP-1 were transfected into RHEK-1 cells. The intracellular EGFR protein half-life was determined by pulse-chase labeling, immunoprecipitation with an anti-EGFR antibody (clone 13; BD-Pharmingen), and autoradiography as described in the legend to Fig. 2. (A) The results are presented as a bar graph representing the mean EGFR protein half-life (+standard deviation) (in hours) for two independent experiments in the presence of the indicated quantities of the lyLMP-1 expression plasmid. These data demonstrate that transfection with clone lyLMP-1 had no significant effect on the EGFR protein half-life. (B) Representative autoradiographs demonstrating no significant difference between the half-life of the EGFR protein in cells transfected with 0 μg of clone lyLMP-1 and that in cells transfected with 75 μg of clone lyLMP-1. In conclusion, expression of lyLMP-1 does not affect the turnover of the EGFR protein, a cell-encoded plasma membrane receptor protein with kinetics similar to those of LMP-1.
In vivo, LMP-1 is expressed during both latent and productive EBV infection from one or both of two upstream promoters, ED-L1 and ED-L1E (Fig. 1) (18, 41). A third promoter, ED-L1A, is located in the first intron of the LMP-1 gene (Fig. 1) and is active in B lymphocytes only during productive EBV infection (25, 44). Most naturally occurring LMP-1 gene sequence variants contain either ATG (methionine) or ATT (isoleucine) at the codon corresponding to amino acid 129, and translation of the ED-L1A message into lyLMP-1 requires the ATG codon (12).
The 45-kDa lyLMP-1 is found in nuclear and intracytoplasmic membranes, intracellular virus particles, and extracellular virions (13, 45). lyLMP-1 does not localize to plasma membranes, oligomerize with LMP-1, activate NF-κB signaling, or transform cells (6, 14, 45, 50, 51). lyLMP-1 functions as a dominant negative regulator of NF-κB and cJun N-terminal kinase signaling by LMP-1, but the mechanism does not involve sequestering TRAF proteins or disrupting LMP-1 oligomerization (13, 14). However, the ability of lyLMP-1 to decrease the LMP-1 half-life could explain the down-regulation of LMP-1 signaling by lyLMP-1. Consistent with the growth factor receptor model, LMP-1 in the plasma membrane is internalized and degraded after signaling (6, 31), likely through the ubiquitin-proteasome pathway (2, 43). lyLMP-1 may act to accelerate LMP-1 turnover, facilitating LMP-1 degradation perhaps even prior to its localization in the plasma membrane and constitutive signaling (40). Expressed early in the EBV infection of a B lymphocyte (13, 14), lyLMP-1 may act to modulate the inhibition of productive EBV replication by LMP-1 (1, 39).
The finding that lyLMP-1 expression decreases the LMP-1 half-life in epithelial cells may explain the observation that the lyLMP-1 ORF is consistently absent from the LMP-1 genes of EBV strains found in patients with NPC (12, 21), despite the apparent transcriptional activity of the ED-L1A promoter in NPC (30). Differences between NPC and B958 LMP-1 signal activation and oncogenicity in vitro (5, 7, 8, 15, 16, 24, 27, 28, 33, 34, 52) may be explained by the absence of the lyLMP-1 ORF in the NPC LMP-1 genes and the consequently longer half-lives of the NPC LMP-1s, although this hypothesis remains to be tested. Nonproductive persistent EBV infection of normal oral epithelium is associated with a unique pattern of replicative and latent EBV gene expression, including the LMP-1 gene (46, 47). If the ED-L1A promoter is transcriptionally active concurrent with the ED-L1 and/or ED-L1E promoters in nonproductive epithelial infection, then EBV strains that lack the lyLMP-1 ORF and that have long LMP-1 half-lives may be selected for the development of NPC.
Acknowledgments
This work was supported by U.S.P.H.S. grant NIH R01-DE12323 to Dennis M. Walling.
We thank Johng S. Rhim for providing the RHEK-1 epithelial cells for this work.
REFERENCES
- 1.Adler, B., E. Schaadt, B. Kempkes, U. Zimber-Strobl, B. Baier, and G. W. Bornkamm. 2002. Control of Epstein-Barr virus reactivation by activated CD40 and viral latent membrane protein 1. Proc. Natl. Acad. Sci. USA 99:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275:23491-23499. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal, V. R., and B. Sugden. 1987. Posttranslational processing of an Epstein-Barr virus-encoded membrane protein expressed in cells transformed by Epstein-Barr virus. J. Virol. 61:866-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 5.Blake, S. M., A. G. Eliopoulos, C. W. Dawson, and L. S. Young. 2001. The transmembrane domains of the EBV-encoded latent membrane protein 1 (LMP1) variant CAO regulate enhanced signalling activity. Virology 282:278-287. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, W. F., III, K. D. Erickson, M. Hoedt-Miller, and J. M. Martin. 2001. The cytoplasmic amino-terminus of the latent membrane protein-1 of Epstein-Barr virus: relationship between transmembrane orientation and effector functions of the carboxy-terminus and transmembrane domain. Oncogene 20:5313-5330. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, C. W., A. G. Eliopoulos, S. M. Blake, R. Barker, and L. S. Young. 2000. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 272:204-217. [DOI] [PubMed] [Google Scholar]
- 8.Dolcetti, R., M. Quaia, A. Gloghini, V. De Re, P. Zancai, R. Cariati, L. Babuin, A. M. Cilia, S. Rizzo, A. Carbone, and M. Boiocchi. 1999. Biologically relevant phenotypic changes and enhanced growth properties induced in B lymphocytes by an EBV strain derived from a histologically aggressive Hodgkin's disease. Int. J. Cancer 80:240-249. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, R. H., F. Seillier-Moiseiwitsch, and N. Raab-Traub. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79-95. [DOI] [PubMed] [Google Scholar]
- 10.Eliopoulos, A. G., S. M. S. Blake, J. E. Floettmann, M. Rowe, and L. S. Young. 1999. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 73:1023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliopoulos, A. G., N. J. Gallagher, S. M. Blake, C. W. Dawson, and L. S. Young. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085-16096. [DOI] [PubMed] [Google Scholar]
- 12.Erickson, K. D., C. Berger, W. F. Coffin III, E. Schiff, D. M. Walling, and J. M. Martin. 2003. Unexpected absence of the Epstein-Barr virus (EBV) lyLMP-1 open reading frame in tumor virus isolates: lack of correlation between Met129 status and EBV strain identity. J. Virol. 77:4415-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson, K. D., and J. M. Martin. 1997. Early detection of the lytic LMP-1 protein in EBV-infected B-cells suggests its presence in the virion. Virology 234:1-13. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, K. D., and J. M. Martin. 2000. The late lytic LMP-1 protein of Epstein-Barr virus can negatively regulate LMP-1 signaling. J. Virol. 74:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fielding, C. A., K. Sandvej, A. Mehl, P. Brennen, M. Jones, and M. Rowe. 2001. Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 75:9129-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, N., B. Kopper, N. Graf, J. R. Schlehofer, F. A. Grasser, and N. Mueller-Lantzsch. 1999. Functional analysis of different LMP1 proteins isolated from Epstein-Barr virus-positive carriers. Virus Res. 60:41-54. [DOI] [PubMed] [Google Scholar]
- 17.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 18.Gilligan, K., P. Rajadurai, L. Resnick, and N. Raab-Traub. 1990. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc. Natl. Acad. Sci. USA 87:8790-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry, S., C. Sacaze, L. Berrajah, H. Karray, M. Drira, A. Hammami, J. Icart, and B. Mariame. 2001. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int. J. Cancer 91:698-704. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi, M., E. Kieff, and K. M. Izumi. 2002. The Epstein-Barr virus latent membrane protein 1 putative Janus kinase 3 (JAK3) binding domain does not mediate JAK3 association or activation in B-lymphoma or lymphoblastoid cell lines. J. Virol. 76:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higuchi, R., B. Krummel, and R. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, L. F., F. Chen, X. Zheng, I. Ernberg, S. L. Cao, B. Christensson, G. Klein, and G. Winberg. 1993. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene 8:1575-1583. [PubMed] [Google Scholar]
- 25.Hudson, G. S., P. J. Farrell, and B. G. Barrell. 1985. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J. Virol. 53:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, R. J., M. Stack, S. A. Hazlewood, M. Jones, C. G. Blackmore, L.-F. Hu, and M. Rowe. 1998. The 30-base-pair deletion in Chinese variants of the Epstein-Barr virus LMP1 gene is not the major effector of functional differences between variant LMP1 genes in human lymphocytes. J. Virol. 72:4038-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, S. N., Y. S. Chang, and S. T. Liu. 1996. Effect of a 10-amino acid deletion on the oncogenic activity of latent membrane protein 1 of Epstein-Barr virus. Oncogene 12:2129-2135. [PubMed] [Google Scholar]
- 29.Mann, K. P., and D. Thorley-Lawson. 1987. Posttranslational processing of the Epstein-Barr virus-encoded p63/LMP protein. J. Virol. 61:2100-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martel-Renoir, D., V. Grunewald, R. Touitou, G. Schwaab, and I. Joab. 1995. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. J. Gen. Virol. 76:1401-1408. [DOI] [PubMed] [Google Scholar]
- 31.Martin, J., and B. Sugden. 1991. The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ. 2:653-660. [PubMed] [Google Scholar]
- 32.Martin, J., and B. Sugden. 1991. Transformation by the oncogenic latent membrane protein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J. Virol. 65:3246-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehl, A. M., N. Fischer, M. Rowe, F. Hartmann, H. Daus, L. Trumper, M. Pfreundschuh, N. Muller-Lantzsch, and F. A. Grasser. 1998. Isolation and analysis of two strongly transforming isoforms of the Epstein-Barr-Virus (EBV)-encoded latent membrane protein-1 (LMP1) from a single Hodgkin's lymphoma. Int. J. Cancer 76:194-200. [DOI] [PubMed] [Google Scholar]
- 34.Miller, W. E., J. L. Cheshire, A. S. Baldwin, Jr., and N. Raab-Traub. 1998. The NPC derived C15 LMP1 protein confers enhanced activation of NF-kappa B and induction of the EGFR in epithelial cells. Oncogene 16:1869-1877. [DOI] [PubMed] [Google Scholar]
- 35.Miller, W. E., R. H. Edwards, D. M. Walling, and N. Raab-Traub. 1994. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J. Gen. Virol. 75:2729-2740. [DOI] [PubMed] [Google Scholar]
- 36.Moorthy, R., and D. A. Thorley-Lawson. 1990. Processing of the Epstein-Barr virus-encoded latent membrane protein p63/LMP. J. Virol. 64:829-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moorthy, R. K., and D. A. Thorley-Lawson. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of Rat-1 fibroblasts. J. Virol. 67:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389-399. [DOI] [PubMed] [Google Scholar]
- 39.Prince, S., S. Keating, C. Fielding, P. Brennen, E. Floettmann, and M. Rowe. 2003. Latent membrane protein 1 inhibits Epstein-Barr virus lytic cycle induction and progress via different mechanisms. J. Virol. 77:5000-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothenberger, S., M. Rousseaux, H. Knecht, F. C. Bender, D. F. Legler, and C. Bron. 2002. Association of the Epstein-Barr virus latent membrane protein 1 with lipid rafts is mediated through its N-terminal region. Cell. Mol. Life Sci. 59:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadler, R. H., and N. Raab-Traub. 1995. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J. Virol. 69:4577-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sylla, B. S., S. C. Hung, D. M. Davidson, E. Hatzivassiliou, N. L. Malinin, D. Wallach, T. D. Gilmore, E. Kieff, and G. Mosialos. 1998. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc. Natl. Acad. Sci. USA 95:10106-10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, W., O. A. Pavlish, V. S. Spiegelman, A. A. Parkhitko, and S. Y. Fuchs. 2003. Interaction of Epstein-Barr virus latent membrane protein 1 with SCFHOS/betaTrCP E3 ubiquitin ligase regulates the extent of NF-kB activation. J. Biol. Chem. 278:48942-48949. [DOI] [PubMed] [Google Scholar]
- 44.Torii, T., K. Konishi, J. Sample, and K. Takada. 1998. The truncated form of the Epstein-Barr virus LMP-1 is dispensable or complimentable by the full-length form in virus infection and replication. Virology 251:273-278. [DOI] [PubMed] [Google Scholar]
- 45.Vazirabadi, G., T. R. Geiger, W. F. Coffin III, and J. M. Martin. 2003. Epstein-Barr virus latent membrane protein-1 (LMP-1) and lytic LMP-1 localization in plasma membrane-derived extracellular vesicles and intracellular virions. J. Gen. Virol. 84:1997-2008. [DOI] [PubMed] [Google Scholar]
- 46.Walling, D. M., C. M. Flaitz, and C. M. Nichols. 2003. Epstein-Barr virus replication in oral hairy leukoplakia: response, persistence, and resistance to valacyclovir treatment. J. Infect. Dis. 188:883-890. [DOI] [PubMed] [Google Scholar]
- 47.Walling, D. M., P. D. Ling, A. V. Gordadze, M. Montes-Walters, C. M. Flaitz, and C. M. Nichols. Expression of Epstein-Barr virus latent genes in oral epithelium: determinants of the pathogenesis of oral hairy leukoplakia. J. Infect. Dis., in press. [DOI] [PubMed]
- 48.Walling, D. M., N. Shebib, S. C. Weaver, C. M. Nichols, C. M. Flaitz, and J. Webster-Cyriaque. 1999. The molecular epidemiology and evolution of Epstein-Barr virus: sequence variation and genetic recombination in the latent membrane protein-1 gene. J. Infect. Dis. 179:763-774. [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 50.Wang, D., D. Liebowitz, and E. Kieff. 1988. The truncated form of the Epstein-Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J. Virol. 62:2337-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, J.-S., H.-C. Tsai, C.-C. Wu, L.-P. Weng, H.-P. Li, P.-J. Chung, and Y.-S. Chang. 2002. Induction of inducible nitric oxide synthase by Epstein-Barr virus B95-8-derived LMP1 in Balb/3T3 cells promotes stress-induced cell death and impairs LMP1-mediated transformation. Oncogene 21:8047-8061. [DOI] [PubMed] [Google Scholar]