Abstract
The interplay between abnormalities in genes coding for proteins and microRNAs (miRNAs) has been among the most exiting yet unexpected discoveries in oncology over the last decade. The complexity of this network has redefined cancer research as these molecules produced from what was once considered “genomic trash”, have shown to be crucial for cancer initiation, progression, and dissemination. Naturally occurring miRNAs are very short transcripts that never produce a protein or amino acid chain, but act by regulating protein expression during cellular processes such as growth, development and differentiation at the transcriptional, post-transcriptional and/or translational level. In this review article we present miRNAs as ubiquitous players involved in all cancer hallmarks. We also describe the most used methods to detect their expression, which have revealed through gene expression studies the identity of hundreds of miRNAs dysregulated in cancer cells or tumor microenvironment cells. Furthermore, we discuss the role of miRNAs as hormones and as reliable cancer biomarkers and predictors of treatment-response. Along with this, we explore current strategies in designing miRNA-targeting therapeutics, as well as the associated challenges that research envisions to overcome. Finally, we introduce a new wave in molecular oncology translational research, the study of long non-coding RNAs.
Keywords: microRNA, diagnosis, therapy, cancer
1. MicroRNAs are strangers in the genomic galaxy
The central dogma of molecular biology is an explanation of the flow of genetic information within a biological system and it is summarized in the fact that “DNA makes RNA, which encodes protein.” However, over the past years DNA segments have been shown to produce RNA transcripts that do not encode proteins. These transcripts were named non coding RNAs (ncRNAs), and they were considered to be part of a “dark” unexplored segment of the human genome. MiRNAs are a class of small non-coding RNAs 19 to 25 nucleotides (nt) in length that can regulate gene expression by various mechanisms still not fully investigated. They represent the most explored side of the “dark” matter of the genome 1, 2, 3, and the full complement of known (cloned) microRNAs present in a genome is named microRNAome (the full complement of microRNAs present in a genome; for a glossary of terms see Table 1).
Table 1.
Glossary of terms used in the microRNA world.
| ASO: An antisense oligonucleotide is a single-stranded, chemically modified DNA-like molecule that is 17 to 22 nt in length and designed to be complementary to a selected messenger RNA or ncRNA and thereby specifically inhibit expression of that gene. |
| Exome sequencing (targeted exome capture) is an efficient strategy to selectively sequence the coding regions of the genome. |
| Messenger RNA (mRNA): The form of RNA that mediates the transfer of genetic information from the DNA in the cell nucleus to ribosomes in the cytoplasm, where it serves as a template for protein synthesis. |
| MicroRNAome: the full complement of known (cloned) microRNAs present in a genome. Due to the multiple cloning efforts this is growing constantly and by May 2014 contains 2578 mature human miRNAs (release 20 of miRBase at http://www.mirbase.org/index.shtml) |
| Noncoding RNAs (ncRNAs): Any RNA molecule that is not translated into a protein. |
| Oncogenic microRNA: a microRNA that when expressed at higher levels as normally, initiate or favor the development of a tumor. |
| Open Reading Frame (ORF): A section of a mRNA that begins with an initiation (methionine ATG) codon and ends with a nonsense codon. ORFs all have the potential to encode a protein or polypeptide, however many may not actually do so. |
| Pol II: RNApolymerase II (also called RNAP II) catalyzes the transcription of DNA to synthesize precursors of mRNA and most small nuclear RNA. |
| Pol III: RNA polymerase III (also called RNAP III) transcribes DNA to synthesize ribosomals RNA, tRNA and other small RNAs. The genes transcribed by RNA Pol III fall in the category of “housekeeping” genes whose expression is required in all cell types and most environmental conditions. |
| Pseudogene: A copy of a gene that usually lacks introns and other essential DNA sequences necessary for function. A pseudogene has been mutated into an inactive form over the course of evolution, but contains the majority of interactor sites with microRNAs as the original functional gene. |
| Sense/Antisense: Referring to the strand of a nucleic acid that directly specifies the product or referring to the strand of a double-stranded molecule that does not directly encode the product but is complementary to it. |
| The Cancer Genome Atlas (TCGA): a project started in 2005, to catalogue genetic mutations responsible for cancer, using gene expression profiling, copy number variation profiling, SNP genotyping, genome wide DNA methylation profiling, microRNA profiling, and exome sequencing. |
| Transcription: The process whereby RNA is synthesized from a DNA template. |
| Translation: The process of ribosome-mediated production of a protein whereby the primary structure of the protein is determined by the codon nucleotide sequence in mRNA. |
| Tumor suppressor microRNA: a microRNA that when expressed normally block the initiation or development of a tumor; at lower levels of expression this braking effect disappear and the tumor can develop. The same miRNA can behave as an oncogene in one type of tissue and as tumor suppressor in another type. |
| Untranslated Region (UTR): 5′UTR is the portion of an mRNA from the 5′end to the position of the first codon used in translation. The 3′ UTR is the portion of an mRNA from the 3′end of the mRNA to the position of the last codon used in translation. |
Initially, the DNA segments containing miRNA-coding genes are transcribed by an RNA polymerase II or III (RNA Pol II and III) to initiate their biogenesis. The primary transcript (pri-miRNAs) can be hundreds or thousands of nt long, but it is further processed, forming a 100 nt precursor transcript (pre-miRNAs) that folds upon itself (Figure 1). The precursor sequence is then exported to the cytoplasm, where it undergoes a series of catalytic steps prior to achieving maturation. In the cytoplasm, mature miRNAs are integrated into a number of proteins that compose the RNA-induced silencing complex (RISC), and thereafter they are incorporated to the messenger RNA (mRNA) 4 (for more details see Figure 1).
Figure 1. miRNA processing.
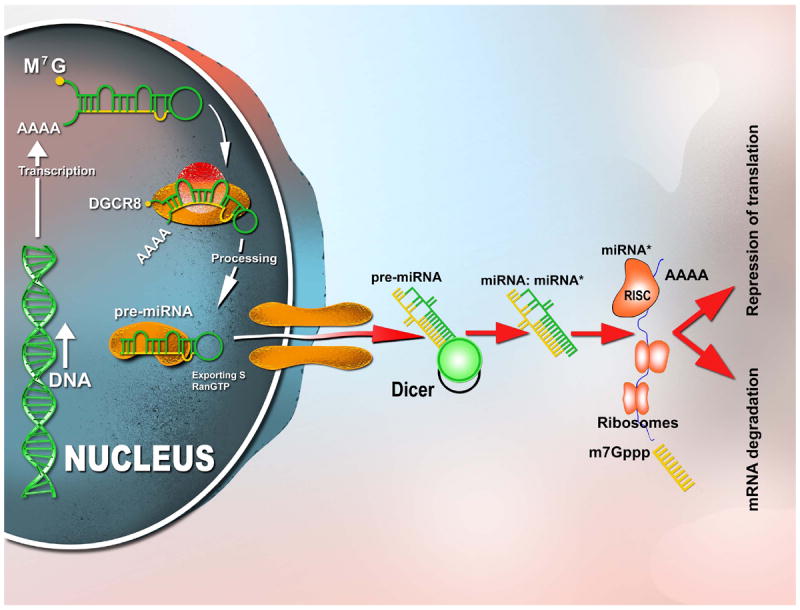
RNA polymerase II is responsible for the initial transcription of the miRNA gene into a long, capped and polyadenylated (poly-A) precursor, called primary (pri)-miRNA [13,14]. A dsRNA-specific ribonuclease, Drosha, in conjunction with its binding partner DGCR8 (DiGeorge syndrome critical region gene 8 or Pasha), further processes pri-miRNA into a 70 to 100 nucleotide hairpin RNA precursor (pre-miRNA) [15]. Pre-miRNA is translocated from the nucleus to the cytoplasm by Exportin 5/RanGTP, and cleaved into an 18- to 24-nucleotide duplex by a ribonucleoprotein complex composed of ribonuclease III (Dicer) and TRBP (HIV-1 transactivating response RNA-binding protein). Finally, the duplex interacts with the RNA-induced silencing complex (RISC), which includes proteins of the Argonaute family (Ago1 to Ago4 in humans). One strand of the miRNA duplex remains stably associated with RISC and becomes the mature miRNA, which primarily, but not exclusively, guides the RISC complex to the 3’-UTR (3’-untranslated region) of target mRNAs. Although the interaction between miRNA and mRNA usually results in translation inhibition, some cleavage of target mRNAs has also been observed
As part of the new ncRNA-based dogma, miRNAs have been known to bind to messenger RNA (mRNA) at the 3’-untranslated region (UTR) and cause the downregulation of protein-coding genes (PCGs, named targets) in the cytoplasm. They do so by inhibiting the ability of the ribosome to “translate” the mRNA. Alternatively, miRNAs can increase mRNA degradation, also reducing the possibility of the mRNA being translated into proteins 5. The level of complementarity between miRNA and the target mRNA may determine the mechanism by which the translation of mRNA to protein is blocked. Perfect or near-perfect complementarity has been found to induce mRNA degradation by RISC, and partial complementarity has been found to repress mRNA translation by blocking ribosomal access to the mRNA 6. Due to the fact that each miRNA has hundreds or thousands of mRNA targets, a broad segment of the protein coding genome is under their control. The regulation of miRNA-coding genes is of critical interest since they might be involved in any type of pathophysiological process and/or pathway, such as B-cell survival (miR-15a and miR-16-1), B-cell lineage fate (miR-181), brain patterning (miR-430), pancreatic cell insulin secretion (miR-375), adipocyte development (miR-145), cell proliferation control (miR-125b and let-7), or cell survival (let-7 family) 7.
The understanding of the mechanisms of action of miRNAs has significantly expanded in the last few years, with discoveries demonstrating unexpected complexities of their regulative manner, such as promoter binding, protein binding or direct interaction with other ncRNAs (Figure 2). miRNAs can be re-localized in the nucleus – for example, human miR-29b has been found predominantly in the nucleus demonstrating that despite their small size, specific miRNAs contain specific nucleotide sequences that control their subcellular localization 8. This localization supports the already proven hypothesis that miRNAs can alternatively regulate transcriptional processes at a DNA level. For example, human miR-373 binds to the E-cadherin (CDH1) promoter, thereby inducing its expression 9. In addition to mRNAs, miRNAs target other different types of ncRNAs, some of them being highly conserved among species such as the ultraconserved genes 13 or poorly conserved such as the pseudogenes 14 (see Glossary for the definition of terms).
Figure 2. Mechanisms of action of microRNAs.
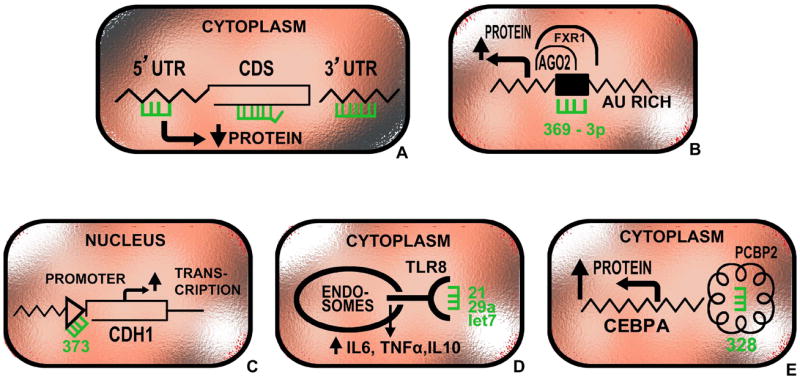
miRNAs can work through a variety of mechanisms: (a) binding by direct complementarity to multiple regions at the mRNA including the 3’-UTRs, 5’-UTRs and coding regions and decreasing protein expression (majority of miRNAs); (b) positively regulation of translation (miR-369-3p) through increased recruitment of processing proteins; (c) direct interaction with promoter sequences (miR-373); (d) direct agonists of receptors such as Toll-like receptors (miR-21, miR-29a and let-7); (e) direct interaction with protein and enhancing protein expression through a decoy mechanism (miR-328). For gene abbreviations see text and http://www.ncbi.nlm.nih.gov/gene.
In the past few years, miRNAs have also been found to favor protein expression (in addition to downregulating it). MiR-369-3p was shown to interact with AU-rich elements in the tumor necrosis factor alpha (TNF-alpha)-mRNA, consequently recruiting proteins that increased the process of translation during cell cycle arrest 15. Similarly, miR-328 was revealed to increase translation by interacting with hnRNP E2, a translational regulator, leading to the release of CCAAT/enhancer binding protein alpha (CEBPA) mRNA. On the other hand, it decreased the translation of PIM1 kinase by binding specifically to its mRNA. Both mechanisms are active during the acute transformation of the chronic phase of chronic myeloid leukemia (CML) and are independent of patients’ response to imatinib. Together, these data reveal that miRNAs possess the ability to control the fate of protein coding genes by attaching through base pairing complementary to mRNA sequences or by interfering with regulatory proteins directly 16.
MiRNAs also work as secreted molecules that trigger a receptor-mediated response in a different cell or tissue. They have the ability to be released into the extracellular environment within exosomes (cell-derived vesicles originated by the inward budding in the plasma membrane generating multivesicular bodies (MVBs)), that are present in many and perhaps all biological fluids. In this way, they can act as “hormones” 17. For example, in breast cancer it has been shown that macrophages influence cell invasion through exosome-mediated delivery of oncogenic miR-223; moreover, pre-treatment of mice with tumor-derived exosomes accelerates lung metastasis formation 18.
Similarly, exosomes have been shown to modulate tumor microenvironments by releasing miRNAs in a coordinated manner. In this regard, exosomes derived from hypoxic cultures of a leukemic cell line in vitro, were found to carry and release miR-210 among other angiogenic miRNAs, increasing tube formation by endothelial cells 19. Along this line, it was also shown that miR-21 and miR-29a can be transported through exosomes and can act as direct agonists of Toll-like receptors. By binding as ligands to receptors of the Toll-like receptor (TLR) family in immune cells, these miRNAs were shown to trigger a TLR-mediated prometastatic inflammatory response (such as secretion of interleukins) that could favor tumor growth and metastasis 20, 21. Understanding such newly-recognized ways the miRNAs are working are important for both scientists and physicians as novel therapeutic approaches can be designed, such as blocking of miR-328 interaction with hnRNP E2 in CML patients or blocking the miR-21 and miR-29a agonistic effects on TLRs in patients with metastatic diseases.
2. MicroRNAs as cancer ubiquitous players
MicroRNA alterations have been identified in many human diseases such as autoimmune, cardiac disorders, schizophrenia and cancer (where they have been found to be highly dysregulated)22. MiRNAs have been found to be differentially expressed between all types of analyzed human tumors and normal tissues including benign and malignant tumors 23, such as leukemias, lymphomas, lung cancers, breast cancers, colorectal cancers, papillary thyroid carcinoma, glioblastoma and other brain tumors, hepatocellular carcinomas, pancreatic tumors, cervical cancers, prostate cancers, kidney and bladder cancers, or pituitary adenomas 23, 24.
MicroRNAs can act as oncogenes and/or tumor suppressors
MiRNAs have been proven to work as oncogenes (see Glossary), such as miR-21 or miR-155, both among the most frequently overexpressed miRNAs in human cancers 23. For example, in a transgenic miR-155 mouse model, the rodents exhibit initially a pre-leukemic pre-B cell proliferation evident in spleen and bone marrow, followed by frank B cell malignancy 25. Overexpression of miR-21 in mice similarly leads to a pre-B malignant lymphoid-like phenotype, demonstrating that this gene is also a genuine oncogene. When miR-21 was inactivated in vivo, the tumors regressed completely in a few days, partly as a result of enhanced apoptosis 26.
MiRNAs can act also as tumor suppressors, such as miR-15a/16-1 cluster, frequently deleted in chronic lymphocytic leukemia (CLL) 27 and prostate cancers 28. The induced deletion of these miRNAs in a knockout mouse model causes development of indolent B cell-autonomous, clonal lymphoproliferative disorders, recapitulating the spectrum of CLL-associated phenotypes observed in humans. The miR-15a/16-1-deletion has been demonstrated to accelerate the proliferation of both human and mouse B cells by modulating the expression of genes controlling cell-cycle progression 29.
In some instances the same miRNA can have dual activities, thereby acting as an oncogene in one specific cell type and as a tumor suppressor in another. For example, miR-221 is overexpressed in liver cancers where it targets the PTEN tumor suppressor, and, in this way, it promoted liver tumorigenicity in a miR-221 mouse transgenic model 30. Similarly in colorectal cancer this same miRNA promotes cell invasion and metastasis by targeting RECK (which normally inhibits invasion and metastasis) 31. However in other tumor types such as gastrointestinal stromal tumors (GIST), miR-221 is downregulated and the consequent derepression of c-KIT and ETV1 (target oncogenes) promotes this malignancy 32.
The genetic basis for abnormal microRNA expression in cancer cells is complex
The widespread disruption of miRNA expression in malignant cells is only beginning to be understood, and a variety of abnormalities could contribute to the microRNAome expression profile in each tumor. Transcription factors are involved in the regulation of specific miRNAs, such as the miR-34 family and miR-15/16 cluster modulated by TP53 33, 34, the miR-17-92 cluster regulated by MYC 34, 35, or miR-210 by HIF-1alpha 36. The aberrant expression causes abnormal levels of mature and/or precursor miRNAs in comparison with the corresponding levels in normal tissues 37. At least three different mechanisms (that could act independently or together) have been described to cause this: the location of miRNA-coding genes at cancer associated genomic regions frequently deleted or amplified 38; epigenetic regulation of miRNA expression by methylation (which adds methyl groups, suppressing genetic expression) or histone modification (which are proteins that package and order the DNA) 39 and finally, abnormalities in miRNA processing genes or proteins (required for miRNA biogenesis and maturation) 40, 41.
Germline and somatic mutations in miRNAs
Variations in the sequences of miRNAs located in the mature, precursor or primary transcript, may contribute to cancer predisposition and initiation 42. For example, germline (passed from parental cell) or somatic (acquired by the cell) mutations of some miRNA genes were found in patients with CLL. In the initial analyses of sequence variations in miRNAs, it was reported that in two patients diagnosed with CLL, a nucleotide substitution (C for T) was associated with lower levels of mature miR-16 (a tumor suppressor miRNA) 43. This mutation proved to affect SRp20, which is an RNA-splicing protein implicated in processing the primary miR-16 transcript; and this was hypothesized to be contributing to the development of CLL 44. Findings such as this one point out that some CLL patients may have a genetic predisposition for this type of cancer. Furthermore, a mouse model also supported the role of certain miRNAs in the pathogenesis of CLL. Similarly, these mice harbor a point mutation (one nucleotide) adjacent to miR-16which results in its reduced overall expression 45.
On a separate note, polymorphisms of a single nucleotide (SNP) in the protein coding mRNAs targeted by miRNAs have shown to influence cancer risk as well. For example, let-7, a tumor suppressor miRNA known to target the mRNA of the KRAS oncogene, has been proven unable to bind to a KRAS SNP-variant, found to be disproportionately enriched in non-small cell lung carcinoma patients (present in 18-20%) 46. This same KRAS variant was also associated with increased risk for developing epithelial ovarian cancer (EOC), consistent between three independent cohorts and two case-control studies46. Moreover, it was present in 61% of patients with a hereditary breast and ovarian cancer (HBOC) history who were previously genetically uninformative (BRCA1/2 negative)46. This suggests that the SNP-variant may be a new independent cancer risk biomarker for HBOC families. To this date, most miRNA binding site SNP studies have followed the case-control study design, thus they are centered on cancer risks. and so the majority of our knowledge, thus far, centers on cancer risk. For more information regarding miRNA binding site SNPs considered to be cancer risk biomarkers see the recent review by Preskill and Weidhaas 46.
MiRNAs as hormones
A large amount of data has accumulated over the last few years describing the participation of miRNAs and microenvironment cells in malignant processes47, 48. Just as hormones, miRNAs are released by a donor cell as ‘free’ molecules or in secreted vesicles, and delivered from body fluids into receptor cells located in other parts17. For example, the tumor suppressor miR-143 was found to be released by normal epithelial prostate cells inducing growth inhibition of prostate cancer cells in vitro and in vivo49. Similarly, intercellular transfer of miR-142 and miR-223 from immune cells to malignant ones (hepatocellular carcinoma cells) inhibited proliferation and destabilized microtubule regulation in vitro 50. Additionally, in a breast cancer study miR-210 was shown to be released from tumor cells to endothelial cells promoting angiogenesis and metastasis through an exosomal-mediated transfer 51. Mir-210 was also proven to be trafficked through exosomes in a leukemia cell line (under hypoxic conditions) 52. Along with other oncogenic miRNAs, the delivery of miR-210 resulted in increased angiogenesis through the promotion of tube formation by endothelial cells in vitro 19.
3. MicroRNAs are involved in all cancer hallmarks
The hallmarks of cancer comprise the biological capabilities acquired during the stepwise process of developing human tumors (Figure 3). MicroRNAs have the potential to influence these hallmarks, and the recognition of the applicability of these interactions will increasingly influence the development of new therapeutic alternatives for cancer patients53, 54. Herein are some representative examples of miRNAs that act as regulators of tumor biology; more detailed information on the roles of miRNAs in cancer can be found in several reviews 22, 24, 40, 55, 56.
Figure 3. Examples of miRNA involved in the eight cancer hallmarks.
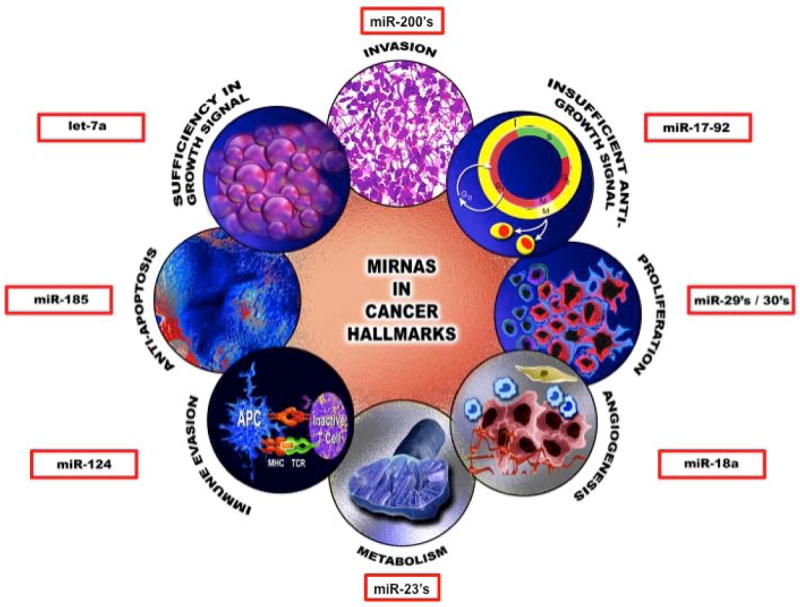
The six biological capabilities acquired during the multistep development of human tumors include sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, reprogramming of energy metabolism and evading immune destruction. For each one representative example of miRNA is presented. For more details see text.
1) Self-sufficiency in growth signals
Activation of the RAS oncogene is a common event that allows tumor cells to escape growth factor dependency and become “oncogene addicted”. All of the three RAS genes (K-, N- and H-) have been proven to be directly modulated by the tumor suppressor miRNA let-7 in a post-transcriptional manner57. Alternatively, let-7 also targets high mobility group A2 (HMGA2), a pleiotropic transcription factor. The characteristic down-regulation of let-7 accompanying tumor development results in an increased expression of the RAS oncogenes along with their downstream effects. It also de-represses HMGA2, thereby facilitating anchorage independent growth in cell cultures 58, as well as cell proliferation and differentiation 59. Evidently, both of these mechanisms have significant relevance regarding tumorigenesis and cancer development. Furthermore, reconstituting the levels of let-7 in an experimental approach, demonstrated that this precursor miRNA family can inhibit cell proliferation resulting in tumor regression as shown in lung cancer models in mice60.
2) Insensitivity to anti-growth signals
E2F is a group of genes that encode a family of transcription factors that tightly regulate cell cycle progression and DNA synthesis. Three of them, E2F1, 2 and E2F3a, are known as the cell cycle “activators”, and they represent attractive factors to target in cancer because they contribute to uncontrolled cell growth. Several miRNAs have been demonstrated to have the potential to modulate the translation of the mRNAs of these transcription factors. For example, miR-20a, miR-17-5p, miR-93 and miR-106b have been shown to negatively regulate E2F161-62. Moreover, the miR-17-92 cluster has been shown to decrease the levels of E2F1-3 63. So, in this sense, it is likely that the downregulation of these miRNAs in different cancer types could favor a proliferative transcriptional network contributing to tumorigenesis. Thus, reconstituting the basal levels of expression of these miRNAs (in E2Fs-dependent tumors) could serve as a future therapeutic alternative with clinical relevance. Some of these miRNAs are part of positive or negative feedback mechanisms, and thus the end result of modulating their levels still remains to be explored.
Finally, in separate studies, the transcription factor FOXO1 (tumor suppressor that controls proliferation and regulates apoptosis) was found decreased in classical Hodgkin lymphoma (cHL) cases. In cHL-cell lines, the levels FOXO1 were proven to be repressed by three up-regulated microRNAs: miR-96, miR-182, and miR-183 64. This repression highly increased proliferation, and at the same time inhibited apoptosis in vitro.
3) Evasion from apoptosis
Apoptosis is a physiological self-destructive cellular mechanism that leads to removal of unwanted cells. MiR-25 was identified as elevated in cholangiocarcinoma cell lines as well as patient samples, and its increased levels where shown to contribute to the evasion of TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. In experiments that reduce levels of miR-25 (in vitro), cells in culture were sensitized to apoptotic death; results that implicated the miRNA in controlling tumor cell apoptosis65.
The cancer associated genomic region 1p36, (frequently lost or rearranged in many types of leukemias), contains a crucial tumor suppressor, miR-34a. In neuroblastoma, loss of miR-34a synergizes with MYCN oncogene amplification, and miR-34a has been shown to be a MYCN negative regulator 66. In addition, miR-34a is known to induce cell cycle arrest and subsequent caspase-dependent apoptosis by repressing the antiapoptotic protein Bcl2 66 and the transcription inducer of cell cycle progression E2F367. The overexpression of this same miRNA exhibits a transcriptome expression similar with that observed with p53 (widely known tumor suppressor) induction, being highly enriched for genes regulating apoptosis, cell-cycle progression, DNA repair and angiogenesis 68, 69, 70. This along with other observations points to the fact that the loss of this miR-34a sidesteps typical apoptotic pathways. Recently, another miRNA was also found to influence pathways leading to apoptotic evasion. In a gastric cancer study, a novel mechanism whereby miR-185 directly targets apoptosis repressor with caspase recruitment domain (ARC) was revealed. The role of this miRNA was studied in vitro and further validated in a gastric tumor xenograft model 71.
4) Limitless replicative potential
Cellular senescence is a physiological withdrawal from the cell cycle in response to a variety of stressful stimuli and involves telomerase (an enzyme that prevents the loss of important DNA from chromosome ends) deregulation. MiRNAs have been linked to premature senescence, and their relevance has been addressed through the generation of a genetic miRNA-screening library. The miR-29 and miR-30 microRNA families are up-regulated during induced replicative senescence and their high levels influence the repression of the B-Myb oncogene inhibiting cellular DNA synthesis 72.
Moreover, miR-373 and miR-372 were identified as capable of allowing proliferation and tumorigenesis of primary human cells harboring oncogenic RAS and wild-type p53 (a functional tumor suppressor). These miRNAs neutralized p53-mediated cyclin dependent kinase (CDK) inhibition, possibly through direct inhibition of the expression of a tumor suppressor called LATS2. This evidence most definitely implicates both of the miRNAs as oncogenic, particularly in the development of human testicular germ cell tumors.
5) Angiogenesis
Tumor cells activate an “angiogenic switch” producing high amounts of pro-angiogenic factors that promote neovascularization. The most important one, vascular endothelial growth factor (VEGF), is highly expressed in most tumors, both solid and hematologic, and has been proven to be induced by hypoxia. During tumor progression, hypoxia has been found to contribute to the modulation of miRNA expression, in part by a direct transcriptional activation of specific miRNAs (such as miR-26, miR-107 and miR-210) triggered by hypoxia-inducible factor-1 (HIF-1) 36. These miRNAs have dual functions: on one hand they participate in angiogenesis, but on the other they potentiate the ability of cells to engage in anti-apoptotic mechanisms to sustain survival. For example, miR-27a restrains the zinc finger gene ZBTB10, a negative regulator of the Specificities Protein (SP) transcription factors, and through this repression it induces an SP dependent transcription of both survival and angiogenesis related genes (i.e. survivin, VEGF, VEGFR) 74. Furthermore, miR-210, through direct modulation of the tyrosine kinase receptor ligand Ephrin A, represents a component of the circuitry controlling endothelial cell chemotaxis and tubulogenesis 75. In addition, the downregulation of miR-18a has also been recently linked to angiogenesis. MiR-18a is known to inhibit the phosphorylation of two substrates of the mTOR pathway. This results in an inactivation of the pathway and a consequent downregulation of factors that stimulate blood vessels production such as HIF-1α and VEGF76.
6) Invasion and metastasis
The process of metastasis begins with the acquisition of invasive properties that allow cells to detach from the primary tumor, enter the blood or lymphatic vasculature and spread to distant organs. Up-regulation of miR-10b has been demonstrated to promote invasion and metastasis by targeting HOXD10, a homeobox transcription factor that promotes or maintains a differentiated phenotype in epithelial cells 77, 78. Recently, a new miRNA that is involved in migration and metastases of lung cancer cells was identified 79. In lung cancer positive transgenic mice, researchers found that miR-136, -376a and -31 were prominently overexpressed. Among these, the antagonization of miR-31 suppressed tumor growth, suggesting causation. This same group also identified LATS2 (a gene encodes a tumor suppressor protein important for cytoskeleton function) and PPP2R2A (a phosphatase that catalyzes the removal of phosphate groups and is therefore considered an “inactivator-protein”) as targets of miR-31 in mouse and human lung cancer. In the attempt to identify miRNAs associated with lymph node metastasis in tissue samples from patients of lung adenocarcinoma, a genome-wide next-generation miRNA-sequencing and a training-validation approach identified and validated miR-31 up-regulation as significantly associated with the presence of lymph node metastasis as well as poor patient survival. In addition, miR-31 was shown to be able to modulate the migratory, invasive, and proliferative behavior of lung adenocarcinoma cell lines in culture by stimulating the oncogenic ERK1/2 signaling-pathway 80.
7) Reprogramming energy metabolism
Malignant cells tailor metabolic pathways to meet their energy requirements 81. Glutamine and glucose are the two major nutrients that fuel cellular metabolism and the pathways utilizing these nutrients are often altered in cancer 82. Various studies have exposed the fine interplay between metabolic pathways orchestrated by protein-coding genes and by miRNAs. For example, glutamine metabolism (glutaminolysis) was shown to be modulated by the MYC oncogene via miR-23a/b in prostate cancer and also in B cell lymphoma 83. Additionally, it can also be modulated by a p65-mediated activation, that downregulates miR-23a levels84. Finally, glycolysis has been proven to be modulated by a series of different miRNAs, including miR-378-star 85 and miR-143 86.
8) Evading immune destruction
Signal transducer and activator of transcription 3(STAT3) regulates a key pathway mediating immunosuppression in the tumor microenvironment. Recently, the roles of miRNAs in tumor-mediated immunosuppression began to be discovered. MiR-124 was found to be strongly downregulated in all grades and pathologic types of gliomas in comparison with normal brain tissue, and it was identified as an important modulator of STAT3 signaling 87. Upregulation of miR-124 in glioma cancer stem cells (gCSC) has been shown to inhibit STAT3 pathway. This resulted in the reversed induction of forkhead box P3 regulatory T cells (Treg) and also it also reversed the gCSC-mediated immunosuppression of T-cell proliferation. Furthermore, systemic treatment of miR-124 administration of mature miRNA (by intratumoral or intravenous infection) demonstrated to have anti-glioma therapeutic effects in engineered murine models of glioblastoma. The resulting effects indicated that miR-124 depends on the presence of a T-cell-mediated antitumor immune response 87.
4 Methods to detect the expression of miRNAs
Multiple platforms are available to identify and quantify miRNAs
An ideal method to use miRNAs as biomarkers for human disease should be easy to carry out and would not require expensive reagents or equipment; furthermore it would be specific enough to detect only the miRNA of interest without detecting closely related miRNAs. It should also be sensitive enough to provide a quantitative expression analysis, even with low amounts of starting material from small clinical samples, and it would need to possess the ability for processing multiple samples in parallel 88, 89. The gold standard meeting these requirements for miRNA detection in clinical laboratories is quantitative reverse transcription PCR (qRT-PCR). MiRNA microarrays are more expensive than qRT-PCR and usually are used for the discovery step of biomarker identification (Table 2). These techniques, however, all use miRNAs extracted from a patient tumor sample that includes malignant cells as well as non-neoplastic stromal and inflammatory cells. If the detection aims to analyze miRNA specifically in malignant cells, it is advisable to use cell sorting with flow-cytometry for liquid tumors or laser-capture microdissection for solid tumors. Another option is to perform in situ hybridization (ISH) 90, as the miRNA of interest can be detected within the different types of cells that compose the tumor (malignant versus microenvironment), and this approach provides additional information on the subcellular localization of the miRNA (Table 2). The newest method to discover and measure miRNA expression is next generation RNA sequencing. This technique is highly sensitive, highly specific, can be used for high-throughput analysis and also discovers ‘de novo’ miRNAs. RNA sequencing generates a massive amount of complex data that needs to be analyzed by a well-trained bio-informatician. For this reason, as well as the high costs of a single RNA-sequencing run, this method is not yet appropriate for diagnostic purposes, but it can still be considered an alternative for screening.
Table 2.
Established profiling methods to quantify miRNAs.
| Advantages | Limitations | References | |
|---|---|---|---|
| qRT-PCR |
|
|
187, 188 |
|
| |||
| Microarray |
|
|
101,189 |
|
| |||
| In situ Hybridization |
|
|
190, 191 |
|
| |||
| RNA sequencing |
|
|
101, 102 |
Understanding the reproducibility of profiling data
There are some technical challenges that make it difficult to compare results from similar profiling platforms used in different places. For example, the different primer design for measurements by qRT-PCR and microarrays is an important factor 91, 92. Another factor is the use of different protocols for sample preparation: some studies use an enriched small RNA fraction whereas others use total RNA 93. Furthermore, technical and biological variability has been assessed by a range of different normalization processes, mostly based on the expression of reference genes. Specifically for cell-free microRNA analysis, no known extracellular reference RNA is currently suitable for a proper normalization 91. Commonly used reference genes, such as U6 small nuclear RNA (RNU6B) and 5S ribosomal RNA, were found to be less stably expressed than others or degraded in serum samples 94. In addition, the significant differences in choice of reference genes to use represent a major obstacle in comparing expression levels between normal tissue and tumors. Finally, the assessment of sample quantity and quality is more challenging for microRNAs than for mRNAs of protein-coding genes, for which sizes and relative abundance of ribosomal RNAs can be used to check RNA integrity.
5 MicroRNAs: the mix of cancer biomarkers and of hormones
Characteristics of a good biomarker
The measurement of miRNAs in body fluids including plasma and serum may represent a gold mine of noninvasive biomarkers for cancer 17, 95. An “ideal” biomarker should have a unique expression profile in the diseased compared to healthy individuals and should show highly increased or decreased expression levels in the diseased organ or tissue as compared to non-diseased. Moreover, the biomarker should be reliable in detecting disease initiation or development before clinical symptoms appear in the aims of enabling early detection, persistence of minimal residual disease, or recurrence after treatment. Furthermore, ideal biomarkers should be accessible through non-invasive methods, should have a long half-life in clinical samples, and should be rapidly detectable by simple, accurate and inexpensive methods 96. MicroRNAs are very stable, even in body fluids such as plasma, serum, urine and saliva 17, their expression is specific to tissues, or organs and their expression level can be easily measured by methods such as quantitative PCR and miRNAs microarrays 91. It has been shown consistently that serum miRNAs remain stable after being subjected to severe conditions that would normally degrade most mRNAs, such as very low or high pH levels, boiling, extended storage, and 10 freeze-thaw cycles. These features make them very suitable as biomarkers.
MicroRNA profiling of tumor versus normal tissues shows significant changes and define common altered miRNAs in human cancers
MicroRNA profiling by various methods has allowed the identification of signatures associated with diagnosis, staging, disease progression, prognosis and response to treatment of human tumors (Table 3). MicroRNA expression profiles allow nowadays the classification of human cancers 23. To date, every type of tumor analyzed by miRNA profiling has shown significantly different miRNA profiles (for mature and/or precursor miRNAs) compared with normal cells from the same tissue type (Table 3). For example, it was shown that profiling a few hundred miRNAs has a significantly better predictive power of diagnosing cancer of unknown primary site (CUP) compared to profiling several tens of thousands of mRNAs 97.
Table 3.
Examples of malignant cells or body fluids microRNAs profiles with clinical significance for cancers patient.
| Cancer type | No patients | miRNA | Role | Ref |
|---|---|---|---|---|
|
| ||||
| Multiple Cancers | 1809 patients (pts) | miR-210 | Predictive effect on survival of patients with studied cancer types as indexed by disease-free survival, progression-free survival and relapse-free survival. | 192 |
| 7 different types of cancers: breast ca., primary head and neck squamous cell carcinoma, renal cancer, soft-tissue sarcoma, pediatric osteosarcoma, bladder ca. and glioblastoma | ||||
|
| ||||
| Multiple Cancers | 174 pts/ 39 controls (ctr): 50 breast ca., 30 gastric ca., 31 lung ca., 31 esophageal ca, 31 colorectal ca. |
miR-21 | Potential broad-spectrum serum-based diagnostic marker for the detection of solid tumors. | 193 |
|
| ||||
| Breast ca. | 120pts/40 ctr | 6 circulating miRNAs: miR-10b, miR-17, miR-34a, miR-93, miR-155, and miR-373 | Known to be relevant for tumor development and progression. | 194 |
| Serum concentrations of deregulated microRNAs may be linked to a particular biology of breast carcinomas favoring progression and metastatic spread. | ||||
|
| ||||
| Colon ca. | 102 pts/54 ctr | miR-141 | Highly correlated with TNM in CRC patients. Elevated levels associated with liver metastasis in CRC. | 195 |
|
| ||||
| Colon ca. | 100 CRC pts/37 adenomas/ 59 ctr | miR-29a | Association with CRC TNM stage. | 196 |
|
| ||||
| Colon ca. | 103 CRC pts/ 37 ctr | miR-221 | Potential non-invasive molecular marker for the diagnosis of CRC. | 197 |
|
| ||||
| Colon ca. | I phase: 12 stage I and IV CRC pt. | miR-200c | Independent predictor for lymph node metastasis and tumor recurrence, emerging as an independent prognostic marker for CRC. | 198 |
| II phase: 182 CRC pt/ 24 ctr. | ||||
| III phase: matched 156 tumor tissues from the phase II CRC cohort + independent set of 20 matched primary CRC with corresponding liver metastases. | ||||
|
| ||||
| Colon ca. | exosome-enriched serum samples from 88 primary CRC pts/ 11 healthy controls/ | let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a | The serum exosomal levels of seven miRNAs were significantly higher in primary CRC patients, even those with early stage disease, than in healthy controls, and were significantly down regulated after surgical resection of tumors. | 199 |
| 29 paired samples from post-tumor resection pts | ||||
|
| ||||
| Gastric ca. | 124 GC non cardia, 36 GC cardia adenocarcinomas/ 160 ctr | miR-16, miR-25, miR-92a, miR-451 and miR-486-5p | Detection of the early-stage GC. | 200 |
|
| ||||
| Gastric ca. | 104 pts/ 65 ctr | miR-18a | Diagnostic power (high sensitivity and specificity). | 201 |
|
| ||||
| HCC | 101 pts/ 89 ctr + 48 pt Hepatitis B | miR-21, miR-122 and miR-223 | They have showed to be elevated in patients with HCC or chronic hepatitis, with strong potential to serve as novel biomarkers for liver injury but not specifically for HCC. | 202 |
|
| ||||
| Lung ca. | Training set: 64 ADC pts. | miR-31 | Predictor of survival in a multivariate cox regression model even when checking for cancer staging. Exploratory in silico analysis showed that low expression of miR-31 is associated with excellent survival for T2N0 patients. | 203 |
| TCGA dataset: 223 ADC pts. | ||||
|
| ||||
| Lung ca. | 25 paired NSCLC, paracancerous tissues and serum, 103 control sera, 201 NSCLC pts | miR-19a | High serum miR-19a expression may be an independent poor prognostic factor for survival in NSCLC patients. | 204 |
|
| ||||
| Lung ca. | 30 and 75 pts tumor nontumor | Exosomes isolated in plasma and BAL | In tumor plasma higher percentage of miRNAs with increased levels than in tumor BAL or in nontumor plasma. The data reveal differences between BAL and plasma exosome amount and miRNA content. | 205 |
|
| ||||
| Glioblastoma | I phase:122 untreated WHO III–IV/123 ctr serum samples. | miR-15b*, miR-23a, miR-133a, miR-150*, miR-197, miR-497 and miR-548b-5p, and the seven-miRNA panel | The 7-serum miRNA panel demonstrated a high sensitivity and specificity for prediction of malignant astrocytomas. | 206 |
| II phase: 55 WHO II, 15 WHO I, 11 astrogliosis, 42 other primary brain tumors serum samples, 8 WHO II–IV astrocytomas tumor tissues. | A marked difference in serum miRNA profile was observed between high-grade astrocytomas and normal controls. | |||
|
| ||||
| Prostate ca. | Pre-biopsy serum samples of 170 pts collected in three study centers; | miR-26a-1 and miR-141 | The analysis of circulating microRNAs does not seem to help identify patients with cancer undergoing prostate biopsy. However, their levels may be useful to identify patients with high-risk prostate cancer. | 207 |
| 133 pts prostate cancer n=54, non-malignant n=79 | ||||
|
| ||||
| Ovarian ca. | 360 EOC pts/200 ctr from two institutions | miR205, let-7f | Plasma miR-205 and let-7f are biomarkers for ovarian cancer detection that complement CA-125; let-7f may be predictive of ovarian cancer prognosis. | 208 |
MiRNAs in body fluids as biomarkers of therapeutic response, residual disease and overall survival in cancer patients
Clinical studies have demonstrated the potential of using miRNAs as predictors of radio-therapeutic and anticancer agent sensitivity (Table 3). For instance, serum miR-21 levels were higher in hormone-refractory prostate cancer patients whose disease was resistant to docetaxel-based chemotherapy when compared to those with chemosensitive disease 98. In a separate study regarding lung adenocarcinoma and squamous cell carcinoma, eleven serum miRNAs proved to separate patients with longer survival vs. shorter. Furthermore, the levels of four of these miRNAs were significantly associated with overall survival and this same signature was consistently an independent predictor of overall survival 99. Likewise, upregulated and downregulated miRNAs have been detected in sensitive or resistant cell lines, in order to predict patient response to anticancer agents. One of these studies identified a miRNA chemosensitivity profile from a set of 59 human cancer cell lines derived from diverse tissues (NCI-60 cell lines). Downregulation of miR-34, miR-17, and let-7a was correlated with sensitivity to drugs commonly used as cancer treatments, such as 5-fluorouacil, Adriamycin, and cyclophosphamide, respectively 100. These findings suggest that several miRNA-based models are efficient diagnostic tools, and they can furthermore be considered as useful when predicting patterns of resistance/ sensitivity to drugs used as cancer treatment.
5 MiRNA profiling represents a new and useful clinical tool
The identification of non-invasive biomarkers still represents the most promising strategy for early detection and accurate diagnosis of many malignancies. Due to their tissue-specific profiles and stability in body fluids, miRNAs rank among the top candidate biomarkers. Because of the exponential increase in the number of publications related to miRNA profiling in human tumors, we will highlight the significance of recent studies reporting miRNA signatures published mainly in the last three years 101. Many in-depth reviews cover publications from the last decade, including profiling topics in multiple types of cancers 102, 103such as lung cancers 104, breast cancers 96, gastroesophageal cancers 105, ovarian cancers 106,107, hepatocellular carcinomas 108, chronic lymphocytic leukemia (CLL) 109 or B-cell lymphomas 110. A compilation of recent publications reporting diagnostic, prognostic or therapeutic associations in studies with over 100 patients is presented in Table 3. Such reports offer the scientific basis for the rational design of prospective large scale studies needed to confirm the multiple biological and clinical associations.
Lung cancer (LC)
In both early and late stages, there is an urgent need of finding reliable discriminators for disease diagnosis and prognosis. A large study of 165 adenocarcinomas (ADC) and 125 squamous cell lung carcinomas (SQ) analyzed by miRNAs array, identified a set of five miRNAs (let-7b, miR-103, miR-107, miR-181a, and miR-191), whose combination was able to distinguish ADC from SQ (Pglobal <0.0001)111. In addition, among male smokers with early-stage SQ, the higher expression of five miRNAs (miR-25, miR-34c-5p, miR-191, let-7e, and miR-34a) was predictive of a low mortality risk (HRs ranged between 0.38 and 0.51; P values ranged between 0.0001 and 0.011) 111. These data suggest that miRNA signatures can discriminate between cancer histotypes and correlates with the most important clinical parameter, e.g. the mortality risk.
In 3 independent cohorts from different countries (Maryland, United States, n=89; Norway, n=37; Japan, n=191) higher expression of miR-21 was found significantly associated with cancer-related mortality (HR 2.06, 95%CI: 1.13–3.75, P=0.018; HR 2.78, 95%CI: 1.22–6.31, P=0.014; HR 2.82 95%CI: 1.57–5.07, P<0.0005, respectively).112 The higher levels were detected in more advanced-stage tumors, consistent with the findings that miR-21 is involved in the disease progression. Importantly, Multivariable Cox regression analysis for just the TMN stage I cases showed that higher miR-21 expression levels were associated with poor cancer specific mortality in the Maryland/Norway cohorts (HR 2.16, 95%CI: 1.11–4.21, P=0.025) and worse Relapse-Free Survival in the Japan cohort (HR 3.40, 95%CI: 1.57–7.36, P=0.001) independently of all clinical covariates, suggesting miR-21 as an early-stage prognostic biomarker for TMN stage I stage patients at high risk of metastases. 112.
An interesting study evaluated the potential of a previously identified 24 plasma -MiRNA Signature Classifier (MSC) 113 to work as non-invasive screening tool for lung cancer in the Multicenter Italian Lung Detection (MILD) cohort of current and former smokers. Plasma samples from 69 patients with lung cancer and 870 disease-free individuals (652 who had been screened with low-dose computed tomography (LDCT) and 287 from an unscreened observation group) were analyzed by RT-qPCR. All the patients were categorized as belonging to one of the three risk-of-cancer groups (low, intermediate, and high) on the basis of predefined cut points of positivity for four different expression ratio signatures of 24 miRNAs, defined as risk of disease, risk of aggressive disease, presence of disease, and presence of aggressive disease. The study confirmed the diagnostic, prognostic and predictive value of this MSC. The diagnostic performance of MSC for lung cancer detection was 87% sensitivity (SE), 81% specificity (SP), 27% positive predictive value (PPV), and 99% negative predictive value (NPV) in lung cancer patients, 88% SE, 80% SP, 31%PPV, and 99% NPV in the LDCT arm, and 82% SE, 83% SP, 16% PPV, and 99% NPV for the observational arm. For all patients, MSC had a NPV of 99% and 99.86% for disease detection and disease related-death, respectively, while LDCT had a 79% SE and a 81%SP with a false-positive rate of 19.4%. The combination of both MSC and LDCT provided a five-fold reduction (compared with LDCT alone) in the false-positive rate to 3.7%, which means that MSC might complement LDCT screening especially for individuals with a low MSC and good prognosis, avoiding unnecessary invasive follow-ups and thereby optimizing health care costs114. Similarly, a combined 17-miRNA risk score derived from serum samples (n=391) of advanced Non-Small Cell Lung Cancer (NSCLC) patients was found to accurately identify those at the highest risk of death. In particular, individuals with a high-risk score had a 2.5-fold increased risk of death, corresponding to a 7.8-months decrease in median survival time (P=9.5×10-14), when compared to those with a low risk score (95%CI: 1.8–3.4, P=1.1× 10-7) 115. Thus, consistently with the data reported above, circulating miRNAs represent diagnostic and prognostic biomarkers which could be efficiently implemented in the clinical setting for lung cancer management.
Breast cancer (BC)
Early stages are curable by surgery and by a combination of hormonal treatment, chemotherapy and radiation. However, late relapses are frequent events after many years. MiRNA-based tests could potentially improve the prediction of Overall Survival (OS) as well as treatment response. An integrated miRNA/gene signature was recently reported in a group of 466 breast cancer patients from The Cancer Genome Atlas (TCGA) (for glossary see Table 1) 116. The final signature, including 30 mRNAs and 7 miRNAs (miR-103, miR-148b, miR-328, miR-484, miR-874, miR-93 and miR-1307), proved to significantly predict OS in a multivariable model independent of other clinical pathological characteristics of the tumors, and showed the highest prognostic value of distant Relapse-Free Survival in early stage I and II tumors (Receiver-Operator Characteristic (ROC) Area Under the Curve (AUC) = 0.77, P < 0.001). Furthermore the validation of this signature on eight independent BC cohorts, comprising a total of 2,399 patients, demonstrated its superior performance for risk stratification with respect to other RNA predictors, including the mRNAs used in MammaPrint and Oncotype DX assays 116. Separately, in a series of 173 triple negative breast cancers (TNBCs), a 4-miRNA signature (miR- 27a, miR-30e, miR-155, and miR-493) allowed the subdivision of TNBCs not only into Core Basal (CB, EGFR and/or CK5, 6 positive) or five negative (5NP) (when all markers are negative) subgroups (75% SE, 56% SP 0, AUC=0.74), but also into high risk and low risk of death groups 117. In particular, the median OSs for the high vs. low risk miRNA signature groups were 75.5 vs. 82 months (HR 2.46; 95% CI: 1.43-4.12; P=0.001). This panel also showed the ability in predicting outcomes of patient treatment with the two most commonly chemotherapy regimens used in TNBC, i.e. anthracycline or anthracycline plus taxanes.
Recently, in the attempt to identify circulating miRNA signatures in BC, a microarray analysis of fresh-frozen preoperative sera as well as normal and BC paired tissues samples was performed in a large cohort of Asian-Chinese BC patients 118. The miRNA expression profiles between sera and matched tumor samples were largely dissimilar. confirming the highly tissue specificity of miRNAs and their selective releasing from the primary tumor. Notably, among the resulting 23 BC-associated serum miRNAs, the ROC curves derived from the combinations of the overexpressed miR-1, miR-92a, miR-133a, and miR-133b exhibited AUCs of 0.90-0.91, confirming that miRNA models are robust diagnostic tools to apply in the clinic.
In an independent study on 48 ER-positive early stage Caucasian BC patients (24 lymph node-positive and 24 lymph node-negative), a novel circulating 9-miRNAs profile enabled for the first time the ability to distinguish the serum of a BC patient from the one of a healthy control (24 age-matched women) 119. This signature included both overexpressed (miR-15a, miR-18a, miR-107, and miR-425) and under-expressed (miR-133a, miR-139-5p, miR-143, miR-145, and miR-365) miRNAs. The miRNA profile validation on a cohort of 111 serum samples as well as independent datas from 3 publicly available databases further confirmed the discriminatory power with an AUC=0.665, 95%CI: 0.562-0.768 (ROC-test) and a P= 0.012. Using a probability cutoff of 0.48, the SE was 83.3%, the SP 41.2%, the PPV (Positive Predictive Value) 62.5%, and the NPV (Negative Predictive Value) 67.4%.
If compared, in the studies reported above, only one miRNA was common between the two patient populations analyzed 118, 119. Thus, there is evidently a high variability of miRNA expression in body fluids which underlines the need of conducting large consortium studies that include samples from as many parts of the world as possible.
Colon Cancer (CC)
Genetic profiling has been found somewhat useful when determining prognosis for intermediate stages, although to date it is not widely used 120. With regard to miRNA profiling, a microarray study of two colon adenocarcinoma patient cohorts (84 from Maryland test cohort and 113 from a Hong Kong independent validation cohort) compared tumors with adjacent non-tumorous tissue121. In both cohorts, high miR-21 levels were predictive of poor survival in both the training (HR 2.5; 95%CI: 1.2-5.2) and the validation cohorts (HR 2.4; 95%CI: 1.4-3.9), independently of clinical stage. Moreover, in stage II and III patients treated with fluoracil-based adjuvant chemotherapy (either intravenous 5-fluorouracil or oral drugs including tegafur with uracil [UFT], with or without generics), increased expression of miR-21 correlated with poor response and worse OS (HR 4.3; 95%CI: 1.1-16.4, P= 0.03 and HR 3.5; 95%CI: 1.1-11.6, P= 0.04 for the Maryland and Hong Kong populations, respectively). miR-21 was also found overexpressed both in colorectal carcinoma (46 CRC, P <0.0001) and related liver metastases (30 CLM, P <0.0001) compared to normal paired tissues and correlation with a worse Disease-Free Interval (DFI) in the CRC sample set was reported (P=0.0026). Finally, two other miRNAs have been proposed to be good predictive markers for therapeutic response in patients with CRC: miR-126 123 and miR-150 124. Higher levels of miR-126 were predictive of a good response to first line capecitabine and oxaliplatin (XELOX) in patients with metastatic colorectal cancer (mCRC): the median miR-126 expression level was significantly higher in the responding patients, with an area per image 3629 μm2 (95%CI: 2566-4846) compared to the non-responding patients (area per image 1670 μm2, 95%CI: 1436-2041, P=5 × 10-6). Accordingly, lower expression levels of miR-126 were predictive of a worse Progression-Free Survival (PFS), and OS, along with the association with the number of metastatic sites. Specifically, the median PFS for patients with high miRNA-126 expression levels was 11.5 months (95%CI: 9.0-12.7) compared to 6.0 months (95%CI: 4.8-6.9) for patients with low expression levels. The median OS in the group with high miRNA-126 expression was 26.2 months (95%CI: 21.8-32.8) compared to 16.8 months (95%CI: 13.8-19.1) in the group with low miRNA-126 expression. Conversely, low expression levels of miR-150 were found associated with worse prognosis (HR 0.57; 95%CI: 0.33-0.97; P=0.037) and poor therapeutic outcome in stage II and III CRC patients following fluoracin-based adjuvant therapy with or without generics (cohort 1 n=289, HR 0.44; 95%CI: 0.20-0.93, P=0.032; cohort 2 n=185, HR 0.38; 95%CI: 0.19-0.79, P=0.009).
To date, the early detection of adenomas is based on invasive screening approaches (such as colonoscopy and biopsies), and on non-invasive approaches (such as fecal occult blood tests or stool DNA tests based on mutations and methylation) with limited diagnostic accuracy. Thus, there is a great need for new non-invasive methods for day-to-day investigation. Recently, researchers embarked on a pilot study with the challenge of identifying specific biomarkers to detect precancerous lesions, colorectal adenomas and CRCs 125. The results of the investigation lead to a panel of 8 plasma miRNAs that distinguished CR adenoma patients (n=16) from controls (n=26) with high accuracy (AUC=0.868, 95%CI: 0.76–0.98, SE 88%; SP 64%), and a panel of 3 plasma miRNAs that could distinguish stage IV CRC (n=15) from controls (AUC=0.868, 95%CI: 0.76–0.98, SE 93%, SP 74%). 126. A further six miRNA-profile of upregulated miRNAs (miR-15b, miR-18a, miR-19a, miR-19b, miR-29a, and miR-335) was achieved when analyzing a total of 196 plasma samples from 123 patients newly diagnosed with sporadic colorectal neoplasia (63 with CRC and 60 with Advanced Adenomas, AA) and 73 healthy individuals (controls). The panel showed a high accuracy in discriminating CRCs from healthy controls (AUC ranging from 0.80, 95%CI: 0.71– 0.89 to 0.70 95%CI: 0.59–0.80), whereas only miR-18a was confirmed to be significantly up-regulated in patients with AAs (AUC 0.64, 95%CI=0.52–0.75)126.
As underlined above for breast cancer, the last two studies reported for colon cancers exclusively share the clinical relevance of miR-15b. This again highlights the importance of performing wider trials to confirm these data in larger cohorts and patient populations of different ethnicity.
Ovarian cancer (OC)
Since the majority of ovarian-cancer patients are diagnosed with advanced-stage disease, identifying miRNA biomarkers capable of detecting early stage disease would have great clinical value. Nevertheless, there are a limited number of patients taken into account when determining overall conclusive results, therefore the certainty of the results obtained with microarray-based profiling studies hinges on this fact. Even so, some of them exhibit great potential to address some clinical issues including characterization of different histologic and genetic subtypes, identification of markers for diagnosis and screening, prediction of clinical outcome, and better individualization of therapies.127. Recently, the miRNA expression analysis performed in a large multicenter cohort of 198 patients (86 patients as training set, and 112 patients as validation set) demonstrated that the downregulation of three miRNAs (miR-484, miR-642, and miR-217) was predictive of resistance to platinum-based chemotherapy when responder and non-responder individuals were compared (P=0.0007, P=0.04, P=0.046, respectively)128. In this context, additional studies on miR-484 demonstrated its capability to interfere with the VEGFB and VEGFR2 pathways, thus indicating new putative targets for therapeutic approaches targeting the tumor vasculature 128.
Separate studies 129, 130 showed the clinical relevance of miR-200c in ovarian cancer. Leskelä et al showed that women with FIGO (Federation of Gynecology and Obstetrics) stages III and IV serous ovarian carcinoma (n=57) without a complete response (CR) to paclitaxel–carboplatin regimen have tumors with significantly lower miR-200c levels when compared to the ones who achieved a complete response (HR 1.43, 95%CI 1.02–1.99, p=0.037)129. Downregulation of miR-200c was also identified associated to OS (HR 0.094, 95%CI: 0.012–0.766, P=0.0272) and Progression Free Survival (PFS) (HR 0.035, 95%CI: 0.004–0·311; P=0.0026), in a multivariable analysis of 144 stage I epithelial ovarian cancer samples 130. Finally, the integrated analysis of miRNA and transcriptome profiles from ovarian cancer TCGA dataset revealed that the expression levels of the miR-200 family members are able to be clearly differentiate between two clinically relevant subtypes, i.e mesenchymal and epithelial phenotypes of Serous Ovarian Cancer131.
Gliomas
MiRNAs seem to have a promising potential in this clinical setting where they might play an informative role regarding their efficacy in discriminating malignant phenotypes as well as predicting the outcome of surgical resection. Expression profiles of 261 gliomas from the TCGA dataset identified 121 miRNAs able to segregate tumor samples into five genetically and prognostically different subclasses 132. Another comprehensive analysis in 160 glioma samples from Chinese patients (63 WHO grade II, 33 grade WHO III, and 64 GBMs) revealed that the combination of 21 miRNAs (so-called “hub miRNAs” because of regulating more than 30 target mRNAs) was able to predict the survival of patients with all types of gliomas (HR 1.097, 95%CI: 1.068-1.127; R2=0.093; P=5.64×10-12; HR 1.076, 95%CI: 1.044-1.109, R2=0.074, P=1.12×10-6, from univariable and multivariable analyses respectively). Among the 21 relevant miRNAs, miR-524-5p (HR 0.849 95%CI: 0.721-0.999, R2-0.164, P=0.044) and miR-628-5p (HR 0.679, 95%CI: 0.470-0.982, R2 -0.387, P=0.036) expression levels represented protective factors whereas miR-938 (HR 1.113 95%CI: 1.029-1.204, R2 0.107, P=0.007), miR-595 (HR 1.200 95%CI: 1.059-1.360, R2 0.183, P=0.004), and miR-346 (HR 1.631 95%CI: 1.043-2.552, R2 0.489, P=0.029) represented risk factors133.
The possibility of detecting miRNAs in patient body fluids such as serum and /or cerebrospinal fluid (CSF) represents a novel, minimally-invasive experimental procedure that could possibly identify and characterize gliomas without requiring a surgical intervention. For instance, pilot studies on a limited number of patients have shown that miR-21 and miR-10b 134, 135, 136 are significantly increased in the CSF of patients with GBM or with brain metastasis of breast and lung cancer, when compared with CSF from patients whose tumors in remission and those with non-neoplastic conditions. Interestingly, since miR-10b is generally expressed in extracranial tissues but undetectable in the brain and CSF of non-cancer patients, probably because of the blood-brain membrane, its specific abundance in the CSF from GBM patients and brain metastases could be referred as a specific signature of brain pathology136. Conversely, members of the miR-200 family (miR-200a, miR-200b, miR-200c, and miR-141) are highly elevated in the CSF of patients with brain metastases but not in GBM (p<0.0001) or other primary brain tumors, suggesting they could serve as specific markers of metastatic brain tumors 136. Additionally, through a genome-wide serum miRNA analysis by next-generation sequencing, serum samples from 122 patients with untreated astrocytomas and 122 normal controls were evaluated, and a panel of seven miRNAs (miR-15b*, miR-23a, miR-133a, miR-150*, miR-197, miR-497 and miR-548b-5p) significantly decreased (P< 0.001) between patients with tumor grades in the range of WHO II–IV and the control group was identified 137 The AUC for the combination of the seven miRNAs was 0.972 (95% CI 0.954–0.990) for malignant astrocytomas (all grades) and controls, and with an optimal cut off of 5.6085, the sensitivity was of 88% and the specificity of 97.87%. Moreover, a marked increase in the levels of expression of the same group of miRNAs was detected after tumor resection (P< 0.001) 137.
A study of exosomes isolated from patient serua demonstrated that the expression levels of RNU6-1, miR-320, and miR-574-3p proved to effectively discriminate GBM patients from healthy individuals, suggesting their significance as a novel potential diagnostic tool 138. In particular, ROC curve analyses revealed that the expression levels of either RNU6-1 alone or in combination with miR-320, and miR-574-3p, were useful and robust biomarkers for differentiating GBM patients (n=50) from healthy controls (n=30), with an AUC of 0.722 (95%CI: 0.60–0.84; P=0.0007) for RNU6-1 and 0.775 (95%CI: 0.65–0.90; P=0.0001) for the 3 markers together. Importantly, at a cutoff value of 0.372 for RNU6-1, the sensitivity was 66% and the specificity was 68%., whereas at a cutoff value of 0.374 for the - miRNA signatures, sensitivity was 70% and specificity was 71%.
All together, these data suggest alternative non-invasive approaches for prediction of malignant progression and detection of residual disease in these types of tumors.
Chronic lymphocytic leukemia (CLL)
CLL has become the paradigmatic disease of miRNA involvement in cancer as the downregulation of miR-15a/miR-16 cluster in the majority of CLL patients was the first report linking miRNAs to cancer 27. A unique miRNA signature composed of 13 genes including miR-15a, miR-16, miR-155 as well as members of the miR-181 family possesses the ability to distinguish aggressive from indolent CLL 43. More recently, a scoring method combining miR-21 expression with the routine cytogenetic procedures fluorescence in situ hybridization (FISH) and KARYO (21FK score) was developed in order to stratify patients with CLL according to survival 139. When compared with the classical prognostic factors, including B2M, ZAP-70, IgVH, and CD19+CD38+, the 21FK score showed to be the only rating able to distinguish good-prognosis (low miR-21 expression, low score) and poor-prognosis (high miR-21 expression, high score) subpopulations both in a homogenously selected cohort of 104 CLL 17DEL patients and non-homogenous sets of 80 cases with either one chromosomal abnormality (13qDEL,11qDEL, T12, 17pDEL) or normal karyotype. Indeed, from the multivariable analysis by using of the 21FK score, IgVH, and ZAP-70, the 21FK score resulted that the only significant variable in both sets of patients (HR 3.514; 95%CI: 1.409-8.764, P=0.007, and HR 5.217; 95%CI: 1.408-19.327; P =0.013, respectively). In the same study, when patients with 17p deletions were compared to ones with normal karyotypes, miR-21, miR-34a, miR-155 and miR-181b were differentially expressed as well. In particular, miR-181b was specifically downregulated in patients refractory to their treatment regimen with a mean Treatment Free Survival (TFS) of 5.35 months versus 13.80 months of patients with high miR-181b expression (HR 2.77; 95%CI: 1.295-5.91, P=0.006). 139.
An interesting study successfully demonstrated the potential of using circulating miRNAs for both detection and stratification of CLL patients 140. In a cohort of CLL plasma samples a number of detectable circulating miRNAs including miR-17, miR-19b, miR-92a, miR-150, miR-223, miR-320, and miR-484 were found to be highly expressed when compared with normal control plasma. Among them, several miRNAs showed distinct profiles between CLL and other hematological malignancies such as hairy cell leukemia or multiple myeloma. Circulating miR-20a (AUC 0.920) and miR-195 (AUC 0.951), as well as miR-29a, miR-195, and miR-222 combination pattern (AUC 0.982) were found to be the best classifiers clearly separating CLL patients from healthy individuals.
Finally, with the aim of investigating the mechanisms underlying the transition from a clinical premalignant lesion named monoclonal B lymphocytosis (MBL) to CLL, Ferrajoli et al evaluated the differential expression of previously mentioned miR-155 in 224 samples from 2 independent validated cohorts of patients with CLL and MBL 141. MiR-155 expression was higher in isolated and purified B cells from MBL individuals when compared with normal B cells (P<0.0001) and CLL (P<0.0001). Increased miR-155 plasma levels, measured before treatment, were found associated with poor overall survival (P=0.01), and subsequent resistance to therapy with either fludarabine, cyclophosphamide, and rituximab (FCR) (P=0.02) or lenalidomide (P=0.03) in non-responder patients as compared to individuals with complete responses. These data support the ability of miR-155 to predict prognosis and response to treatment independent of the type of therapy administered 141.
Acute myelogenous leukemia (AML)
The profiling findings with AML have highlighted the existence of a complex network comprising gene mutations, along with aberrantly expressed genes and miRNAs. All of them collectively define the AML phenotype associated with clinically aggressive disease. High levels of miR-10a-5p in NPM1mut-AML patients were found to be associated with complete remission (CR) after chemotherapy with idarubicin and cytarabine (OR 1.33 P=0.019) 142. When NPM1 mutation status and miR-10a-5p were included in a multivariable model, they show a significant interaction correlating with CR status (P=0.01), suggesting a synergistic effect facilitating the tumor to acquire sensitivity to treatment 142. Furthermore, another study including a large cohort of complex karyotype AML patients (CK-AML) with known TP53 status demonstrated that low levels of miR-34a expression like TP53 alterations were predictive of chemotherapy response (OR 0.78 95%CI: 0.42–1.44 P=0.42) and poor clinical outcome (OS: HR 1.47 95%CI: 1.06–2.03 P=0.02; Relapse Free Survival (RFS): HR 1.90 95%CI: 1.14–3.18 P=0.01) 143. As in CLL, high levels of miR-155 has also been shown to be significantly correlates with a shorter OS (HR 1.62 95%CI: 1.25- 2.09 P<0.001) and less than a 50% of CR rate (OR 0.46 95%CI=0.27-0.81 P=0.007) independently of other strong clinical and molecular predictors144.
Recently a global screening of 670 miRNAs in 238 intermediate-risk cytogenetic AML patients (IR-AML) identified that high levels of miR-644 and miR-196b were other independent miRNAs associated with shorter overall survival (OR 0.51, 95%CI: 0.51–0.85; P=0.004, and OR 0.42, 95% CI: 0.19–0.94; P=0.036, respectively), and, in addition, increased levels of miR-135a and miR-409-3p were correlated with a higher risk of relapse at 5 years(OR: 0.23 95%CI: 0.0863–0.624, p=0.003; OR: 0.62 95%CI: 0.3762–1.033 p=0.067, respectively)145. This four-miRNA scoring scale generated an independent prognostic rate allowing a finite stratification of IR-AML patients into different prognostic categories and facilitating the proper assignment of post-remission therapy 145.
Regarding circulating miRNAs in AML, less is known about their potential clinical application. Advancements have been made in the past year by analyzing 140 newly diagnosed AML patients and 135 normal adult donors. New data set analyses have led to the identification of a 6-miRNA panel consisting of miR-10a-5p, miR-93-5p, miR-129-5p, miR-155-5p, miR-181b-5p and miR-320d which are able to differentiate between AML patients and normal controls (AUC ranging from 0.8129 to 0.9531) 146. Even more, high levels of miR-181b-5p in patients’ sera were found to be significantly associated with a worse OS (P=0.002) 146. Overall, these data demonstrated that the expression patterns of circulating miRNAs might be systematically altered in AML and detecting their levels could provide clinically useful diagnostic and prognostic classifiers for AML.
Acute lymphoblastic leukemia (ALL)
One of the first large-scale miRNA profiling analyses in a total of 72 cases of ALL, 98 cases of AML, and 13 normal bone marrow controls, identified a signature of 27 differentially expressed miRNAs able to separate ALL and AML. Six (i.e., miR-128a, miR-128b, miR-151*, j-miR-5, miR-130b, and miR-210) were expressed at a significantly higher level in ALL than in AML 147. More importantly, four miRNAs (i.e., miR-128a, miR-128b, let-7b, and miR-223) revealed the strongest diagnostic performance: the combination of any two or three of these miRNAs showed to separate ALL from AML cases with an overall diagnostic accuracy of 97–99% 147.
When referring to ALL, the pediatric population becomes one of clinical significance since ALL is the most common malignancy diagnosed in children, representing over a quarter of all pediatric cancers. In pediatric ALL bone marrow samples it was shown that miR-100, miR-196b and let-7e, are downregulated, while miR-128a and miR-181b are upregulated when compared to normal pediatric control samples148. The subsequent correlation analysis with the clinical and biological features showed that miR-196b expression was significantly elevated in the T-cell (P = 0.01) ALL phenotype and miR-100 expression was elevated in patients with a white blood count<50,000/mm3 at diagnosis, as well as in the presence of t(12;21) (P=0.04) and the absence of a hyperdiploid karyotype (P = 0.04) 148. Furthermore, ALL patients with reduced miR-335 are likely to have a poorer 5-year OS (OS=74.2%) compared to patients with high miR-335 (OS=96.9%, p=0.009). ROC curve analysis using the expression level of miR-335 resulted in a highly significant AUC of 0.73 (p=0.003). In vitro studies suggested that the poor prognosis would be linked to the development of resistance to glucocorticoid (GC) therapy mediated by the de-repression of MAP1K 149. Indeed, after restoring miR-335 levels, cancer cells proved to acquire sensitivity to a prednisolone-based therapy because of the miR-335-mediated direct inhibition of MAPK1. These findings provide suggestion for the development of new strategies that might work, for example, against the GC resistance arising in treated ALL patients 149.
Multiple myeloma (MM)
Because of the inherent heterogeneity of multiple myeloma, several groups have attempted to classify the disease based on gene expression profiling. One of the largest profiling studies was performed on 163 bone marrow samples from MM patients and revealed distinct miRNA signatures associated to the TC (Translocation/Cyclin D)-classified MM subgroups. This study demonstrated the miRNA pattern correlation with clinical outcome in the different TC prognostic groups of patients, including both the unfavorable 4p16 and MAF, (v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)), and the favorable 11q13 one. In particular, the specific upregulation of the miR-99b/let-7e/miR-125a cluster on 19q was detected in the 4p16 group, and miR-150, miR-155, and miR-34a were upregulated in the MAF group. Upregulation of miR-1275 and downregulation of miR-138 were observed in the 11q13 cases. Furthermore, the upregulation of three clusters of miRNAs, (miR503~424, miR17HG, and miR106A~363), were found significantly associated with decreased OS. More importantly, after a stepwise selection in a multivariable analysis, miR-886-5p and miR-17 expression levels were used to construct a miRNA-based risk predictor able to stratify patients into three risk groups (median OS 19.4, 40.6 and 65.3 months, P=0.001). This scale was demonstrated to ameliorate the classification of patients according to OS by improving the predictive power of conventional ISS/FISH approach (p= 0.0004). Indeed, the expression value of this miRNA risk estimator identified a separate class of patients with a significantly worse prognosis within a group defined as “low-risk” by the ISS/FISH classification system 150.
Dysregulated patterns of circulating serum miRNAs in comparison with healthy controls have also been identified in MM. When using a multivariate logistical regression model on a set of 103 newly diagnosed patients, 18 patients in relapse, 57 with monoclonal gammopathy of undetermined significance (MGUS) and 30 healthy donors, a combination of two miRNAs, miR-34a and let-7e, proved to be able to distinguish MM and MGUS from healthy donors (AUC 0.898, SE 80.6% and SP 86.7%; AUC 0.976, SE 91.1% and SP 97.6% respectively)151. In same study, lower levels of miR-744 and let-7e were associated with shorter OS and remission: 1-year mortality rate for miR-744 and let-7e was 41% (95%CI 28.8%-57.9%) and 34.6% (95%CI 23.4%-49.2%) respectively in the low miR-expressing patients and 3.3% (95%CI:0.8%-12.7%) and 3.9% (95%CI: 1.0%-14.8%) in the highly expressing patients. The Cox model also showed their prognostic impact on time to progression (TTP) for both (miR-744 HR: 0.690 95%CI 0.584-0.817, p<0.0001; let-7e HR: 0.552, 95%CI: 0.424-0.718, p<0.0001), overall suggesting they might be useful as a determinant of patient survival rates 151. It was also proven that miR-29a is upregulated in the serum of MM patients, and that this dysregulation could potently discriminate MMs from healthy donors with a sensitivity of 88% and a specificity of 70% 152.
Carcinoma of an unknown primary (CUP)
Metastatic cancer of unknown primary site (CUP) represents one of the 10 most frequent cancer diagnoses worldwide (3–5% of newly diagnosed cancer) 153 Patients with CUP present with metastases (late stage disease), without an identified primary tumor. MiRNAs are of particular importance in the identification of the site of origin for CUP because their expression levels and profiles reflect with great fidelity the tissue origin, and also because they are highly stable in formalin-fixed paraffin-embedded (FFPE) tissue blocks (the most available specimen type in pathology). MiRNA profiling may be helpful in further discriminating the histological origin of the tumor, as well as its optimal treatment. In an attempt to classify tumors based on their tissue specific miRNA signatures, a miRNA expression profiling was performed in 400 paraffin-embedded and fresh-frozen samples from 22 different tumor tissues and metastases by a custom-made microarray 154. The authors built a miRNA-based classifier including a total of 48 differentially expressed miRNAs, demonstrating to successfully classify the most of the samples with a high confidence (accuracy of 89%) 154. This 48 miRNAs panel was further validated on a cohort of 204 FFPE tumor samples comprising 25 different classes as accurately stratified by the array. Indeed, for 124 out of 204 the samples (66%), the sensitivity of the array was 90% (111/124 of them classifications agreed with the reference diagnosis), and it exceeded 90% for most tissue types. Specificity (negative agreement) in this group ranged from 95% to 100% and averaged above 99% 155. A new enriched miRNA-based assays (second generation array) comprising a higher number of miRNAs (64) able to discriminate a greater number (48) of tumor types has been developed 156. As reported in a cohort of 84 FFPE samples from CUP patients, it has shown to agree with the clinical diagnosis at presentation and after patient management in 70% and 89% of patients respectively; moreover it agreed with the final clinical diagnosis reached with supplemental immunohistochemical stains in 92% of patients, indicating a 22% improvement in agreement from diagnosis at presentation to the final clinical diagnosis 157. In conclusion, these novel microRNA-based approaches show high accuracy in identifying the final clinical diagnosis in CUP patients providing a useful tool for a better management and rational therapeutic intervention of this particularly challenging tumor category.
6 microRNAs as targets for anticancer drug therapy
RNA inhibition with the use of miRNAs
By blocking the function of a messenger RNA, miRNAs could represent a valid option for treating specific cancer patients in the near future 158, 159, 160. MicroRNAs can be separated into two groups: tumor suppressive or oncogenic, depending on their messenger RNA targets. Both groups are very well suited for therapeutic purposes, and they are represented by patients with a negatively correlated expression between a specific miRNA and its experimentally proven target(s) in the tumors. With the aims of recovering tumor suppressor proteins and inhibiting oncogenic ones, the strategy would involve restoring the miRNAs that are down-regulated and silencing the miRNAs that are up-regulated (Figure 4). There are two major advantages of using miRNAs (compared to other gene-silencing therapies explained in detail in Table 4): first, miRNAs are “natural” molecules produced in human cells and, second they can target multiple genes from the same pathway and therefore their action can occur at multiple levels in the same pathway significantly reducing the development of resistance as multiple mutations in multiple genes are needed. For example, miR-15a and miR-16-1, both with reduced expression in CLL, have two anti-apoptotic targets, the messenger RNAs encoded by the oncogenes for BCL2 and MCL1 161. Therefore, a personalized therapy based on the identification of patients with reduced miR-15a/miR-16-1 cluster expression and BCL2 and MCL1 protein overexpression in CD5/CD19 positive B-lymphocytes can be envisioned. Notice how these types of therapies are very specific for patients having these molecular characteristics in the same cells. Patients with low levels of miR-15a/miR-16 but normal levels of BCL2 or MCL1 or conversely with normal levels of miRNAs but abnormal levels of these anti-apoptotic oncogenes should be excluded from clinical trials targeting miR15a/miR-16.
Figure 4. MicroRNAs as targets for anticancer drug therapy.
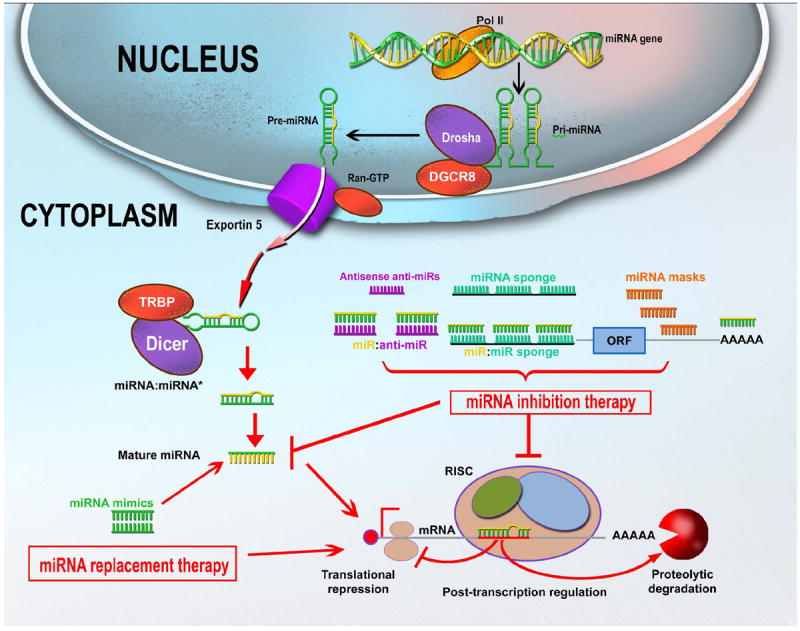
In order to restore tumor suppressor proteins levels, oncomiRs can be targeted with therapies such as antagomirs or LNAs anti-miRs. When aiming to target oncogenic proteins, the levels of tumor suppressor-miRNAs can be restored by using miRNA mimics, or alternatively through siRNA delivery against the oncogenic target. A direct messenger RNA inhibition of oncogenic proteins can be obtained also through a ribozyme-mediated translation inhibition.
Table 4.
The principal types of RNA therapeutic drugs.
| ANTISENSE OLIGONUCLEOTIDES (ASOs) |
|---|
| Definition: |
| A single-stranded chemically modified DNA-like molecule 17 to 22 nts in length designed to be complementary to a selected messenger RNA. |
| Mechanism of action: |
| Specifically inhibit expression of that gene mainly through formation of an mRNA-ASO duplex by sequence complementarity, leading to cleavage of the mRNA of target gene. |
| Example:NCT00030641 209 |
| Oligonucleotide: Oblimersen (Genasense, G3139) Phase II/III |
| Condition: Lung Cancer – Stage IIIB or IV relapsed or refractory disease |
| Molecular Target: BCL-2 |
| Rationale: Oblimersen may increase the effectiveness of docetaxel by making the tumor cells more sensitive to the drug. |
| Purpose: Randomized phase II/III trial to compare the effectiveness of docetaxel with or without oblimersen in treating patients who have relapsed or refractory non-small cell lung cancer that has been previously treated. |
Other examples:
|
|
|
| RIBOZYMES or DNAZYMES |
| Definition: |
| A ribozyme, or RNA enzyme, is an RNA molecule that can catalyze a chemical reaction. A DNAzyme, or dezoxyribozyme, is a catalytic DNA that site specifically cleaving the target RNA. |
| Mechanism of action: |
| Consists of three steps, cyclically repeated: base pairing to a complementary target sequence, followed by site-specific cleavage of the substrate and finally release of the cleavage products. |
| Example: NCT00021021 213 |
| Oligonucleotide: Angiozyme Phase II |
| Condition: Kidney Cancer |
| Molecular Target: VEGF |
| Rationale: RPI.4610 may stop the growth of metastatic kidney cancer by stopping blood flow to the tumor. |
| Purpose: Phase II trial to study the effectiveness of RPI.4610 in treating patients who have metastatic kidney cancer. |
Other examples:
|
|
|
| siRNAs |
| Definition: |
| A double strand (ds) RNA homologous to an mRNA of a target gene. |
| Mechanism of action: |
| The siRNAs are incorporated into a multiprotein RNA-induced silencing complex (RISC), leaving the antisense strand to guide this complex to its homologous mRNA target for endonucleolitic cleavage of messenger RNA. |
| Example: NCT01591356 215 |
| Oligonucleotide: siRNA -EphA2-DOPC Phase I |
| Condition: Advanced Cancers |
| Molecular Target: EphA2 |
| Rationale: siRNA targeting EphA2 may decrease tumor growth in cancer recurrence. |
| Purpose: The goal of this clinical research study is to learn about the safety of siRNA-EphA2-DOPC when given to patients with advanced, recurrent cancer. Researchers also want to learn the highest tolerable dose of this drug that can be given. siRNA-EphA2-DOPC is designed to shut down the activity of a gene that causes tumor growth. |
Other examples:
|
|
|
| MicroRNA mimics |
| Definition: |
| A microRNA mimic is a small single-strand 19 to 24 nt RNA with an identical sequence of the miRNA of interest (to be re-expressed) |
| Mechanism of action: |
| Mimic the effects of an endogenous miRNA with consequent inhibition of protein production by either transcriptional inhibition or translational block or both. |
| Example: NCT01829971 219 |
| Oligonucleotide: MRX34 (miR-34 mimic) Phase I |
| Condition: Primary Liver Cancer, Solid tumors, Lymphoma, AML, ALL, CLL, CML (in Accelerated or Blast Phase) |
| Molecular Target: MYC, MET, BCL2, β-catenin |
| Rationale: MRX34 will block tumor development and progression by targeting a variety of cancer related pathways. |
| Purpose: This is a study to evaluate the safety of MRX34 in patients with primary liver cancer or those with liver metastasis from other cancers. The drug is given intravenously, twice per week for three weeks in a row and then one week off. Additionally patients with hematologic malignancies will be evaluated on a treatment schedule of five days in a row with two weeks off. |
|
|
| ASOs/AMOs, LNAs anti-miR and antagomirs |
| Definition: |
| Small single-stranded oligonucleotides specifically designed to block the function of microRNA. |
| The ASOs/AMOs are single-stranded, chemically modified DNA-like molecule that is 17 to 22 nt in length and designed to be complementary to a selected microRNA and specifically inhibit its expression. |
| The LNAs anti miRNAs represents LNA modified ASOs. |
| The antagomirs are single-stranded 23-nucleotide RNA molecules complementary to the targeted microRNA that have been modified to increase the stability of the RNA and protect it from degradation. |
| Mechanism of action: |
| AMOs are ASOs against miRNAs, and therefore produce ASO - miRNA duplex through sequence complementarity, leading to RNAse-H mediated cleavage of the target miRNA gene. |
| The LNA anti miRNA have the same mechanism as the ASO/AMO. |
| The miRNA/antagomir – duplexes induces degradation of the miRNA and recycling of the antagomir in a way still not completely known. |
| Example: NCT01200420 220 |
| Oligonucleotide: Miravirsen (LNA) Phase 2 |
| Condition: Hepatitis C |
| Molecular Target: miR-122 |
| Rationale: Miravirsen is a novel LNA-therapeutic agent inhibits miR-122. Since the replication of hepatitis C viruse (HCV) is dependent on this miRNA, by blocking it researchers aim to prevent the development of hepatocellular carcinoma. |
| Purpose: The main purpose of this study is to determine the safety and tolerability of multiple dosing of miravirsen in subjects infected with chronic hepatitis C, which are at high risk of developing hepatocellular carcinoma. Another purpose includes assessing the pharmacokinetics of miravirsen and assessment of miravirsen’s effect on HCV viral titer. |
Other examples:
|
Strategies for miRNA-targeting
These are based on either restoring miRNA levels, or blocking miRNA function with oligonucleotide-based strategies (Table 4). A targeted approach to replenish the expression levels of particular miRNAs is restoring the levels and function of one or a limited number of miRNAs, usually located in a cluster (such as miR-15a and miR-16-1 at 13q14.3), either with miRNA mimics or with miRNAs encoded in expression vectors. MiRNA-mimic molecules are double stranded sequences with 100% similarity to the endogenous miRNA, which can be delivered by nanoparticles and thereby expressed in the cells. This approach is particularly attractive since nanoparticles can be coated with antibodies that recognize tumor-specific antigens, therefore allowing a tumor specific delivery of the miRNA of interest. A recent study targeted the delivery of miR-34a using nanoparticles coated with a neuroblastoma- specific anti-disialoganglioside GD2 antibody. With this design, the therapy resulted in the inhibition of neuroblastoma tumor growth in a murine orthotopic xenograft model 162. Moreover, MRX34, a liposome-formulated mimic of the tumor suppressor miR-34 is developed in a clinical phase 1 trial by MiRNA Therapeutix for patients with advanced or metastatic liver cancer. Preclinical studies have already shown that tail vein injection of MRX34 reduced tumor growth and significantly enhanced survival with a favorable safety profile in orthotopic mouse models of hepatocellular carcinoma 163.
Another mechanism has been targeting the new blood vessel formation on tumors. Positively charged liposomes have been investigated to deliver drugs to tumor blood vessels efficiently. The negative charge of the proteoglycans on the endothelial cell membranes can bind and help cells internalize cationic liposome due to their electrostatic interaction 164. This approach has been proven effective in the delivery of miRNA mimics, such as seen in a non-small cell lung (NSCLC) cancer xenograft murine model. MiR-29b has been shown to be significantly reduced in NSCLC, consequently increasing the expressing of its oncogenic target CDK6. Through cationic-lipoplexes (LPs)-based carriers, miR-29b was successfully delivered resulting in inhibited tumor growth by 60% compared to a LP-miR negative control 165. These results demonstrated that cationic LPs target tumor vasculature in a specific manner, and thereby achieve a greater therapeutic potential for lung cancer treatment.
Current strategies for inhibitory miRNA-targeting are mainly based on antisense oligonucleotides (so called anti-miRs), comprised locked nucleic acids (LNA) along with and tiny LNA anti-miR constructs, antagomirs and miRNA sponges (Table 4). Miravirsen (SPC3649), an LNA against miR-122 developed by Santaris Pharma A/S, is the first miRNA-target therapy tested on clinical trial for the treatment of hepatitis C virus (HCV) infection. The recently completed Phase 2a trial showed that SPC3649 exhibited robust anti-viral activity in a dose-dependent manner 166. More impressively, four out of nine patients treated at the highest dose (7 mg/kg) with SPC3649 reached undetectable levels of HCV RNA 167 (consequently decreasing the possibility of developing hepatocellular carcinoma). The effectiveness of antimiR-122 treatment proved the plausibility of the LNA-based therapeutics and encourages the development of other specific miRNA-targeting therapeutic strategies.
Combination approaches with miRNA therapeutics
The introduction of combination chemotherapy by Emil Freireich and Emil Frei in early 1960, contrary to the initial opinion in medical community, not only did not harm children with acute lymphocytic leukemia but moreover represented a huge step forward, responsible for the excellent outcome of this otherwise 100% lethal disease 168. Therefore, we emphasize the need of using combinatorial therapies of various miRNA agents in “cocktails” to be delivered to patients along with chemotherapeutics (therapies with miRNA-mimetic and anti-miRNA agents). Regarding CLL for example, there are two hypotheses that drive our multi-modal approach in which we could achieve the inhibition of messenger RNAs, and therefore protein expression 109. First, a “sandwich RNA inhibition strategy” comprising the use of multiple agents could focus on a major molecular alteration clearly linked to CLL pathogenesis. Given the fact that for this disease the overexpression of anti-apoptotic factor BCL2 has been correlated with poor patient outcome, we could target this protein by designing regimens using a cocktail of anti-BCL2 ASOs and miR-15a and miR-16 (already known to target BCL2 mRNA). Published studies have already proven that Oblimersen sodium (an ASO compound designed to specifically bind to human BCL2 mRNA) has a relative efficacy in treating relapsed or refractory CLL 169; therefore our expectations of this multi-modal approach seems to be promising for this indolent disease. Second, “the multiplex RNA-inhibition strategy” targets various molecular defects that are part of the same pathway (such as apoptosis). In this case, multiple synthetic miRNAs targeting overexpressed regulators of apoptosis such as BCL2 (miR-15a and miR-16) and MCL1 (the miR-29 family) may be more efficient at consistently and robustly reducing the expression levels of these proteins rather than a single therapeutic agent.
Novel approaches can be developed in the near future. One is a combinatorial approach that consists in boosting the therapeutic synergy between miRNA and small interfering RNA (siRNA). The dual inhibition of a specific oncogene by its single-targeting-siRNA and a multi-targeting-miRNA can be utilized to obtain improved therapeutic efficiency. While siRNA-mediated silencing of EphA2, an ovarian cancer oncogene, results in reduction of tumor growth, additional inhibition of EphA2 by a mir-520d-3p further ‘boosts’ its anti-tumor effects 170. Finally, a therapeutic strategy where cells are stimulated to secrete oncogenic miRNA-loaded vesicles and the cancer patient is subsequently treated with dialysis can be envisioned, as a way to “wash-out” non-coding oncogenes and induce the tumor suppressor proteins inside the cancer cells.
8 Instead of conclusion: The new wave – long ncRNAs in clinics
Although the leading role published in research regarding miRNAs as cancer-related non-coding RNAs, new categories of ncRNAs have recently emerged. The size range of ncRNAs is vast (from 200 to tens of thousands nucleotides), and at least 10 times longer than miRNAs (and in some instances, thousands of times larger). They include lincRNAs and ultraconserved genes which were found to be abnormally expressed in cancer and involved in tumorigenic mechanisms 3,171. As the spectrum of ncRNAs is much greater than for miRNAs (the estimates are higher as tens of thousands of ncRNA transcripts versus thousands of microRNAs in the human genome), the impact on any aspect of basic and translational cancer research will be huge.
Long non-coding RNAs (lncRNAs) promise to be a subtype of ncRNA as important (or most likely more important) as miRNAs. They can regulate transcription of target genes by protein binding, interference, hybridization, and epigenetic mechanism as chromatin remodeling, methylation, and histone modification 172. They have also been linked to every hallmark of cancer: sustaining proliferative signaling, evading growth suppressors, enabling replicative immortality, activating invasion and metastasis, inducing angiogenesis and resisting cell death. LncRNAs have strong potential to become biomarkers due to their specific localization in cells and tissues. Tissue specificity of lncRNAs can also enable them as valuable therapeutic agents. Hence, they could serve for diagnostic, prognostic, and monitoring purposes. 173.
Despite this, truth of the fact is that we are only experiencing the beginning of our understanding of the complexity of the lncRNA-world. It is certain that their structures and repertoire of functions exhibit robustness in cancer, and that these functions are adversary to those of the proteomic field. Nevertheless, what is known about cancer is just the tip of the iceberg; lncRNAs have been linked to a wide array of known human pathologies/diseases, especially in the last five years. Some of the pathologies include cardiovascular 174, 175, inflammatory 176, 177, diabetes 178, 179, Alzheimers 180, infectious diseases 181 schizophrenia 182, ischemic disease/stroke 183, neuromuscular disease 184, immune response 185,186 and so on. Certainly, this is an exciting time for the RNA therapeutic arena, and we are looking forward to more results of clinical studies investigating novel strategic approaches.
Acknowledgments
Financial support: Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, Developmental Research Awards in Prostate Cancer, Multiple Myeloma, Leukemia (P50 CA100632) and Head and Neck (P50 CA097007) SPOREs, a SINF MDACC_DKFZ grant in CLL, a SINF grant in colon cancer, a Kidney Cancer Pilot Project, the Duncan Family Institutional Seed Funds, The Blanton-Davis Ovarian Cancer - 2013 Sprint for Life Research Award, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson, Jr. and by the CLL Global Research Foundation. Dr. Berindan-Neagoe’s work was financed by POSCCE 709/2010 grant with title: “Clinical and economical impact of proteome and transcriptome molecular profiling in neoadjuvant therapy of triple negative breast cancer (BREASTIMPACT)”.
Footnotes
Financial disclosures: The authors declare no financial disclosures.
References
- 1.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14(10):1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 8.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 9.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 11.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105(39):14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 16.Eiring AM, Harb JG, Neviani P, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140(5):652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M, Chen J, Su F, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288(48):34343–34351. doi: 10.1074/jbc.M113.480822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehmann SM, Krüger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15(6):827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 22.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 25.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 27.Calin GA, Dumitru C, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 29.Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Callegari E, Elamin BK, Giannone F, et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56(3):1025–1033. doi: 10.1002/hep.25747. [DOI] [PubMed] [Google Scholar]
- 31.Qin J, Luo M. MicroRNA-221 promotes colorectal cancer cell invasion and metastasis by targeting RECK. FEBS Lett. 2014;588(1):99–104. doi: 10.1016/j.febslet.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Gits CM, van Kuijk PF, Jonkers MB, et al. MiR-17-92 and miR-221/222 cluster members target KIT and ETV1 in human gastrointestinal stromal tumours. Br J Cancer. 2013;109(6):1625–35. doi: 10.1038/bjc.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabbri M, Bottoni A, Shimizu M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305(1):59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Psathas JN, Thomas-Tikhonenko A. MYC and the Art of MicroRNA Maintenance. Cold Spring Harb Perspect Med. 2014 doi: 10.1101/cshperspect.a014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15(4):667–671. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 37.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9(4):293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 38.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31(13):1609–1622. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 41.Melo SA, Esteller M. Disruption of microRNA nuclear transport in human cancer. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Calin GA, Croce CM. Genomics of Chronic Lymphocytic Leukemia MicroRNAs as New Players With Clinical Significance. Seminars in Oncology. 2006;33(2):167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Calin GA, Ferracin M, Cimmino A, et al. MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 44.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152(4):844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raveche ES, Salerno E, Scaglione BJ, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109(12):5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preskill C, Weidhaas JB. SNPs in microRNA binding sites as prognostic and predictive cancer biomarkers. Crit Rev Oncog. 2013;18(4):327–340. doi: 10.1615/critrevoncog.2013007254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91(4):431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berindan-Neagoe I, Calin GA. Molecular pathway: microRNAs, cancer cells and microenvironment - Chess principles and patient care. Clinical Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-13-2500. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem. 2012;287(2):1397–1405. doi: 10.1074/jbc.M111.288662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. 2013;191(12):6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288(15):10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 54.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21(3):470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in Cancer. Cell. 2009;137(3):586–586.e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 57.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 61.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 62.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 63.Sylvestre Y, De Guire V, Querido E, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282(4):2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 64.Xie L, Ushmorov A, Leithäuser F, et al. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119(15):3503–11. doi: 10.1182/blood-2011-09-381905. [DOI] [PubMed] [Google Scholar]
- 65.Razumilava N, Bronk SF, Smoot RL, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55(2):465–75. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei JS, Song YK, Durinck S, Chen QR, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27(39):5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 68.Bommer GT, Gerin I, Feng Y, et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr Biol. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 69.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q, Wang JX, He YQ, et al. MicroRNA-185 regulates chemotherapeutic sensitivity in gastric cancer by targeting apoptosis repressor with caspase recruitment domain. Cell Death Dis. 2014;5:e1197. doi: 10.1038/cddis.2014.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci U S A. 2011;108(2):522–527. doi: 10.1073/pnas.1017346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124(6):1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 74.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67(22):11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 75.Fasanaro P, D’Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283(23):15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng Y, Li S, Ding Y, Wang Q, Luo H, Shi Q, Hao Z, Xiao G, Tong S. The role of miR-18a in gastric cancer angiogenesis. Hepatogastroenterology. 2013;60(127):1809–13. [PubMed] [Google Scholar]
- 77.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 78.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120(4):1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meng W, Ye Z, Cui R, et al. MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin Cancer Res. 2013;19(19):5423–5433. doi: 10.1158/1078-0432.CCR-13-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 82.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rathore MG, Saumet A, Rossi JF, et al. The NF-kappaB member p65 controls glutamine metabolism through miR-23a. Int J Biochem Cell Biol. 2012;44(9):1448–1456. doi: 10.1016/j.biocel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 85.Eichner LJ, Perry MC, Dufour CR, et al. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 2010;12(4):352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 86.Fang R, Xiao T, Fang Z, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287(27):23227–23235. doi: 10.1074/jbc.M112.373084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei J, Wang F, Kong LY, et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73(13):3913–3926. doi: 10.1158/0008-5472.CAN-12-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Planell-Saguer M, Rodicio MC. Analytical aspects of microRNA in diagnostics: a review. Anal Chim Acta. 2011;699(2):134–152. doi: 10.1016/j.aca.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 89.Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn. 2013;13(2):183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
- 90.Sempere LF, Korc M. A method for conducting highly sensitive microRNA in situ hybridization and immunohistochemical analysis in pancreatic cancer. Methods Mol Biol. 2013;980:43–59. doi: 10.1007/978-1-62703-287-2_4. [DOI] [PubMed] [Google Scholar]
- 91.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717(1-2):85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics. 2014;15:29. doi: 10.1186/1471-2105-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. doi: 10.1186/1471-2407-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14(5):844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 96.Mulrane L, Klinger R, McGee SF, Gallagher WM, O’Connor DP. microRNAs: a new class of breast cancer biomarkers. Expert Rev Mol Diagn. 2014;14(3):347–363. doi: 10.1586/14737159.2014.901153. [DOI] [PubMed] [Google Scholar]
- 97.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 98.Zhang HL, Yang LF, Zhu Y, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–331. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 99.Hu Z, Chen X, Zhao Y, et al. Serum MicroRNA Signatures Identified in a Genome-Wide Serum MicroRNA Expression Profiling Predict Survival of Non-Small-Cell Lung Cancer. Journal of Clinical Oncology. 2010;28(10):1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 100.Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6(5):1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 101.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferdin J, Kunej T, Calin GA. Non-coding RNAs: identification of cancer-associated microRNAs by gene profiling. Technol Cancer Res Treat. 2010;9(2):123–138. doi: 10.1177/153303461000900202. [DOI] [PubMed] [Google Scholar]
- 103.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121(4):141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 104.Boeri M, Pastorino U, Sozzi G. Role of MicroRNAs in Lung Cancer: microRNA Signatures in Cancer Prognosis. Cancer J. 2012;18(3):268–274. doi: 10.1097/PPO.0b013e318258b743. [DOI] [PubMed] [Google Scholar]
- 105.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143(1):35–47. doi: 10.1053/j.gastro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 106.Zheng H, Liu JY, Song FJ, Chen KX. Advances in circulating microRNAs as diagnostic and prognostic markers for ovarian cancer. Cancer Biol Med. 2013;10(3):123–130. doi: 10.7497/j.issn.2095-3941.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Di Leva G, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Frontiers in Oncology. 2013;3:153. doi: 10.3389/fonc.2013.00153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.Gailhouste L, Ochiya T. Cancer-related microRNAs and their role as tumor suppressors and oncogenes in hepatocellular carcinoma. Histol Histopathol. 2013;28(4):437–451. doi: 10.14670/HH-28.437. [DOI] [PubMed] [Google Scholar]
- 109.Calin GA, Croce CM. Chronic lymphocytic leukemia: interplay between noncoding RNAs and protein-coding genes. Blood. 2009;114(23):4761–4770. doi: 10.1182/blood-2009-07-192740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Troppan K, Wenzl K, Deutsch A, Ling H, Neumeister P, Pichler M. MicroRNAs in diffuse large B-cell lymphoma: implications for pathogenesis, diagnosis, prognosis and therapy. Anticancer Res. 2014;34(2):557–564. [PubMed] [Google Scholar]
- 111.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16(2):430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boeri M, Verri C, Conte D. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sozzi G, Boeri M, Rossi M, et al. Clinical Utility of a Plasma-Based miRNA Signature Classifier Within Computed Tomography Lung Cancer Screening: A Correlative MILD Trial Study. J Clin Oncol. 2014;32(8):768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Y, Gu J, Roth JA, et al. Pathway-Based Serum microRNA Profiling and Survival in Patients with Advanced Stage Non–Small Cell Lung Cancer. Cancer Res. 2013;73(15):4801–4809. doi: 10.1158/0008-5472.CAN-12-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci USA. 2013;110(18):7413–7417. doi: 10.1073/pnas.1304977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gasparini P, Cascione L, Fassan M, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5(5):1174–1184. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chan M, Liaw CS, Ji SM, et al. Identification of Circulating MicroRNA Signatures for Breast Cancer Detection. Clin Cancer Res. 2013;19(16):4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 119.Kodahl AR, Lyng MB, Binder H, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: A case control study. Molecular Oncology. 2014 doi: 10.1016/j.molonc.2014.03.002. http://dx.doi.org/10.1016/j.molonc.2014.03.002. [DOI] [PMC free article] [PubMed]
- 120.Mutch MG. Molecular profiling and risk stratification of adenocarcinoma of the colon. J Surg Oncol. 2007;96(8):693–703. doi: 10.1002/jso.20915. [DOI] [PubMed] [Google Scholar]
- 121.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Gen Cytogen. 2010;200(2):154–160. doi: 10.1016/j.cancergencyto.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 123.Hansen TF, Sørensen FB, Lindebjerg J, Jakobsen A. The predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancer. BMC Cancer. 2012;12:83. doi: 10.1186/1471-2407-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 125.Kanaan Z, Roberts H, Eichenberger R, et al. A Plasma MicroRNA Panel for Detection of Colorectal Adenomas - A Step Toward More Precise Screening for Colorectal Cancer. Ann Surg. 2013;258(3):400–408. doi: 10.1097/SLA.0b013e3182a15bcc. [DOI] [PubMed] [Google Scholar]
- 126.Giráldez MD, Lozano JJ, Ramírez G, et al. Circulating MicroRNAs as Biomarkers of Colorectal Cancer: Results From a Genome-Wide Profiling and Validation Study. Clin Gastronenter and Hepatol. 2013;11(6):681–688. doi: 10.1016/j.cgh.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 127.Gómez-Raposo C, Mendiola M, Barriuso J, Hardisson D, Redondo A. Molecular characterization of ovarian cancer by gene-expression profiling. Gynecol Oncol. 2010;118(1):88–92. doi: 10.1016/j.ygyno.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 128.Vecchione A, Belletti B, Lovat F, et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci U S A. 2013;110(24):9845–9850. doi: 10.1073/pnas.1305472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leskelä S, Leandro-García LJ, Mendiola M, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2010;18(1):85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 130.Marchini S, Cavalieri D, Fruscio R, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12(3):273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 131.Yang D, Sun Y, Hu L, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23(2):186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71(9):3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Y, Xu J, Chen H, et al. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res. 2013;41(22):e203. doi: 10.1093/nar/gkt1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012;14(1):29–33. doi: 10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Akers JC, Ramakrishnan V, Kim R, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Teplyuk NM, Mollenhauer B, Gabriely G, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14(6):689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang C, Wang C, Chen X, et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132(1):116–127. doi: 10.1002/ijc.27657. [DOI] [PubMed] [Google Scholar]
- 138.Manterola L, Guruceaga E, Gállego Pérez-Larraya J, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. doi: 10.1093/neuonc/not218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rossi S, Shimizu M, Barbarotto E, et al. MicroRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116(6):945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moussay E, Wang K, Cho JH, et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2011;108(16):6573–8. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ferrajoli A, Shanafelt TD, Ivan C, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122(11):1891–1899. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Havelange V, Ranganathan P, Geyer S, et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood. 2014;123(15):2412–2415. doi: 10.1182/blood-2013-10-532374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rücker FG, Russ AC, Cocciardi S, et al. Altered miRNA and gene expression in acute myeloid leukemia with complex karyotype identify networks of prognostic relevance. Leukemia. 2013;27(2):353–361. doi: 10.1038/leu.2012.208. [DOI] [PubMed] [Google Scholar]
- 144.Marcucci G, Maharry KS, Metzeler KH, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013;31(17):2086–2093. doi: 10.1200/JCO.2012.45.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Díaz-Beyá M, Brunet S, Nomdedéu J, et al. MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leukemia. 2014;28(4):804–812. doi: 10.1038/leu.2013.281. [DOI] [PubMed] [Google Scholar]
- 146.Zhi F, Cao X, Xie X, Wang B, et al. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One. 2013;8(2):e56718. doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mi S, Lu J, Sun M, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104(50):19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.de Oliveira JC, Scrideli CA, Brassesco MS, et al. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk Res. 2012;36(3):293–298. doi: 10.1016/j.leukres.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 149.Yan J, Jiang N, Huang G, et al. Deregulated MIR335 that targets MAPK1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia. Br J Haematol. 2013;163(1):93–103. doi: 10.1111/bjh.12489. [DOI] [PubMed] [Google Scholar]
- 150.Wu P, Agnelli L, Walker BA, et al. Improved risk stratification in myeloma using a microRNA-based classifier. Br J Haematol. 2013;162(3):348–359. doi: 10.1111/bjh.12394. [DOI] [PubMed] [Google Scholar]
- 151.Kubiczkova L, Kryukov F, Slaby O, et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99(3):511–518. doi: 10.3324/haematol.2013.093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sevcikova S, Kubiczkova L, Sedlarikova L, Slaby O, Hajek R. Serum miR-29a as a marker of multiple myeloma. Leuk Lymphoma. 2013;54(1):189–191. doi: 10.3109/10428194.2012.704030. [DOI] [PubMed] [Google Scholar]
- 153.Ferracin M, Pedriali M, Veronese A, et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225:43–53. doi: 10.1002/path.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 155.Rosenwald S, Gilad S, Benjamin S, et al. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor origin. Mod Pathol. 2010;23(6):814–823. doi: 10.1038/modpathol.2010.57. [DOI] [PubMed] [Google Scholar]
- 156.Zion O, Burnstein I, Chajut A, et al. A Second-Generation MicroRNA-Based Assay for Diagnosing Tumor TissueOrigin. The Oncologist. 2012;17:801–812. doi: 10.1634/theoncologist.2011-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pentheroudakis G, Pavlidis N, Fountzilas G, et al. Novel microRNA-based assay demonstrates 92% agreement with diagnosis based on clinicopathologic and management data in a cohort of patients with carcinoma of unknown primary. Mol Cancer. 2013;12:57. doi: 10.1186/1476-4598-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Melo SA, Kalluri R. Molecular pathways: microRNAs as cancer therapeutics. Clin Cancer Res. 2012;18(16):4234–4239. doi: 10.1158/1078-0432.CCR-11-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105(13):5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tivnan A, Orr WS, Gubala V, et al. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One. 2012;7(5):e38129. doi: 10.1371/journal.pone.0038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bader AG. miR-34 — a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao G, Rodriguez BL. Molecular targeting of liposomal nanoparticles to tumor microenvironment. Int J Nanomedicine. 2013;8:61–71. doi: 10.2147/IJN.S37859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu Y, Crawford M, Mao Y, et al. Therapeutic Delivery of MicroRNA-29b by Cationic Lipoplexes for Lung Cancer. Mol Ther Nucleic Acids. 2013;2:e84. doi: 10.1038/mtna.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199(3):407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 168.Freireich EJ, Frei E., 2nd Recent Advances in Acute Leukemia. Prog Hematol. 1964;4:187–202. [PubMed] [Google Scholar]
- 169.Advani PP, Paulus A, Masood A, Sher T, Chanan-Khan A. Pharmacokinetic evaluation of oblimersen sodium for the treatment of chronic lymphocytic leukemia. Expert Opin Drug Metab Toxicol. 2011;7(6):765–774. doi: 10.1517/17425255.2011.579105. [DOI] [PubMed] [Google Scholar]
- 170.Nishimura M, Jung EJ, Shah MY, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3(11):1302–131. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12(6):433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 174.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res. 2012;111(10):1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 175.Iaconetti C, Gareri C, Polimeni A, Indolfi C. Non-coding RNAs: the “dark matter” of cardiovascular pathophysiology. Int J Mol Sci. 2013;14(10):19987–20018. doi: 10.3390/ijms141019987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111(3):1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cui H, Xie N, Tan Z, et al. The human long noncoding RNA, lnc-IL7R, regulates inflammatory response. Eur J Immunol. 2014 doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55(2):941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 179.Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, Regazzi R. Emerging roles of non-coding RNAs in pancreatic β-cell function and dysfunction. Diabetes Obes Metab. 2012;14(Suppl 3):12–21. doi: 10.1111/j.1463-1326.2012.01654.x. [DOI] [PubMed] [Google Scholar]
- 180.Faghihi MA, Modarresi F, Khalil AM. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14(7):7237–7230. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Scaria V, Pasha A. Long Non-Coding RNAs in Infection Biology. Front Genet. 2013;3:308. doi: 10.3389/fgene.2012.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Barry G, Briggs JA, Vanichkina DP, Poth EM. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19(4):486–94. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 183.Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012;43(10):2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cabianca DS, Casa V, Bodega B, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Carpenter S, Aiello D, Atianand MK. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Imamura K, Imamachi N, Akizuki G. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 187.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50(4):244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 188.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10(5):543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 189.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3(4):563–78. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 190.Kloosterman WP, Wienholds E, De Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3(1):27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 191.Sempere LF, Preis M, Yezefski T, et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res. 2010;16(16):4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang J, Zhao J, Shi M, Ding Y, Sun H, Yuan F, Zou Z. Elevated expression of miR-210 predicts poor survival of cancer patients: a systematic review and meta-analysis. PLoS One. 2014;9(2):e89223. doi: 10.1371/journal.pone.0089223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138(10):1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013;59(10):1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- 195.Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3):e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 197.Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 198.Toiyama Y, Hur K, Tanaka K, et al. Serum miR-200c is a Novel Prognostic and Metastasis-Predictive Biomarker in Patients With Colorectal Cancer. Ann Surg. 2014;259(4):735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PLoS One. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhu C, Ren C, Han J, et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110(9):2291–2299. doi: 10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tsujiura M, Komatsu S, Ichikawa D, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer. 2014 doi: 10.1007/s10120-014-0363-1. [DOI] [PubMed] [Google Scholar]
- 202.Xu J, Wu C, Che X, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50(2):136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 203.Meng W, Ye Z, Cui R, et al. MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin Cancer Res. 2013;19(19):5423–5433. doi: 10.1158/1078-0432.CCR-13-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lin Q, Chen T, Lin Q, et al. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol. 2013;107(7):767–771. doi: 10.1002/jso.23312. [DOI] [PubMed] [Google Scholar]
- 205.Rodríguez M, Silva J, López-Alfonso A, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer. 2014 doi: 10.1002/gcc.22181. [DOI] [PubMed] [Google Scholar]
- 206.Yang C, Wang C, Chen X, et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132(1):116–127. doi: 10.1002/ijc.27657. [DOI] [PubMed] [Google Scholar]
- 207.Westermann AM, Schmidt D, Holdenrieder S, et al. Serum microRNAs as biomarkers in patients undergoing prostate biopsy: results from a prospective multi-center study. Anticancer Res. 2014;34(2):665–669. [PubMed] [Google Scholar]
- 208.Zheng H, Zhang L, Zhao Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One. 2013;8(11):e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Genta Incorporated; Berkeley Heights, New Jersey. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. Randomized Study of Docetaxel Versus Docetaxel Plus Genasense™ (G3139; Bcl-2 Antisense Oligonucleotide) in Patients With Previously Treated Non-Small Cell Lung Cancer. [Internet]. Available from: http://clinicaltrials.gov/show/NCT00030641 NLM Identifier: NCT00030641. [Google Scholar]
- 210.Isis Pharmaceuticals; Carlsbad, California. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. A Phase 1/2 Study of ISIS 481464, an Antisense Oligonucleotide Inhibitor of STAT3, Administered to Patients With Advanced Cancers. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT01563302 NLM Identifier: NCT01563302. [Google Scholar]
- 211.Hoosier Oncology Group; Indianapolis, Indiana. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. The Borealis-2 Clinical Trial: A Randomized Phase 2 Study Comparing Docetaxel Alone to Docetaxel in Combination With OGX-427 in Patients With Relapsed or Refractory Metastatic Urothelial Carcinoma After Receiving a Platinum-containing Regimen: Hoosier Oncology Group GU12-160. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT01780545 NLM Identifier: NCT01780545. [Google Scholar]
- 212.AstraZeneca; London, United Kingdom. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. A Phase I/Ib, Open-Label, Multicentre Study to Assess the Safety, Tolerability, Pharmacokinetics and Preliminary Anti-tumour Activity of AZD9150 in Patients With Advanced/Metastatic Hepatocellular Carcinoma. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT01839604 NLMIdentifier: NCT01839604. [Google Scholar]
- 213.Ribozyome Pharmaceuticals Incorporated; Boulder, Colorado. ClinicalTrials.gov. Bethesda (MD): national Library of Medicine (US); 2000. [2014 May 09]. A Phase II, Open-Label, Multicenter Trial of Angiozyme in Subjects With Metastatic Renal Cell Cancer. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT00021021?term=%22Angiozyme%22&rank=1NLM Identifier: NCT00021021. [Google Scholar]
- 214.Xiangya Hospital of Central South University; Changsha, China. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. Phase I/II Study of EBV-LMP1 Targeted DNAzyme in Nasopharyngeal Carcinoma. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT01449942 NLM Identifier: NCT01449942. [Google Scholar]
- 215.MD Anderson Cancer Center; Houston, Texas. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. EphA2 Gene Targeting Using Neutral Liposomal Small Interfering RNA Delivery (IND# 72924): A Phase I Clinical Trial. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT01591356?term=%22sirna%22+AND+%22Cancer%22&rank=1 NLM Identifier: NCT01591356. [Google Scholar]
- 216.Scott Pruitt, Duke University Medical Center at Durham, North Carolina. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. Phase I Study of Active Immunotherapy of Metastatic Melanoma With Mature Autologous Dendritic Cells Transfected With Tumor Antigen RNA and Small Inhibitory RNAs to Alter Proteasomal Antigen Processing. [Internet]. Available from: http://www.clinicaltrials.gov/ct2/show/NCT00672542 NLM Identifier: NCT00672542. [Google Scholar]
- 217.Dicerna Pharmaceuticals, Inc.; Watertown, Massachusetts. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. Phase I, Multicenter, Cohort Dose Escalation Trial to Determine the Safety, Tolerance, and Maximum Tolerated Dose of DCR-MYC, a Lipid Nanoparticle (LNP)-Formulated Small Inhibitory RNA (siRNA) Oligonucleotide Targeting MYC, in Patients With Refractory Locally Advanced or Metastatic Solid Tumor Malignancies, Multiple Myeloma, or Lymphoma. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT02110563 NLM Identifier: NCT02110563. [Google Scholar]
- 218.Hadassah Medical Center; Kiryat, Hadassah. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. Use of SV40 siRNA Vectors to Treat CML. [Internet]. Available from: http://clinicaltrials.gov/show/NCT00257647 NML Identifier: NCT00257647. [Google Scholar]
- 219.Mirna Therapeutics, Inc.; Austin, Texas. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. A Multicenter Phase I Study of MRX34, MicroRNA miR-RX34 Liposome Injectable Suspension. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT01829971?term=MRX34&rank=1 NLM Identifier: NCT01829971. [Google Scholar]
- 220.Santaris Pharma A/S; Hørsholm, Denmark. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. A Phase 2a, Randomized, Double-Blind, Placebo-Controlled, Ascending Multiple-Dose Study to Assess Safety, Tolerability, Pharmacokinetics and Antiviral Activity of SPC3649 (Miravirsen) Administered to Treatment-Naïve Subjects With Chronic Hepatitis C (CHC) Infection. [Internet]. Available from: https://clinicaltrials.gov/ct2/show/NCT01200420?term=miravirsen&rank=3 NLM Identifier: NCT01200420. [Google Scholar]
- 221.Enzon Pharmaceuticals, Inc.; Piscataway, New Jersey. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. A Phase 1, Open-Label, Dose Escalation Study Evaluating the Safety and Tolerability of EZN-2968, a Locked Nucleic Acid Antisense Oligonucleotide Against Hypoxia-Inducible Factor-1α, Administered as a Weekly 2-Hour Intravenous Infusion in Adult Patients With Advanced Solid Tumors or Lymphoma. [Internet]. Available from: http://clinicaltrials.gov/ct2/show/NCT00466583 NLM Identifier: NCT00466583. [Google Scholar]
- 222.Santaris Pharma A/S; Hørsholm, Denmark. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. An Open-labelled, International, Multicenter, Dose Escalating, Phase I/II Study of SPC2996, an LNA Antisense Molecule Against Bcl-2, in Patients With Relapsed or Refractory Chronic Lymphocytic Leukaemia. [Internet]. Available from: http://clinicaltrial.gov/ct2/show/NCT00285103?term=%22SPC2996%22&rank=1NLM Identifier: NCT00285103. [Google Scholar]
- 223.Enzon Pharmaceuticals, Inc.; Piscataway, New Jersey. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); 2000. [2014 May 09]. T2009-007: A Phase I Study Evaluating the Safety, Tolerability and Biological Activity of EZN-3042, a Survivin mRNA Antagonist, Administered With Re-induction Chemotherapy in Children With Relapsed Acute Lymphoblastic Leukemia (ALL) [IND 108753] [Internet]. Available from: http://clinicaltrial.gov/ct2/show/NCT01186328?term=%22EZN3042%22&rank=1 NML Identifier: NCT01186328. [Google Scholar]


