Abstract
Human APOBEC3G (huAPOBEC3G), also known as CEM15, is a broad antiretroviral host factor that deaminates dC to dU in the minus strand DNA of human immunodeficiency virus type 1 (HIV-1), other lentiviruses, and murine leukemia virus (MLV), thereby creating G-to-A hypermutation in the plus strand DNA to inhibit the infectivity of these viruses. In this study, we examined the antiretroviral function of a murine homologue of APOBEC3G (muAPOBEC3G) on several retrovirus systems with different producer cells. MuAPOBEC3G did not suppress the infectivity of murine retroviral vectors produced from human or murine cells, whereas it showed antiviral activity on both wild-type and Δvif virions of HIV-1 in human cells. In contrast, huAPOBEC3G showed broad antiviral activity on HIV-1 and murine retroviral vectors produced from human cells as well as murine cells. These data suggested that muAPOBEC3G does not possess antiretroviral activity on murine retroviruses and has a different target specificity from that of huAPOBEC3G and that huAPOBEC3G works as a broad antiviral factor not only in human cells but also in murine cells. A functional interaction study between human and murine APOBEC3G supported the former hypothesis. Furthermore, studies on the expression of APOBEC3G in producer cells and its incorporation into virions revealed that muAPOBEC3G is incorporated into HIV-1 virions but not into MLV virions. Thus, muAPOBEC3G cannot suppress the infectivity of murine retrovirus because it is not incorporated into virions. We suggest that murine retroviruses can replicate in murine target cells expressing muAPOBEC3G because they are not targets for this enzyme.
The human immunodeficiency virus type 1 (HIV-1) Vif protein plays a crucial role during the viral life cycle by regulating virion infectivity (27, 28) and in vivo pathogenesis (5). Vif counteracts an anti-HIV-1 cellular factor in nonpermissive cells (16, 26), referred to as APOBEC3G, thereby enhancing virion infectivity (23).
APOBEC3G is a member of the Apobec superfamily, which shares a cytidine deaminase motif and is homologous to Apobec-1 (2, 12, 22, 31), the prototypic protein of this family. Several recent studies revealed that human APOBEC3G (huAPOBEC3G) regulates HIV-1 infectivity by deaminating dC to dU in the newly synthesized minus strand DNA, resulting in G-to-A hypermutation of the viral plus strand DNA (15, 17, 25, 33). They also revealed that huAPOBEC3G has broad antiretroviral activity on other lentiviruses and murine leukemia virus (MLV) with the same mechanism (9, 17). We and others also showed that the enzymatic activity of huAPOBEC3G is essential for the antiviral activity in mutagenesis studies (17, 25, 33). Furthermore, subsequent reports showed that Vif inhibits the incorporation of huAPOBEC3G into virions, thereby facilitating the infectivity of HIV-1 (13, 18, 19, 24, 29, 32). However, the mechanism responsible for this remains controversial. Some groups suggested that Vif induces the ubiquitin-dependent degradation of huAPOBEC3G (19, 24, 32), while others reported that it affects the translation of this protein (13, 18, 29).
In this study, we examined the antiviral function of the murine homologue of APOBEC3G in several retrovirus systems with different producer cells to elucidate more detail on the mechanisms responsible for APOBEC3G regulation of virion infectivity in a variety of retroviruses.
MATERIALS AND METHODS
Expression vectors and molecular clones.
HuAPOBEC3G cDNA was amplified from H9 cDNA by reverse transcription-PCR and cloned into pDON/EGFP and pcDNA3/hygro/HA vectors (Invitrogen, Carlsbad, Calif.) for the expression of enhanced green fluorescent protein (EGFP)-fused and hemagglutinin (HA)-tagged huAPOBEC3G (EGFP- and HA-huAPOBEC3G), respectively. Full-length murine APOBEC3G cDNA (longer form, designated muAPOBEC3Gwt) and spliced cDNA (shorter form, designated muΔexon5 as described in reference 18) were also amplified from murine bone marrow and NIH 3T3 cDNA by reverse transcription-PCR and cloned into the vectors described above for the expression of EGFP-/HA-muAPOBEC3Gwt and EGFP-/HA-muΔexon5, respectively. Firefly luciferase was cloned into murine retrovirus vectors pDON-AI (14) (Takara Bio Inc., Otsu, Japan) and pMSCV/neo (7) (BD Biosciences Clontech, Franklin Lakes, N.J.) to generate pDON/Luc and pMSCV/Luc, respectively. Expression vectors for point mutants (human E259Q and murine E290Q) and human-murine chimera mutants were generated by PCR. pNL43-Luc and pNL43/Δvif-Luc were constructed as previously described (1, 11).
Cell lines.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal calf serum, penicillin, streptomycin, and glutamine (Invitrogen). The retroviral packaging cell lines GP293 (4) and PT67 (21) were purchased from BD Biosciences Clontech and maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, penicillin, streptomycin, and glutamine. M8166 cells were maintained in RPMI 1640 (Sigma-Aldrich Corp., St. Louis, Mo.) containing 10% fetal calf serum, penicillin, streptomycin, and glutamine.
Preparation of HIV-1 and infectivity assay.
Luciferase reporter viruses with or without Vif were prepared in HEK293T cells by cotransfection of pNL43-Luc (wild type) or pNL43/Δvif-Luc (Δvif) together with a mock vector or expression vectors for huAPOBEC3G, muAPOBEC3Gwt, muΔexon5, or their mutants by the calcium phosphate method as previously described (25). Viruses in the supernatant were collected after 48 h of transfection, and virus titers were measured with an enzyme-linked immunosorbent assay kit against p24 antigen (RETRO-TEK; ZeptoMetrix Corporation, Buffalo, N.Y.). An adjusted amount of virus was used to challenge M8166 target cells. Twenty hours postinfection, the cells were lysed in passive lysis buffer (Promega, Madison, Wis.), and luciferase activity was measured with a luminometer (EG & G Berthold, Bad Wildbad, Germany). Values are presented as the percent infectivity relative to the value of the wild-type virus without the expression of APOBEC3G.
Preparation of murine retroviral vectors and infectivity assay.
Murine retroviral reporter viruses produced from human cells were prepared by cotransfection of pDON/Luc, pMSCV/Luc, or pDON/EGFP with pVSV-G into GP293 cells by the calcium phosphate method as previously described (4). An infectivity assay was performed as described above for HIV-1 except that virus titers were measured with a reverse transcriptase assay kit (F. Hoffmann-La Roche Ltd., Diagnostics Division, Basel, Switzerland), and the target cells were HEK293T cells. Values are presented as the percent infectivity relative to the value of murine retrovirus without the expression of APOBEC3G. Murine retroviral reporter viruses produced from murine cells were prepared by transfection of pDON/Luc or pDON/EGFP with pcDNA3/hygro/HA, pcDNA3/hygro/HA-huAPOBEC3G, or pcDNA3/hygro/HA-muAPOBEC3Gwt into PT67 cells by the calcium phosphate method. An infectivity assay was performed as described above 2 days after transfection or 2 weeks after selection with 1 mg of G418 (Nacalai Tesque, Inc., Kyoto, Japan) per ml and 200 μg of hygromycin B (Invitrogen) per ml.
Detection of hypermutation in viral DNA.
Hypermutation in HIV-1 DNA was detected with endogenous reverse transcription as previously described (15, 25). In brief, after being treated with DNase I, viral stocks were incubated at 39°C for 120 min. DNA was purified and amplified with the following primer pairs: op-6.4(CCATGCTCCTTGGGATATTG) and op-29.10(CCTCCTGAGGATTGCTTAAA). The PCR products were cloned into pT7-Blue (Novagen), and the inserts of individual clones were sequenced. Hypermutation in HIV-1 DNA was also detected by sequencing Env DNA integrated into target cells as previously described (15, 17, 33). In brief, DNA was purified from target cells infected with HIV-1 (wild type or Δvif) prepared with huAPOBEC3G or muAPOBEC3Gwt. The Env region was amplified and sequenced as described above. Hypermutation in murine retroviral DNA was detected by sequencing EGFP DNA integrated into target cells as previously described (9). DNA was purified from target cells infected with pDON/EGFP retroviral vectors prepared with or without APOBEC3G. The entire EGFP coding sequence was amplified and sequenced as described above.
Coimmunoprecipitation assay.
To observe protein-protein interactions in vivo, we performed an immunoprecipitation assay as previously described (30). In brief, expression vectors for HA-huAPOBEC3G or HA-muAPOBEC3Gwt were cotransfected with pDON/EGFP, pDON/EGFP-huAPOBEC3G, or pDON/EGFP-muAPOBEC3Gwt into HEK293T cells by the calcium phosphate method. Two days after transfection, the cells were lysed with lysing buffer. Cell lysates were immunoprecipitated with anti-EGFP monoclonal antibody (kindly provided by A. Imura, Kyoto University) and protein G-Sepharose beads and subjected to immunoblotting with anti-HA tag monoclonal antibody (12CA5) (F. Hoffmann-La Roche Ltd.). HA-APOBEC3G was visualized with an ECL detection system (Amersham Biosciences Corp., Piscataway, N.J.).
Protein expression of APOBEC3G in producer cells and its incorporation into virions.
To detect the expression of APOBEC3G protein in producer cells, whole-cell lysates of producer cells were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti-β-actin antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) and anti-HA monoclonal antibody (12CA5). To detect the incorporation of APOBEC3G into HIV-1 and MLV virions, adjusted amounts of the virus supernatants were ultracentrifuged with a Beckman TL-100s ultracentrifuge at 60,000 × g for 10 min. The virion pellets were boiled in SDS sample buffer and subjected to SDS-PAGE and immunoblotting with anti-p24 monoclonal antibody (ZeptoMetrix Corporation) and anti-HA monoclonal antibody (12CA5).
RESULTS
Expression of muAPOBEC3Gwt inhibited the infectivity of wild-type as well as Δvif virions of HIV-1.
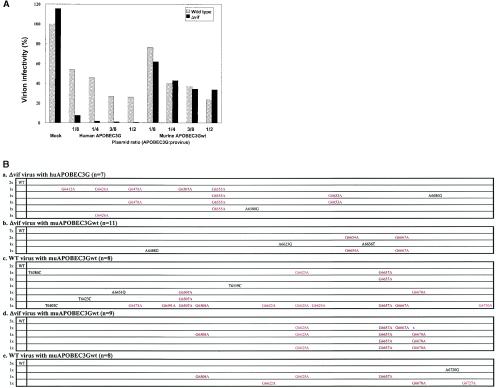
First, we transfected expression vectors for huAPOBEC3G or muAPOBEC3Gwt with pNL43/Luc (wild type) or pNL43/Δvif-Luc (Δvif) into HEK293T cells, prepared viruses, and tested their infectivity. As shown in Fig. 1A, the expression of muAPOBEC3Gwt in HEK293T cells suppressed the infectivity of wild-type as well as Δvif virions to the same extent in a dose-dependent manner, while huAPOBEC3G had a more potent antiviral effect on Δvif virions than wild-type virions, as reported previously (18). A point mutant such as muE290Q lost this suppressive effect on either wild-type or Δvif virions (data not shown), suggesting that the antiviral effect of muAPOBEC3Gwt on wild-type and Δvif virions is specific and dependent upon its enzymatic activity.
FIG. 1.
Effect of muAPOBEC3Gwt on the infectivity of HIV-1 virions. We transfected pNL43-Luc (wild type) or pNL43/Δvif-Luc (Δvif) with pDON-based vectors (increasing amounts of huAPOBEC3G or muAPOBEC3Gwt, with the empty parental vector making up the balance) into HEK293T cells. Viruses from these cells were used to challenge M8166 cells, and productive infection was measured by luciferase activity. Values are presented as the percent infectivity relative to the wild-type virus not expressing APOBEC3G. (A) Expression of muAPOBEC3Gwt suppressed the infectivity of wild-type as well as Δvif virions to the same extent, while huAPOBEC3G showed a more potent antiviral effect on Δvif virions than wild-type virions. (B) muAPOBEC3Gwt introduced G-to-A hypermutation in the viral DNA of wild-type as well as Δvif virus. A DNA sequence analysis of the env region was performed with DNA synthesized in endogenous reverse transcription reactions (a, b, and c) or DNA amplified from target cells (d and e). G-to-A mutations are shown in red, with nucleic acid numbers corresponding to the pNL43 sequence, while other mutations are denoted in black. The numbers before the sequence indicate the number of each clone, while those in parentheses indicate the total number of clones sequenced. WT indicates no mutations in this region. (a) Δvif virions with huAPOBEC3G. (b and d) Δvif virions with muAPOBEC3Gwt. (c and e) Wild-type virions with muAPOBEC3Gwt.
To confirm the antiviral function of muAPOBEC3Gwt on HIV-1, we next examined the occurrence of hypermutation in the viral DNA of HIV-1 induced by muAPOBEC3Gwt. HuAPOBEC3G introduced more G-to-A hypermutation in the viral DNA of Δvif virus than that of wild-type virus, as reported previously (17, 25, 33) (Fig. 1B, a and data not shown). In contrast, muAPOBEC3Gwt introduced G-to-A hypermutation in the viral DNA of both wild-type (Fig. 1B, c and e) and Δvif virus (Fig. 1B, b and d) to a similar extent, indicating that muAPOBEC3Gwt possesses DNA editing as well as antiviral activity on both wild-type and Δvif virus. Interestingly, preferential sites of G-to-A hypermutation in viral DNA induced by muAPOBEC3Gwt were different from those induced by huAPOBEC3G, suggesting that muAPOBEC3Gwt targets different DNA sequences from huAPOBEC3G even for the same virus target.
MuAPOBEC3Gwt did not suppress the infectivity of murine retroviral vectors produced from human cells.
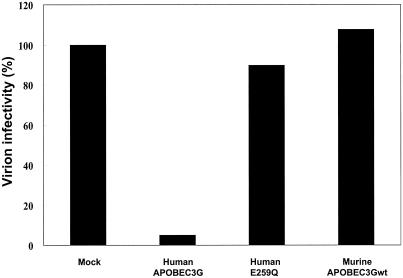
Recent reports revealed that huAPOBEC3G also possesses broad antiviral activity on other lentiviruses and MLV-based vectors (9, 17). Therefore, we tested the antiviral activity of muAPOBEC3Gwt on several murine retroviral vectors. However, as shown in Fig. 2, muAPOBEC3Gwt did not suppress the infectivity of the MLV-based luciferase reporter vector pDON/Luc while huAPOBEC3G showed antiviral activity, as reported previously (9, 17). A point mutant of huAPOBEC3G such as huE259Q lost this antiretroviral activity, suggesting that the effect was specific for huAPOBEC3G. We also obtained similar results for pDON/EGFP and another murine retroviral vector, pMSCV/Luc (data not shown). These data indicate that huAPOBEC3G has broad antiretroviral activity on several murine retroviruses and that muAPOBEC3Gwt does not. A sequencing analysis of integrated EGFP DNA in target cells revealed that huAPOBEC3G induced G-to-A hypermutation in the EGFP sequence and that muAPOBEC3Gwt did not (data not shown), confirming that muAPOBEC3Gwt cannot function on murine retroviral vectors produced from human cells.
FIG. 2.
MuAPOBEC3Gwt did not suppress the infectivity of MLV-based vectors produced from human cells. We transfected pDON-Luc and pVSV-G with pcDNA3/HA-based vectors into GP293 cells. An adjusted amount of virus as determined by reverse transcriptase values was used to challenge HEK293T cells, and productive infection was measured by luciferase activity. Values are presented as the percent infectivity relative to the value with MLV not expressing APOBEC3G.
MuAPOBEC3Gwt acted synergistically with huAPOBEC3G on the infectivity of HIV-1 virions but not of MLV virions.
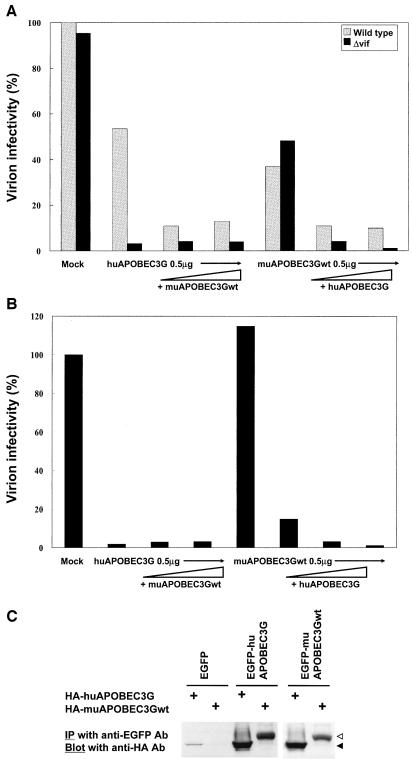
Since our data suggested species-specific target specificity, we next examined the functional interaction between muAPOBEC3Gwt and huAPOBEC3G on HIV-1 or MLV. The expression of increasing amounts of huAPOBEC3G with a fixed amount of muAPOBEC3Gwt showed an additive effect on the infectivity of HIV-1 (both wild-type and Δvif) virions, while the addition of increasing amounts of muAPOBEC3Gwt to a fixed amount of huAPOBEC3G showed an additive effect only on wild-type virions (Fig. 3A). In contrast, the expression of increasing amounts of huAPOBEC3G showed an additive effect on the infectivity of MLV virions, while the addition of increasing amounts of muAPOBEC3Gwt on huAPOBEC3G did not (Fig. 3B). These data indicate that muAPOBEC3Gwt worked synergistically with huAPOBEC3G on the infectivity of HIV-1 virions but not on MLV virions. It suggests that MLV is not a target for muAPOBEC3Gwt. A coimmunoprecipitation assay revealed the physical interaction between muAPOBEC3Gwt and huAPOBEC3G in vivo (Fig. 3C), supporting the hypothesis of a functional interaction between these molecules.
FIG. 3.
MuAPOBEC3Gwt worked synergistically with huAPOBEC3G on the infectivity of HIV-1 virions but not on MLV virions. (A) We transfected pNL43-Luc (wild type) or pNL43/Δvif-Luc (Δvif) with a combination of expression vectors for APOBEC3G (increasing amounts of muAPOBEC3Gwt or huAPOBEC3G with a fixed amount of huAPOBEC3G or muAPOBEC3Gwt, respectively) into HEK293T cells. An infectivity assay was performed, and values are presented as described in the legend to Fig. 1. (B) We transfected pDON/Luc with a combination of expression vectors for APOBEC3G as described in A into GP293 cells. An infectivity assay was carried out, and values are presented as described in the legend to Fig. 2A. (C) A coimmunoprecipitation assay revealed the physical interaction between huAPOBEC3G and muAPOBEC3Gwt. We transfected pcDNA3/HA-based vectors with pDON/EGFP-based vectors. Cell lysates were immunoprecipitated (IP) with anti-EGFP monoclonal antibody (Ab) and subjected to immunoblotting with anti-HA monoclonal antibody. Solid arrow, huAPOBEC3G; open arrow, muAPOBEC3Gwt.
HuAPOBEC3G suppressed the infectivity of MLV-based vectors produced from murine cells, but muAPOBEC3Gwt did not.
We also suspected that the inability of muAPOBEC3Gwt to suppress the infectivity of MLV was due to cell type specificity and therefore tested the antiviral function of muAPOBEC3Gwt in murine producer cells. We prepared MLV-based vectors from the murine packaging cell line PT67 with huAPOBEC3G or muAPOBEC3Gwt and tested their infectivity. MuAPOBEC3Gwt did not suppress the infectivity of MLV-based vectors produced from murine cells, while huAPOBEC3G showed similar antiviral activity even in murine cells (Fig. 4A), although protein expression of APOBEC3G was confirmed by Western blotting (Fig. 4B). A sequence analysis of EGFP DNA also revealed that the DNA editing activity of huAPOBEC3G was retained, while muAPOBEC3Gwt showed no activity (data not shown), again confirming this phenomenon. These data indicate that muAPOBEC3Gwt could not inhibit the infectivity of murine retroviruses regardless of virus-producing cell type, while huAPOBEC3G showed broad antiretroviral activity not only in human cells but also in murine cells. This suggests that muAPOBEC3Gwt has a different target specificity from that of huAPOBEC3G and that huAPOBEC3G works as an antiviral factor independent of producer cell type.
FIG. 4.
HuAPOBEC3G suppressed the infectivity of MLV-based vector produced from murine cells, but muAPOBEC3Gwt did not. (A) We transfected pDON-Luc with pcDNA3/HA-based vectors into PT67 cells. An infectivity assay was performed 2 days after transfection or 2 weeks after selection with G418 and hygromycin B, and values are presented as described in the legend to Fig. 2A. (B) The protein expression of APOBEC3G in PT67 cells was confirmed by Western blot analysis with anti-HA monoclonal antibody. Lane 1 is a positive control, and lanes 2 to 6 correspond to the lanes in panel A. Solid arrow, huAPOBEC3G; open arrow, muAPOBEC3Gwt.
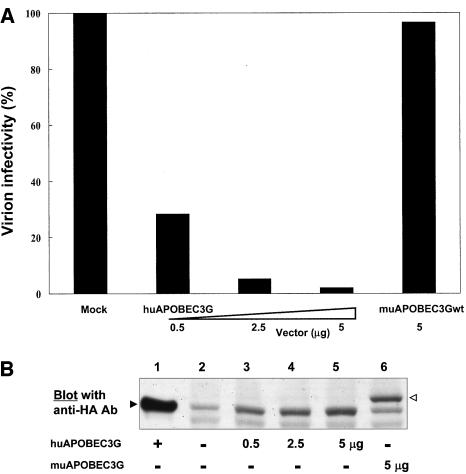
MuΔexon5 and muAPOBEC3Gwt showed similar antiviral effect.
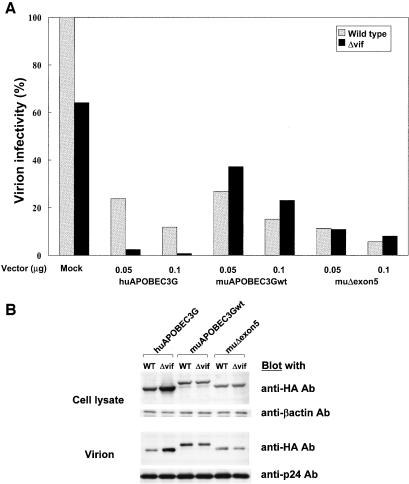
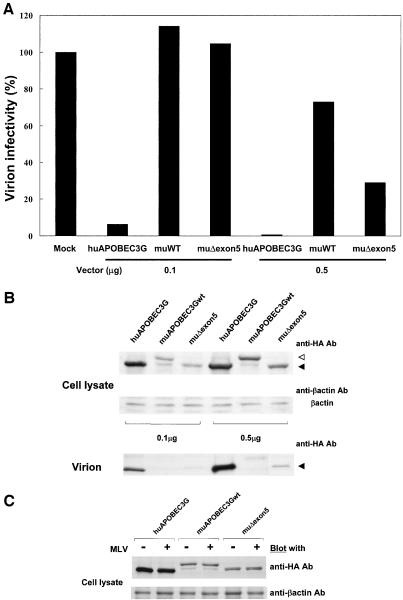
In the experiments described above, the full-length form of muAPOBEC3G (muAPOBEC3Gwt) was used. Because Mariani et al. reported that muΔexon5 possesses a more potent antiviral activity on HIV-1 (18), we next tested the function of muΔexon5 on HIV-1 and MLV to compare it with that of muAPOBEC3Gwt. As shown in Fig. 5A, muΔexon5 showed a slightly more potent activity on HIV-1 than muAPOBEC3Gwt, but the activity was comparable. Furthermore, the antiretroviral activity of muΔexon5 on MLV was also comparable to that of muAPOBEC3Gwt (Fig. 6A), suggesting that muΔexon5 possesses a similar antiviral effect on both HIV-1 and MLV compared with muAPOBEC3Gwt.
FIG. 5.
MuΔexon5 and muAPOBEC3Gwt showed similar antiviral effects. (A) We transfected pNL43-Luc (wild type) or pNL43/Δvif-Luc (Δvif) with pcDNA/HA-based vectors into HEK293T cells with the indicated amount of plasmid. An infectivity assay was carried out, and values are presented as described in the legend to Fig. 1. MuΔexon5 and muAPOBEC3Gwt showed similar antiviral effects. (B) Protein expression of APOBEC3G in producer cells (upper column) and incorporation of APOBEC3G into virions (lower column) were detected by Western blot analysis with anti-HA monoclonal antibody. The expression of β-actin and the incorporation of p24 were used as internal controls for protein expression in the cell lysate and protein incorporation into virions, respectively. The expression of HIV-1 Vif protein reduced the protein expression of huAPOBEC3G in producer cells as well as the incorporation of huAPOBEC3G into HIV-1 virions, while it did not affect the protein expression or incorporation of muAPOBEC3Gwt and muΔexon5 into virions.
FIG. 6.
MuAPOBEC3G (muAPOBEC3Gwt or muΔexon5) did not show antiviral effects on MLV because it was not incorporated into MLV virions. We transfected pDON-Luc and pVSV-G with the indicated amount of pcDNA3/HA-based vectors into GP293 cells. An infectivity assay was performed, and values are presented as described in the legend to Fig. 2A. (A) HuAPOBEC3G suppressed the infectivity of MLV, but muAPOBEC3G did not. (B) Protein expression of APOBEC3G in producer cells (upper column) and the incorporation of APOBEC3G into virions (lower column) were detected by Western blot analysis with anti-HA monoclonal antibody. MuAPOBEC3G was not incorporated into MLV virions, although it was expressed in producer cells at a level comparable to huAPOBEC3G. Solid arrow, huAPOBEC3G or muΔexon5; open arrow, muAPOBEC3Gwt. (C) The expression of muAPOBEC3G was not impaired by the expression of MLV.
MuAPOBEC3G was incorporated into HIV-1 virions but not into MLV virions.
Since several recent studies showed that huAPOBEC3G was not incorporated into HIV-1 virions in the presence of Vif protein, we tested the incorporation of muAPOBEC3G into HIV-1 and MLV virions. The upper column in Fig. 5B shows that the expression of Vif protein reduced the expression level of huAPOBEC3G in producer cells but not that of muAPOBEC3Gwt or muΔexon5. The lower column shows that the incorporation of huAPOBEC3G into HIV-1 virions was impaired in the presence of Vif, while muAPOBEC3Gwt and muΔexon5 showed similar levels of incorporation into HIV-1 virions. On the other hand, muAPOBEC3Gwt or muΔexon5 was not incorporated into MLV virions as efficiently as huAPOBEC3G, although it was expressed in producer cells at a level comparable to that of huAPOBEC3G (Fig. 6B). Protein expression of muAPOBEC3G in producer cells was not impaired by the expression of MLV (Fig. 6C). These data suggest that muAPOBEC3G did not show antiretroviral activity on MLV because it was not incorporated into MLV virions.
DISCUSSION
In this study, we showed that the target virus specificity of APOBEC3G is different between species. MuAPOBEC3G did not suppress the infectivity of murine retroviruses produced from human cells or murine cells, whereas it showed antiviral activity on both wild-type and Δvif virions of HIV-1 in human cells. In contrast, huAPOBEC3G showed broad antiviral activity on HIV-1 Δvif and murine retroviral vectors produced from human cells as well as murine cells. Our data suggest several important findings. First, muAPOBEC3G does not show antiretroviral activity on murine retroviruses and has a different target virus specificity than huAPOBEC3G. Studies on the functional interaction between huAPOBEC3G and muAPOBEC3Gwt support this finding regarding the target specificity of APOBEC3G. This is the first description of the target virus specificity of APOBEC3G, although the antiviral activity of APOBEC3G on MLV is still controversial.
Harris et al. and Mangeat et al. first reported that huAPOBEC3G possesses antiretroviral activity on MLV by deaminating dC (9, 17), while Mariani et al. showed that neither huAPOBEC3G nor muAPOBEC3Gwt possesses antiretroviral activity on MLV (18). We showed that huAPOBEC3G introduces G-to-A hypermutation to inhibit the infectivity of several murine retroviral vectors in human cells as well as murine cells, but muAPOBEC3G does not. The reason for this discrepancy is unknown. It might be due to differences in the ratio of APOBEC3G to virus vector in each study. We tried to extend this study to examine the antiretroviral function of APOBEC3G on HIV-1 produced from murine cells with the expression of cyclin T1 but could not obtain enough virus to test for infectivity.
Our findings may help to answer a question raised by previous reports regarding how retroviruses without Vif manage to replicate in the presence of this antiviral enzyme (6, 8, 10); that is, murine retrovirus replicates in murine target cells expressing APOBEC3G because it is not a target for murine homologue of this enzyme. Second, our data also indicate that huAPOBEC3G works as a broad antiretroviral factor not only in human cells but also in murine cells. This suggests that huAPOBEC3G might not need human cofactors such as another cytidine deaminase, Apobec-1, which requires the cofactor Apobec-1 complementary factor to act as an RNA-editing enzyme (3, 20). Another possibility is that huAPOBEC3G might share murine cofactors that enable it to function in murine cells. Further studies are ongoing to answer this question. Third, the G-to-A hypermutation profile in HIV-1 induced by muAPOBEC3G was different from that induced by huAPOBEC3G, suggesting target DNA sequence specificity even for the same virus target. This finding differs from a report by Mariani et al. in which mutation profiles with an HIV-1 substrate were similar to each other. This discrepancy might come from differences in the fragments sequenced. We sequenced a fragment of the Env region to detect mutations, while they used a long terminal repeat region. Further study is necessary to clarify this discrepancy.
Interestingly, the hypermutation profiles in HIV-1 induced by muAPOBEC3G also differed for samples prepared in the presence or absence of Vif protein only in an assay with endogenous reverse transcriptase reaction (Fig. 1B, b and c). This might suggest another function of Vif protein on APOBEC3G other than ubiquitin-dependent degradation (19, 24, 32) and impaired synthesis of APOBEC3G (13, 29), as described below. Vif protein might directly affect the deaminating process of the viral DNA by APOBEC3G during reverse transcription, although it cannot exclude muAPOBEC3G from HIV-1 virions. Since this difference is only seen in an endogenous reverse transcription reaction assay, this might be due to an artificial phenomenon induced by the endogenous reverse transcription reaction assay. Further study is also necessary to elucidate this novel function of Vif protein.
In this study, we examined the function of the full-length form of muAPOBEC3G (muAPOBEC3Gwt). Since Mariani et al. showed that the spliced form of muAPOBEC3G (muΔexon5) has more potent activity (18), we compared the function of this isoform with that of muAPOBEC3Gwt. Our results indicate that both forms possess similar antiviral activity on HIV-1 and MLV, although muΔexon5 is more active, as reported previously (18). The reason why the mouse possesses these two different isoforms remains unclear.
The reason why muAPOBEC3G does not function on murine retroviral DNA and the basis for the target specificity of APOBEC3G were determined. Mariani et al. also showed that huAPOBEC3G and muAPOBEC3G are incorporated into Δvif virions of HIV-1 and that Vif protein bound to only huAPOBEC3G excludes this enzyme from wild-type virions (18). Furthermore, recent studies showed that HIV-1 Vif induces the ubiquitin-dependent degradation of APOBEC3G (19, 24, 32), while others suggest impaired protein synthesis of huAPOBEC3G in Vif-expressing cells (13, 29). In this study, we showed that muAPOBEC3G is not incorporated into MLV virions, while it is incorporated into HIV-1 virions even in the presence of Vif protein. On the other hand, huAPOBEC3G is incorporated into MLV virions. We could not detect differences in the expression of muAPOBEC3G between control cells and virus-producing cells, suggesting that nonincorporation of muAPOBEC3G into MLV virions is not due simply to the degradation of this protein in virus-producing cells, as shown with huAPOBEC3G in the presence of Vif protein. Furthermore, it is important to determine the site on huAPOBEC3G that is responsible for the functional interaction with Vif protein. However, a human-murine chimera study on APOBEC3G showed that all chimeric mutants lost antiviral activity on wild-type as well as Δvif virions (data not shown) and did not reveal the site responsible for this functional interaction with Vif protein. Human-murine chimeras might affect the protein structure itself, causing a loss in antiviral activity as well as enzymatic activity, as we reported earlier for deletion mutants of huAPOBEC3G (25).
Finally, it remains unclear why muAPOBEC3G is not incorporated into MLV virions. Elucidating this will yield more detailed mechanisms on how this enzyme is regulated by different retrovirus systems and allow us to develop novel therapeutic strategies against HIV-1 infection.
Acknowledgments
This study was partly supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
We are very grateful to Akihiro Imura for providing us with anti-EGFP monoclonal antibody. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institutes of Health: pNL4-3 from Malcom Martin and pNL4-3.Luc.E−R− from Nathaniel Landau.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anant, S., A. J. MacGinnitie, and N. O. Davidson. 1995. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J. Biol. Chem. 270:14762-14767. [PubMed] [Google Scholar]
- 3.Blanc, V., and N. O. Davidson. 2003. C-to-U RNA editing: mechanisms leading to genetic diversity. J. Biol. Chem. 278:1395-1398. [DOI] [PubMed] [Google Scholar]
- 4.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desrosiers, R. C., J. D. Lifson, J. S. Gibbs, S. C. Czajak, A. Y. Howe, L. O. Arthur, and R. P. Johnson. 1998. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 72:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff, S. P. 2003. Death by deamination: a novel host restriction system for HIV-1. Cell 114:281-283. [DOI] [PubMed] [Google Scholar]
- 7.Grez, M., E. Akgun, F. Hilberg, and W. Ostertag. 1990. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 87:9202-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu, Y., and W. I. Sundquist. 2003. Good to CU. Nature 424:21-22. [DOI] [PubMed] [Google Scholar]
- 9.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 10.Harris, R. S., A. M. Sheehy, H. M. Craig, M. H. Malim, and M. S. Neuberger. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immunol. 4:641-643. [DOI] [PubMed] [Google Scholar]
- 11.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 13.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, S. H., S. S. Yu, J. S. Park, P. D. Robbins, C. S. An, and S. Kim. 1998. Construction of retroviral vectors with improved safety, gene expression, and versatility. J. Virol. 72:994-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 16.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 18.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 19.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 20.Mehta, A., M. T. Kinter, N. E. Sherman, and D. M. Driscoll. 2000. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 20:1846-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, A. D., and F. Chen. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 70:5564-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navaratnam, N., J. R. Morrison, S. Bhattacharya, D. Patel, T. Funahashi, F. Giannoni, B. B. Teng, N. O. Davidson, and J. Scott. 1993. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 268:20709-20712. [PubMed] [Google Scholar]
- 23.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 25.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 26.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 27.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon, J. H., A. M. Sheehy, E. A. Carpenter, R. A. Fouchier, and M. H. Malim. 1999. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J. Virol. 73:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 30.Takaori-Kondo, A., T. Hori, K. Fukunaga, R. Morita, S. Kawamata, and T. Uchiyama. 2000. Both amino- and carboxyl-terminal domains of TRAF3 negatively regulate NF-kappaB activation induced by OX40 signaling. Biochem. Biophys. Res. Commun. 272:856-863. [DOI] [PubMed] [Google Scholar]
- 31.Teng, B., C. F. Burant, and N. O. Davidson. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260:1816-1819. [DOI] [PubMed] [Google Scholar]
- 32.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]