Abstract
By analysis of a single, variable, and short DNA sequence of 447 bp located within open reading frame 22 (ORF22), we discriminated three major varicella-zoster virus (VZV) genotypes. VZV isolates from all six inhabited continents that showed nearly complete homology to ORF22 of the European reference strain Dumas were assigned to the European (E) genotype. All Japanese isolates, defined as the Japanese (J) genotype, were identical in the respective genomic region and proved the most divergent from the E strains, carrying four distinct variations. The remaining isolates carried a combination of E- and J-specific variations in the target sequence and thus were collectively termed the mosaic (M) genotype. Three hundred twenty-six isolates collected in 27 countries were genotyped. A distinctive longitudinal distribution of VZV genotypes supports this approach. Among 111 isolates collected from European patients, 96.4% were genotype E. Consistent with this observation, approximately 80% of the VZV strains from the United States were also genotype E. Similarly, genotype E viruses were dominant in the Asian part of Russia and in eastern Australia. M genotype viruses were strongly dominant in tropical regions of Africa, Indochina, and Central America, and they were common in western Australia. However, genotype M viruses were also identified as a minority in several countries worldwide. Two major intertypic variations of genotype M strains were identified, suggesting that the M genotype can be further differentiated into subgenotypes. These data highlight the direction for future VZV genotyping efforts. This approach provides the first simple genotyping method for VZV strains in clinical samples.
Varicella-zoster virus (VZV) is a human herpesvirus that commonly causes chicken pox (varicella), usually in young children. Following primary infection a lifelong latent infection is established, and the virus often reactivates in adulthood or senescence to cause shingles (zoster). The VZV genome consists of 125 kb of linear, double-stranded DNA comprising one long and one short unique region, each flanked by inverted repeats (10), and five internal repeat regions (R1 to R5) have been identified. The VZV genome contains at least 71 open reading frames (ORFs), and the functions of many of the proteins they encode have been characterized (10).
During the past 2 decades, several groups attempted to evaluate VZV phylogeny. Early efforts in VZV typing used DNA restriction fragment length polymorphism (RFLP) analysis (13, 37, 38), an approach that confirmed the identity of the VZV strain that causes varicella on primary infection and later reactivates to cause zoster. Relatively consistent restriction enzyme digestion profiles for different VZV strains were observed, providing the first evidence that VZV has a highly conserved genome. Intrastrain variation in restriction enzyme fragment profiles among wild-type VZV isolates was observed (22, 36, 37). However, the most prominent differences were linked to variation in the number and composition of VZV genome repeat elements (23, 39, 40, 41). A number of RFLP strategies to distinguish VZV isolates from different geographic regions and to differentiate between the live-attenuated Oka vaccine strain and wild-type viruses have been proposed (1, 5, 23-28, 39, 41). It has since been determined that neither the repeat regions R1 to R5 nor the combination of ORF38 and ORF54 single-nucleotide polymorphisms (SNPs) reliably discriminate between VZV wild-type strains and the Oka vaccine strain (20, 30).
Nevertheless, two restriction enzyme cleavage sites corresponding to these SNPs have served as powerful reference points for characterizing both epidemiologic and geographic strain variation in VZV. Most VZV isolates in the United States contain a PstI restriction enzyme site in ORF38 that distinguishes them from a subset of Japanese strains, such as the Oka vaccine strain, that lack this restriction site (24). In addition, a C-to-T change at position 95241 leads to the elimination of a BglI restriction enzyme site in ORF54 that is found in all Japanese strains and that is absent in most U.S. VZV isolates (1, 28-31). As such, Japanese strains are either PstI+ BglI+ or PstI− BglI+, while most isolates in the United States are PstI+ BglI−. The latter genotype clearly predominates in eastern Australia, the United Kingdom, and the United States (21, 29, 36, 38). Strains bearing the Japanese PstI+ BglI+ genotype are also common in countries with a history of European colonization, and this is the predominant genotype in equatorial Africa, India, Bangladesh, China, and western Australia (29-31, 33-34). Furthermore, in eastern London, in which a large number of Indian and Bangladashi immigrants reside, the percentage of BglI+ VZV strains increased from 10% in the 1980s to more than 30% in the 1990s (21). At first, it was believed that the PstI− BglI+ (Oka vaccine strain) genotype was no longer circulating, but it has recently been identified among isolates from Japan, the western United States, and Hawaii (30, 31, 40). VZV strains with a PstI− BglI− profile have never been observed. In summary, RFLP analysis of VZV strains using the above approach identified two major groups. The first group includes strain Dumas and the typical European, North American, and Australian PstI+ BglI− isolates, whereas the second group harbors Asian (including Japanese), African, and western Australian PstI− BglI+ isolates.
RFLP analysis has also been performed using long-distance PCR products, facilitating the classification of 14 Japanese clinical specimens into nine subgroups (41). A separate study of VZV strains isolated from patients in Thailand used restriction endonuclease analysis with BglI, PstI, EcoRI, SmaI, and BamHI to show that VZV strains circulating in Thailand are distinct from those circulating in Japan. Sixteen different genotypes were evident, including those of strains with the BglI restriction site characteristic of Japanese (42) and African-Indian (30, 34) isolates. The high number of variants identified using this approach undoubtedly resulted from its reliance on genome repeat element copy number and the use of multiple endonucleases. Nevertheless, Japanese and Thai VZV strains displayed both common and distinctive markers. A milestone finding obtained by using antipeptide antibodies to discriminate Japanese and European strains was also reported by Kinchington and Turse (26).
RFLP studies of VZV mutability in serial culture have produced inconsistent observations. While some investigators have documented changes in VZV restriction profiles on serial passage (11, 12, 20, 45, 47), others failed to observe any differences after as many as 33 passages in cultured cells (20, 45). The serial passage of the Oka parental virus in tissue culture resulted in a well-documented accumulation of mostly single point mutations (20). The completion of the genomic sequencing for VZV Oka vaccine and parental strains (20) has opened new horizons not only for the definitive differentiation of VZV vaccine strains from wild-type viruses but also for the genotypic analysis of wild-type VZV strains. The direct comparison of these data with the published genome sequence for the European Dumas strain has yielded important information. For example, the sequences that vary between the Oka vaccine and parental strains are concentrated in a single ORF, ORF62 (20). In addition, sequence polymorphisms were shown to be extremely rare among VZV wild-type strains and to be generally limited to individual point mutations distributed across the entire genome. Last, the differences between the Japanese strain (Oka) and the European strain (Dumas) provided a blueprint of mutations valuable for genotyping. Given these observations, it is not surprising that conventional RFLP methodology was ineffective at distinguishing these minor variations. Almost all of the sequence changes in the VZV Oka vaccine strain genome are SNPs, the detection of which requires the use of a series of specific restriction enzymes.
To date, three research groups, including our own, have advanced the development of a strategy for practical VZV genotyping (14, 33, 34, 44; V. N. Loparev and D. S. Schmid, Abstr. 26th Int. Herpesvirus Workshop, abstr. 3.15, 2001). As a first step toward a refined strain surveillance methodology, we evaluated VZV genome variability to establish a basis for the stable classification of wild-type strains into major viral genotypes. Genotype-based classification schemes are feasible even in the absence of distinct phenotypes, as has been amply demonstrated for other human herpesviruses (1, 4, 7, 9, 32, 43, 46). Nevertheless, careful genotypic analysis could lead to the identification of virulence factors and hence to an improved understanding of VZV biology (35). Another important issue that may be addressed using this approach is the assessment of the direction and predisposition of virus phylogeny in VZV-vaccinated and unvaccinated populations. We report here the development of a novel test for the simple typing of wild-type strains of VZV into three major circulating genotypes, which we have designated E (European), J (Japanese), and M (mosaic), using SNPs identified in a 447-bp region of ORF22. Altogether, 323 globally distributed VZV isolates obtained from recent clinical infection and representing currently circulating strains in Europe, Asia, Africa, Oceania, and the Americas could all be readily assigned to one of three genotypes. Furthermore, regional specificity for each genotype was evident from this analysis. These data shed light on evolutionary relationships among these major genotypes and their geographic clustering. We anticipate that these observations will help to predict future evolutionary patterns and to track the changes in VZV epidemiology that inevitably accompany active vaccination programs.
MATERIALS AND METHODS
VZV strains and isolates.
Forty-five low-passage isolates and five laboratory VZV strains were propagated in human lung fibroblast cells, MRC5 cells (Biologics Branch, Scientific Resources Program, Centers for Disease Control and Prevention [CDC], Atlanta, Ga.), or MeWo melanoma cells (gift from C. Grose, Iowa City, Iowa) by cocultivation of trypsinized uninfected cells and VZV-infected cells at a ratio of 1:20 to 1:120 for 5 days. Material from 267 clinical specimens of vesicular fluid air dried onto glass slides or cotton swabs and/or dried scabs was also collected for testing by CDC, general practitioners, and infectious disease physicians between 1976 and 2002. The isolates originated from a variety of geographic locations, including the United States, Canada, Mexico, Nicaragua, Chile, Democratic Republic of Congo (DRC), Chad, Morocco, Iceland, Germany, Czech Republic, Poland, the Russian Federation and other former Soviet republics, Jordan, Bangladesh, India, Nepal, China, Japan, and Australia. The VZV strains and isolates used in this study are listed in Table 1. VZV DNA from cells infected with Oka vaccine VZV and the laboratory strains Webster, Ellen, and VZV 11, all of which have long histories of tissue culture passage, were also examined. Low-passage Oka parental strain virus was also examined. In this study, only data from isolates obtained from individual cases of zoster or varicella were analyzed. Where clinical isolates were obtained from outbreak investigations, only a single specimen from each outbreak was included for analysis in this study.
TABLE 1.
VZV strains analyzed for this study
| Origin | No. of samples
|
No. of isolates
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Clinical | Tissue culture | From patients with
|
With site for:
|
Of genotype:
|
|||||
| Varicella | Zoster | PstI (ORF38) | BglI (ORF54) | E | M (M1, M2) | J | ||||
| USA | 52 | 27 | 25 | 44 | 8 | 49 | 14 | 38 | 11 (5, 6) | 3b |
| USA (Hawaii) | 3 | 3 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 3b |
| Canada | 50 | 50 | 0 | 18 | 32 | 50 | 12 | 38 | 10 (0,10) | 2 |
| Mexico | 5 | 5 | 0 | 5 | 0 | 5 | 5 | 0 | 5 (0, 5) | 0 |
| Chile | 2 | 2 | 0 | 2 | 0 | 2 | 1 | 1 | 1 (1, 0) | 0 |
| Nicaragua | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 (1, 0) | 0 |
| DRC | 19 | 19 | 0 | 19 | 0 | 19 | 19 | 0 | 19 (19, 0) | 0 |
| Chad | 5 | 5 | 0 | 5 | 0 | 5 | 5 | 0 | 5 (5, 0) | 0 |
| Morocco | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 (1, 0) | 0 |
| Germany | 21 | 12 | 9 | 21 | 0 | 21 | 0 | 21 | 0 | 0 |
| Iceland | 17 | 17 | 0 | 5 | 12 | 17 | 1 | 16 | 1 (0, 1) | 0 |
| Czech Republic | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Poland | 4 | 4 | 0 | 4 | 0 | 4 | 0 | 4 | 0 | 0 |
| Russia (Moscow) | 20 | 20 | 0 | 20 | 0 | 20 | 0 | 20 | 0 | 0 |
| Belarus | 5 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 0 |
| Ukraine | 6 | 6 | 0 | 6 | 0 | 6 | 0 | 6 | 0 | 0 |
| Lithuania | 5 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 0 |
| Latvia | 9 | 9 | 0 | 9 | 0 | 9 | 1 | 8 | 1 (0, 1) | 0 |
| Moldavia | 7 | 7 | 0 | 7 | 0 | 7 | 2 | 5 | 2 (2, 0) | 0 |
| Estonia | 3 | 3 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | 0 |
| Jordan | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Kazakhstan | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Russia (Novosibirsk) | 12 | 12 | 0 | 12 | 0 | 12 | 0 | 12 | 0 | 0 |
| Japan | 20 | 0 | 20 | 16 | 4 | 18 | 20 | 0 | 0 | 20 |
| Oka vaccine | 2 | 0 | 2a | 2 | 0 | 0 | 2 | 0 | 0 | 2 |
| Nepal | 5 | 5 | 0 | 5 | 0 | 5 | 5 | 0 | 5 (1, 4) | 0 |
| India | 16 | 16 | 0 | 16 | 0 | 16 | 16 | 0 | 16 (6, 10) | 0 |
| Bangladesh | 7 | 7 | 0 | 7 | 0 | 7 | 7 | 0 | 7 (7, 0) | 0 |
| South China | 3 | 3 | 0 | 3 | 0 | 3 | 3 | 0 | 3 (0, 3) | 0 |
| Western Australia | 10 | 10 | 0 | 10 | 0 | 10 | 7 | 3 | 7 (3, 4) | 0 |
| Eastern Australia | 10 | 10 | 0 | 10 | 0 | 10 | 0 | 10 | 0 | 0 |
| Lab strains | 3 | 0 | 3a | 3 | 0 | 3 | 0 | 3 | 0 | 0 |
| Total | 326 | 267 | 59 | 269 | 57 | 316 | 125 | 201 | 95 | 30 |
High-passage virus strain.
J genotype strains all displayed the PstI− BglI+ markers; all M strains were PstI+ BglI+; all E strains were PstI+ BglI−.
DNA isolation, primer sequences, and PCR and FRET probe hybridization.
DNA was purified from lysates of VZV-infected cells by using the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) or the genomic DNA purification kit (Promega, Madison, Wis.) according to the manufacturer's instructions. DNA from uninfected human lung fibroblast cells was used as a negative control in all experiments. Total DNA was isolated from vesicular fluid and scabs by use of NucleoSpin tissue kits (Clontech Laboratories Inc., Palo Alto, Calif.), which were used according to the manufacturer's instructions. DNA from individual lesions was recovered in a final volume of 200 μl of molecular-grade water or 10 mmol of Tris-HCl (pH 8.0)/liter.
All samples initially were characterized by the use of conventional PCR targeting VZV-specific sequences in ORF38, -54, and -62 in a 50-μl reaction mixture, followed by restriction enzyme analysis as previously described (28, 29) or by fluorescence resonance energy transfer (FRET) probe-based annealing and melting curve evaluation (30). A PCR/RFLP method for detecting SNPs in ORF38 and -54 was adapted to FRET-based real-time PCR, using the published primers for amplification (28). Ten-microliter aliquots of the resulting amplicon were mixed with 10 μl of LightCycler probes (CDC) in a mixture of 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate; Difco Laboratories, Detroit, Mich.), 1× bovine serum albumin (New England Biolabs, Beverly, Mass.), and 3 mM MgCl2 (Applied Biosystems, Foster City, Calif.), and the melting point for each probe was detected. The probes 5′-GGA CTT GAA GAT GAA CTT AAT GAA GCC CGT GA-3′ (anchor probe; positions 69393 to 69362) and 5′-ACG ATA TAT ACC GCA GTT GTT GCG GTA-3′ (sensor probe; positions 69360 to 69334) were used for sequence evaluation of the PstI site region, in which the melting curve peak differed from that of the PstI+ VZV strain Webster by 3°C. The probes 5′-CAT TTT GCA TAC ACT CAA CTA GGC TTG TGA-3′ (anchor probe; positions 95201 to 95230) and 5′-AAC CGC CGC TCC TCT GG-3′ (sensor probe; positions 95232 to 95248) were used for evaluating the BglI site region, with a 4°C difference between the melting curve peaks of the BglI+ VZV Oka vaccine strain and the BglI− VZV strain Webster. The five pairs of FRET probes for genotyping are depicted in Table 2. The anchor probes were 5′ labeled with LC640 dye and 3′ phosphorylated. The sensor probes were 3′ labeled with fluorescein, and the resulting data were used for preliminary genotyping (Fig. 1). All nucleotide positions for the sequence polymorphisms, primers, and genes indicated in this paper correspond to the reference Dumas strain sequence (GenBank accession number gi: 9625875). For all melting curve analyses with FRET probes, the LightCycler (Roche) real-time PCR instrument was used.
TABLE 2.
Probes used for genotyping
| Oligonucleotide | Probe typea | Sequence (5′-) | Peak melting temp (°C) for:
|
Difference (°C) | |
|---|---|---|---|---|---|
| Webster (E) | Oka (J) | ||||
| R1P1M1 | S | CGC GCC ATG TAT TTA ACG GG | 63.84 | 68.5 | 4.66 |
| R1P2M1 | A | AG AAC TGG CAC GGT TGC GAG ATA TGG | |||
| aR1P1M2 | A | GCC GTT ACG GCA TTA GAT ACT GTG | 60.11 | 69.0 | 8.89 |
| sR1P2M2 | S | TC GCC ACA ATC CAC ATA CC | |||
| R1P1M3 | S | AAT ATT CCA CCA CCT TTG G | 58.83 | 61.80 | 2.97 |
| R1P2M3 | A | TG TTA AGA GGG TTA ACA YGG TTT G | |||
| R1P1M4 | A | GTA TTC ACC GTT ATG TTT CCA GGT | 55.80 | 63.30 | 7.50 |
| R1P2M4 | S | TA GTA TTG AAG GAC TCC TTC | |||
| R1P1M-Mex | S | CTA CAG CCG TTA CGG CA | 69.98 | 69.98 | 3.00b |
| R1P2M-Mex | A | TA GAT ACT GTG TTT CGC CAC AAT CCA | |||
S, sensor probe; A, anchor probe.
A difference in annealing temperatures is seen only in selected Mexican M strain variants (annealing temperature for the Mexican sample was 66.98°C).
FIG. 1.
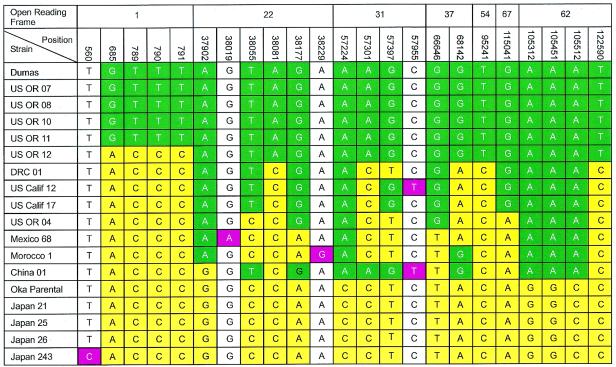
Analysis of genotypic variation using data from multiple VZV ORFs. The alignment of SNP in ORF1, -22, -31, -37, -54, -67, and -62 was performed for clinical isolates of VZV from around the world. RFLP of Pst+ Bgl−, Pst+ Bgl+, and Pst− Bgl− isolates was also performed. Numbers in the top rows indicate ORF and position of the SNPs in the genome, with the European VZV strain Dumas as a reference for the base pair numbers. The ORF54 SNP is the site that has been used in some protocols to discriminate the Oka vaccine strain from most wild-type strains. Green indicates mutations common to the E genotype reference strain; yellow indicates mutations in common with the J genotype Oka parental strain; red indicates strain-specific individual mutations; white indicates bases common to both the J and E reference strains.
PCR amplification of the ORF22 447-bp target region was performed by using 100 ng of total DNA or 5 μl of DNA extracted from skin lesions of chicken pox or zoster patients. Reactions with 100 pmol of each of the 20-mer primers and 2.5 U of Taq DNA polymerase (Applied Biosystems) were run under the following conditions: 95°C for 2 min, followed by 25 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 3 min and ending with a 10-min extension at 72°C. The PCR forward primer p22R1f (5′-GGG TTT TGT ATG AGC GTT GG-3′; positions 37837 to 37856) and the reverse primer p22R1r (5′-CCC CCG AGG TTC GTA ATA TC-3′; positions 38383 to 38356) were designed to amplify a 447-bp fragment (positions 37837 to 38264) representing amino acids 1252 to 1400 of the protein encoded by ORF22. DNA amplification reactions were performed with a GeneAmp PCR System 9700 (Applied Biosystems) in 50-μl reaction volumes, using AmpliTaq Gold PCR Master Mix (0.025 U of GoldTaq DNA polymerase enzyme, 1× PCR buffer II, 2.5 mM MgCl2, 200 μM [each] deoxynucleotide triphosphate [Applied Biosystems]), 0.2 μM (each) primer, and 1 to 5 μl of VZV DNA extract. Thermal cycling included an initial hot start at 96°C for 15 min for enzyme activation, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s and a final extension at 72°C for 4 min. PCR products were separated on 4 to 20% precast gradient polyacrylamide gels prestained with ethidium bromide (Invitrogen) and visualized under UV irradiation alongside a 100-bp ladder (Invitrogen).
For DNA purification, PCR products were electrophoresed on 2% precast agarose gels (BMA, Rockland, Maine); the 447-bp band was cut out, and DNA was extracted with a gel extraction kit (QIAGEN, Valencia, Calif.). To prepare amplicons for the sequencing procedure without gel purification, we used ExoSAP-IT reagent (USB Corporation, Cleveland, Ohio).
DNA sequencing and analysis.
Automated DNA sequencing was performed with the ABI PRISM 377 genetic analyzer (Perkin-Elmer, Foster City, Calif.). Sequencing reactions of gel-purified PCR products were performed by using the BigDye Terminator, version 3.0, cycle sequencing ready reaction mixture (Applied Biosystems) according to the manufacturer's instructions. Either the p22R1f or the p22R1r primer (3.2 pmol) and 10 ng of PCR product as a template were used. The GeneAmp 9700 thermal cycler (Applied Biosystems) was set to 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. The resulting products were purified on spin columns (Princeton Separations, Adelphia, N.J.), dried down, and resuspended in template suppression buffer (Applied Biosystems). Cycle sequencing reaction products were fractionated on an ABI 377 analyzer (Applied Biosystems). Primary DNA sequence assembly and analysis were performed with Sequencher (Gene Codes Corp., Ann Arbor, Mich.), and the sequence was compared with that of VZV strain Dumas.
Altogether for this study, 326 strains were used to amplify and sequence the 447-bp fragment, for a total 145,722 bp (Table 1), and about 10,000 bp was sequenced in various ORFs for 16 strains (160,000 bp), for a grand total of 305,722 bp worth of DNA sequence data used in these analyses. This is the equivalent of 2.446 complete VZV genomes. Targeted sequences included those of ORF1 (positions 915 to 592), ORF22 (positions 34083 to 42371), ORF31 (positions 57008 to 59611), ORF37 (positions 66074 to 68596), ORF54 (positions 95984 to 93678), ORF67 (positions 114496 to 115560), and ORF62 (positions 109133 to 105204).
The obtained DNA sequences were compared with the Dumas and Oka parental strain sequences (data not shown) in order to locate SNPs. The sample-derived sequences were aligned with the corresponding regions of the reference VZV strains by the PileUp program of the Wisconsin GCG Sequence Analysis Package, version 10 (Genetics Computer Group, Madison, Wis.) or MegAlign (DNAstar package; DNASTAR Incorporated) into pairwise and multiple sequence alignments of DNA with the Clustal V method. MegAlign was also used to create phylogenetic trees. The Clustal method groups sequences into clusters by examining sequence distances between all pairs. Clusters are aligned as pairs and then collectively as sequence groups to produce the overall alignment. After the multiple alignment is completed, a neighbor-joining method is employed to reconstruct phylogeny for the putative alignment. A neighbor-joining tree was calculated to illustrate the distribution of intergenotypic differences.
In a final short version of VZV genotyping we described VZV genotypes as lineages of VZV distinguished by differing in least two out of four reference mutations in the 447-bp region of ORF22.
RESULTS
Evaluation of ORF38 (PstI) and ORF54 (BglI) single-nucleotide polymorphisms among globally distributed VZV isolates.
Direct sequence analysis is the most accurate and informative approach to identifying SNPs that are useful for the classification of virus strains. We used this approach in tandem with PCR amplification of selected regions directly from clinical samples for these studies. On the basis of RFLP analysis data, the primary targets for sequencing were selected; the most divergent patterns were presumed to accurately reflect the level of strain divergence. We performed targeted ORF sequencing for a variety of globally distributed isolates, beginning with a comparison of the published PstI site in ORF38 and the BglI site in ORF54.
Preliminary analysis indicated that Pst+ BglI− viruses represent most (75 to 90%) strains collected in United States and Canada (Table 1). In a representative subset of 52 U.S. strains presented in Table 1, 86% were determined to have the PstI+ BglI− profile. This genotype was also the most commonly isolated genotype in Canada (76%), Europe (95%), and the Asian portion of the Russian Federation (100%). All specimens collected from eastern Australia were also PstI+ BglI− strains.
In contrast the PstI+ BglI+ genotype represented a minority of U.S. isolates (8%). As illustrated in Table 1, this genotype was common among isolates from India (100%), south China (100%), Bangladesh (100%), Nepal (100%), DRC (100%), and Chad (100%). Seventy percent of the strains collected in western Australia were also of this genotype.
We also confirmed earlier reports (28, 29) that most modern Japanese strains have the PstI+ BglI+ restriction enzyme site profile (100% of 20 isolates examined). Of interest, three wild-type VZV strains with the PstI− BglI+ profile of the Japanese Oka vaccine strain genotype were isolated in Hawaii in 2002. All PstI/BglI genotyping was performed in parallel, by both RFLP analysis and melting point analysis with FRET probes, with 100% observed agreement between the two methods. Given that 326 specimens of diverse geographic origins were evaluated, this study confirms the validity of using LightCycler-based FRET analysis to identify SNPs.
Targeted sequence analysis in multiple VZV ORFs for selected representative VZV isolates.
From the CDC repository of more than 400 VZV isolates obtained from countries all over the world, a subset of 16 VZV isolates representing all of the PstI/BglI genotypes (identified through RFLP analysis) were selected for extensive targeted sequence analysis. Direct comparison of newly acquired and previously reported VZV DNA sequence data (14, 20) documented very limited variation in genes encoding the envelope glycoproteins gB, gE, gI, gH, and gL and in ORF1, -4, -10, -22, -33, and -62. However, some common patterns became apparent and are displayed in Fig. 1.
Most VZV strains collected in the United States had the greatest overall similarity to the reference European strain Dumas. In a like manner, all Japanese strains exhibited maximum similarity to other Japanese isolates and differed the most from Dumas and most U.S. isolates. This was true for both Pst− and Pst+ Japanese strains.
A number of strains isolated from patients in various countries, particularly Africa and Indochina, carry a combination of Japanese (J) genotype-like and European (E) genotype-like mutations alternately across the entire genome. Thus, they feature a mosaic of J genotype-like and E genotype-like SNPs. This pattern was consistently observed for specific mutations analyzed in ORF22, -31, -37, and -62 (Fig. 1) and for 27 additional mutations in different VZV genomic loci (data not shown). ORF1 sequences were identical for Japanese and African-Chinese strains (Fig. 1) and, in addition, invariably shared the J-genotype-specific SNP (Bgl+) in ORF54. In contrast, the ORF62 sequence of those same African-Chinese strains had closest homology to E genotype isolates.
Several strain-specific mutations were also observed (positions 560, 38019, and 38229) and are presented in Fig. 1. Further analysis of VZV strains and additional targeted sequence analysis will be required to clarify how useful these more restricted markers will be for characterizing regional genotypic variation.
VZV genotyping using sequence variation in ORF22.
Genomic analysis of mutations located in seven VZV ORFs permitted the categorization of all of our clinical isolates into three distinct genotypes: E, J, and M (mosaic, combined type). Given that the quantity of DNA recovered from clinical isolates is often limited and that assessing such a large number of SNPs is laborious, we sought a simplified approach to performing the preliminary genotyping of VZV strains.
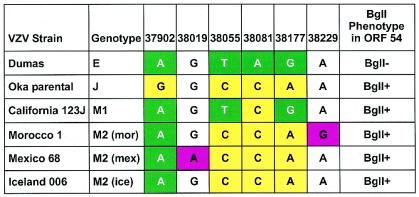
We identified four polymorphic positions in a short 447-bp fragment of ORF22 that could be used to robustly categorize circulating strains of VZV into one of the three genotypes. In a manner similar to the expanded-genotyping approach used in Fig. 1, a simplified protocol using a single ORF22 amplicon unambiguously segregated strains into the E, J, and M major genotypes (Fig. 2). All of the VZV isolates that appear in this study were analyzed by this method. Single representatives of every variation that we observed are listed in Fig. 2. Several variants of M genotype were also identified by this method. As illustrated in Fig. 2, the M genotype strain California123 contained more E type variations (three of four) than J type variations (one of four). By contrast, the Mexico_68, DR (Morocco), and Iceland_006 strains contained more J type variations (three of four) and only one of four E type variations in the analyzed region. Additional strain-specific mutations at position 38019 and position 38229 in Mexico_68 and Iceland_006, respectively, may provide the basis for further differentiation into M subtypes.
FIG. 2.
Analysis of genotypic variation using the 447-bp ORF22 site. Nucleotide diversity found in the 447-bp fragment of ORF22 was used to type all 326 VZV isolates displayed in Table 1. Dumas and Oka parental strains are included as the reference standard for E and J genotypes, respectively. All E and J strains displayed the same SNP patterns. Some variability was apparent among M genotype strains. The examples displayed represent all of the VZV variations that were observed; the figure has been condensed to make it more readable. The color scheme for the boxes is as for Fig. 1.
Analysis of globally distributed VZV isolates using ORF22-based (447-bp fragment) sequence data.
To map the worldwide distribution of genotypes, we used a simplified VZV genotyping protocol to analyze all 321 VZV isolates that were included in our collection at the time of this study; a representative subset of these strains is shown in Fig. 3.
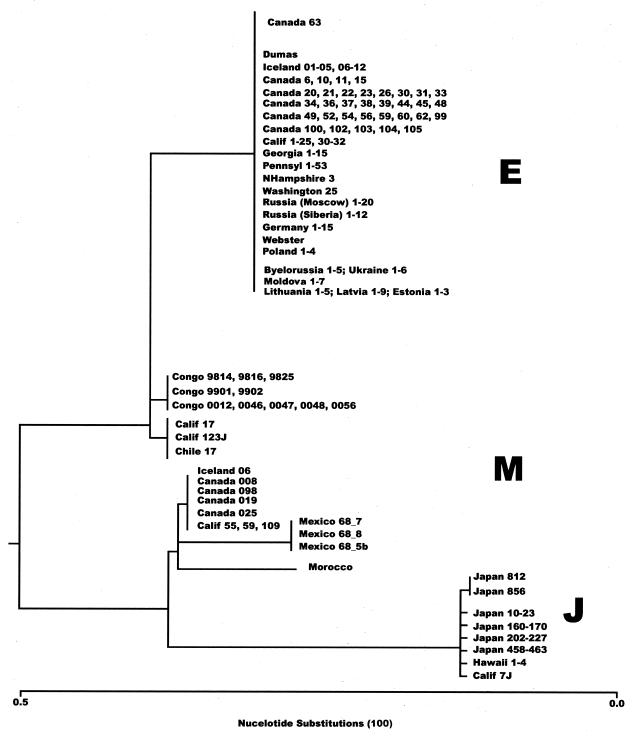
FIG. 3.
Genetic relatedness of global VZV strains. Phylogenetic analysis of ORF31 and -22 of VZV strains was performed by the neighbor-joining method (Clustal). All different structural variants detected in this study are included (16).
This small set of ORF22 markers derived from the 447-bp amplicon resolved distinct genotypes that correlate with our understanding of VZV variability and are consistent with the BglI− typing results and geographic origins of strains. All BglI+ strains collected in Japan with the PstI+ and PstI− profiles were completely identical in the 447-bp fragment and fall into the J genotype. Most strains obtained from Germany, Iceland, Czech Republic, Poland, European Russia, Belarus, Ukraine, Lithuania, Latvia, Moldova, and Estonia were identified as E strains.
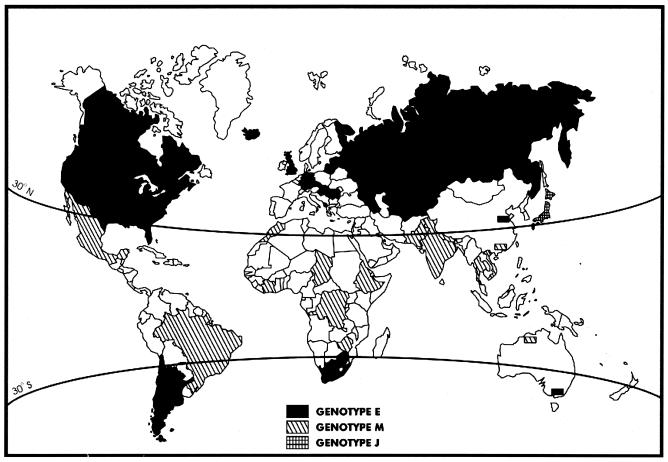
The E genotype was also most prevalent in Asian Russia, Kazakhstan, eastern Australia, Canada, and the United States. E type viruses were also isolated in Jordan, Chile, and Nicaragua (Fig. 1). On the basis of analysis of numerous isolates obtained from widely separated geographic locales, the E genotype was shown to form a conservative group based on the profile of ORF22 SNPs, and all E strains also displayed the PstI+ BglI− marker pattern. One surprising observation was the discovery of geographic dominance of M genotype strains in tropical and subtropical areas such as Africa (DRC, Chad, Morocco) and in Asia (India, Bangladesh, Nepal, and China). For this aspect of the study, we added a number of additional strains more recently obtained from Brazil, Argentina, Cote D'Ivoire, Ethiopia, Zimbabwe, South Africa, Thailand, Vietnam, north China, Great Britain, Tajikistan, Kazakhstan, Uzbekistan, Kyrgyzstan, Turkmenistan, and Azerbaijan. Above 30oN and below 30oS, E strains and J strains represent the vast majority of circulating VZV strains, whereas in the midlatitudes framed by those lines of latitude M strains predominate (Fig. 4). A substantial fraction of western Australian isolates also had the M genotype, exclusively in the portion of Australia that lies above the 30th parallel. All of these M strains contained the BglI site in ORF38.
FIG. 4.
Global distribution of the three major genotypes. Marked areas indicate the major circulating genotype (>70%) for the region. The 326 strains used in previous analysis (Table 1) are all represented; additionally, a number of other more recently acquired strains were analyzed only by ORF22 genotyping: Brazil, 10 isolates (8 M, 2 E); Argentina, 6 E isolates; Cote D'Ivoire, 5 M isolates; Ethiopia, 5 M isolates; Zimbabwe, 3 M isolates; South Africa, 3 E isolates; Thailand, 1 M isolate; Vietnam, 2 M isolates; north China, 3 E isolates; Great Britain, 5 E isolates; Tajikistan, 2 E isolates; Kazakhstan, 2 E isolates; Uzbekistan, 2 E isolates; Kyrgyzstan 2 E isolates; Turkmenistan, 2 E isolates; Azerbaijan, 2 E isolates. (Modified with permission, Corel Corp., copyright [copy] 2004.)
As expected, contagious VZV is actively disseminated across international borders, particularly into countries attractive to immigrants and tourists. During the course of this study, we detected a considerable number of J and M strains in the United States, Canada, and Australia.
All amplicons from the targeted region in ORF22 were tested with the FRET LyghtCycler probes. As a result of analyzing 326 samples (Table 1), we identified 201 E type, 88 M type, and 28 J type strains and confirmed the practicality of VZV genotyping without resorting to DNA sequencing.
DISCUSSION
Novel strategy for VZV genotyping.
By evaluating DNA sequences of clinical VZV isolates at eight genetic loci, we observed SNPs that shed light on the evolutionary pathways of VZV. More than 300 globally distributed VZV strains were sorted into three major genotypes E, J, and M. E and J strains were most divergent, whereas the M type comprises VZV isolates carrying variable assortments of E and J type SNPs.
Genotyping is a useful epidemiologic tool for many human herpesviruses, although it has not generally led to correlations between sequence variation and virus pathology (7, 8, 9, 17, 32, 43, 46). Genotyping protocols intended for general laboratory use must be simple to perform. They should also be parsimonious and able to yield results from the limited amount of DNA typically recovered from clinical specimens, e.g., vesicular swabs or scabs. An ideal format permits reliable VZV genotyping using a single short amplicon or, if necessary, a very limited number of them.
We searched for short sequences containing multiple SNPs that consistently identified the E, J, and M genotypes observed by using broader SNP data (ORF1, -22, -31, and -62). The VZV genome is extremely stable; all three genotypes of VZV differ in DNA sequence by only about 0.2%. The biological properties of VZV help to confer genomic stability. VZV is highly infectious and causes large outbreaks in naive populations, a characteristic that flattens the molecular divergence of VZV strains. VZV establishes lifelong latency, but subclinical reactivation is believed to occur less frequently than with other alphaherpesviruses (14). The low number of replicative cycles in the infected host (14) is thus thought to restrict opportunities to introduce mutations. Reactivated VZV is sometimes transmitted to susceptible individuals, causing varicella and periodically reintroducing older strains into the population.
Fortuitously, we identified a 447-bp sequence proximal to the carboxy terminus-encoding portion of ORF22 that satisfied our need for detecting viral DNA in low-titer clinical specimens and for reliably discriminating the three major genotypes. Both the multiple-ORF SNP analysis and the ORF22 fragment SNP patterns unerringly resolved the same stable VZV genotypes, all three of which are likely to have emerged in the distant past (19). We also noted that the ORF22 region selected for genotyping was stable in highly passaged European (Webster) and Japanese (Oka) strains.
Geographic distribution of the three ORF22 genotypes.
Regional VZV strain dominance could reflect the immunogenetic composition of the local human population in which it originally evolved (2, 6, 19). This might explain the occurrence of a uniform genotype in relatively isolated and conservative populations, e.g., Japan and the former Soviet Union. In geographic locations with populations in which large-scale immigration from multiple regions occurred, such as the United States, western Europe, and Australia, the distribution of VZV genotypes reflects that demographic quality. In support of this view, we have isolated J strains in North America exclusively from areas with the highest rates of immigrants from Asia.
We could also differentiate European and North American strains from Japanese, Asian, and African strains based on the ORF54 BglI site alone. Thus, ORF54-based sequence analysis could be useful for preliminary genotyping in the United States, Canada, much of Europe, eastern Australia, and South America, where M and J VZV strains are not as common. The routine differentiation of J and M strains requires DNA sequencing, RFLP analysis, or real-time hybridization methods with fluorescent probes, as described in this report.
In addition to discriminating the major circulating genotypes of VZV, the analysis of the SNPs in the 447-bp fragment also permitted further subtyping of M genotype strains. Further analysis of genomic sequence is likely to reveal additional regions that will enable finer discrimination of regionally distributed VZV strains, a process that should be accelerated by our recent determination of a genomic sequence for an M strain.
Correlation with other polymorphic genomic markers.
Genotypic analysis of the N-terminal part of the VZV gB gene (ORF31) revealed a highly conserved amino acid sequence and 99.88% homology at the nucleotide sequence level, in agreement with previously published results (7). This has held true for the 212 isolates we have examined for sequence variation in gB thus far. Comparison of the Dumas (E) and Oka parental (J) strains identified three prominent SNPs in ORF31 (Table 1): one was at codon 73 (position 57224), resulting in an amino acid substitution (T versus P), and one silent mutation each was at codons 99 (position 57301) and 131 (position 57397). A curious finding was that all ORF22-classified E and J strains were allocated to the same genotypic clusters by analysis of the ORF31 5′-terminal region. Two of the M genotypic clusters could be resolved with the ORF22 fragment SNPs, specifically, a cluster with more J type variations (M1; African Morocco_01 and DRA_1 strains) or more E type variations (M2; USOR_12 and USOR_17) identified through ORF31 SNPs. Analysis using ORF62 variations yielded the same results (Fig. 1).
We also determined that the J and E gB alleles were each significantly linked with specific allelic forms of ORF22. All these data provide additional confirmation for the robustness of the J, E, and M genotyping. The extent to which these observed differences in selected fragments of the VZV genome are stabilized by environmental or host-related selective pressures remains to be investigated.
Toward a common strategy for VZV genotyping.
The limited number of VZV SNPs are disseminated evenly over the genome but can nonetheless be used to classify strains into genotypes having a specific regional distribution. Evaluation of phylogenetic trees produced high bootstrap values, confirming the integrity of the genotypic analyses used in several reports (14, 33, 34). However, the use of this approach can be misleading, particularly for the identification of subgenotypes within the major circulating types, and must be confirmed by evaluating large numbers of clinically isolated specimens obtained from diverse geographic locales.
Although other investigators have attempted to describe the phylogenetic origin of currently circulating VZV strains, no consensus for a genotyping system has been reached. Here, we propose a simple methodology for elucidating the phylogenetic relationship between the VZV strains responsible for recent outbreaks and their evolutionary and/or transmission history. To prevent unnecessary confusion arising from the differing genotyping strategies and nomenclatures adopted by different laboratories, it is useful to summarize and correlate those findings here.
Two broad, nonintersecting studies focused on multiple SNPs positioned in various loci, comprehensively representing several thousand base pairs of sequence. Recent publications on ORF62 (tegument, transactivator), ORF31 (gB), ORF67 (gI), ORF68 (gE), ORF60 (gL), and ORF37 (gH) indicate that all are well conserved, and no single small region of the genome displays a genetic signature common to all VZV isolates in the United States (20, 33, 34).
Faga et al. and Wagenaar et al. (14, 44) proposed the division of VZV strains into four groups: (i) an Asian cluster (chiefly Japanese), (ii) a U.S. west coast genotype, (iii) a Dumas-like European cluster, and (iv) a central U.S. cluster. This represented the only effort previous to ours to characterize U.S. VZV genotypes but was limited by its examination of a relatively small number of strains and by a heavy reliance on laboratory-cultured strains.
Muir et al. and Quinlivan et al. (33, 34) examined a large number of VZV isolates from the United Kingdom, Asia, and Africa and, using sequence data obtained from ORF1, -21, -38, -50, and -54, identified three major genotypes, designated A, B, and C. B and C isolates were most commonly isolated in the United Kingdom and the United States, and A isolates were most common in Asia and Africa. However, the data in Fig. 1 indicate that all four of the targets in ORF1 selected for the Breuer study (33) coincidentally carry mainly J genotype-like SNPs at positions common to both J and M strains and as such could not clearly differentiate African strains from Asian.
Virtually all of our specimens were clinical isolates or low-passage strains, with only limited resort to established laboratory strains. It was observed (14) that laboratory strains of VZV can establish selected sequence variations in culture. For example, the T-to-C change at position 106262 that we have used to discriminate the vaccine strain from the wild type (30, 31) has been found only in the extensively cultured VZV Ellen and Oka vaccine strains (14). European genotype strains were most commonly isolated in Europe and in countries with a history of European colonization. The Japanese genotype was almost exclusively found in Japan, with the exceptions of Hawaii, the west coast of North America, and eastern Australia. Isolates from Africa and South America, as well as western Australia were of the M genotype.
The SNPs in the 447-bp fragment were able to discriminate several subtypes of M in a geographically distinctive fashion. These results refine the current understanding of VZV sequence variation and should permit both practical strain surveillance and the identification of the probable geographic origins of clinically isolated strains.
Speculation on mechanisms for establishing the M genotypes.
VZV genomes characterized by alternating J-like and E-like regions detected in M strains can be explained by at least two mechanisms, neither of which is excluded or favored by the present state of knowledge. One mechanism is strain recombination in dually infected cells, and the other mechanism involves independently arising, nonrandom point mutations.
Mixed infections with two different strains have been reported for other human herpesviruses, notably human cytomegalovirus (7), Epstein-Barr virus (15), and human herpesvirus type 6 (8). Second-episode varicella has been observed in recent years, increasing the possibility of intrastrain recombination in the host (3, 18, 21). However, there is as yet no direct evidence that VZV recombination can occur in coinfected cells. A proposed recombinant genotype from Brazil described by Muir et al. (33) is likely an M genotype strain. Of 10 independently obtained isolates from Brazil tested in our laboratory, 8 strains were M (Fig. 4) and had SNP patterns similar to those described by Muir et al. (33). Since the VZV genome is so stable and observed DNA sequence variation is virtually all individual point mutations, it just as plausible that M strains reflect a small number of favored variations that have occurred independently among wild-type strains in more than one geographic location. We also cannot rule out the possibility that M virus occurred first in tropical regions and mutated following the migration of humans into temperate climates. J and E strains may have arisen independently from M as a result of the isolation of the Japanese islands and the broad explorations of the Europeans.
In conclusion, this study demonstrates that VZV genotypes can be classified into subgenotypes on the basis of limited DNA sequence variation in ORF22. Most important, this study introduces a new approach to the molecular epidemiology of VZV infection that should be especially advantageous for the study of the host-VZV interaction. The amplification of ORF22 sequences directly from infected tissues produces a broader image of VZV heterogeneity than do other methods described thus far. The application of this tool to the analysis of large collections of worldwide isolates should help to elucidate the mechanism or mechanisms through which VZV variability develops. Last, the method should provide a useful tool for the detection of VZV coinfection and could provide new insights into the mechanisms of VZV persistence.
Acknowledgments
This work was supported by grants from the CDC National Vaccine Program Office (United States), the Deutsche Forschungsgemeinschaft (Bonn), and the Varicella Research Foundation (New York).
We thank colleagues who provided samples for this study, including Neli N. Maltceva, Richard Garceau, Inger Damon, Michaela Schmidt, Herman Meyer, Mikhail Sousloparov, Karin Galil, Naoki Inoue, Michiko Takayama, Ann Arvin, Leigh Zerboni, John Zaia, and Sharka Nemechkova.
REFERENCES
- 1.Adams, S. G., D. E. Dohner, and L. D. Gelb. 1989. Restriction fragment differences between the genomes of the Oka varicella vaccine virus and American wild-type varicella-zoster virus. J. Med. Virol. 29:38-45. [DOI] [PubMed] [Google Scholar]
- 2.Agostini, H. T., R. Yanagihara, V. Davis, C. F. Ryschkewitsch, and G. L. Stoner. 1997. Asian genotypes of JC virus in native Americans and in a Pacific island population: markers of viral evolution and human migration. Proc. Natl. Acad. Sci. USA 94:14542-14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvin, A. M., C. M. Koropchak, and A. E. Wittek. 1983. Immunologic evidence of reinfection with varicella-zoster virus. J. Infect. Dis. 148:200-205. [DOI] [PubMed] [Google Scholar]
- 4.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 5.Casey, T. A., W. T. Ruyechan, M. N. Flora, W. Reinhold, S. E. Straus, and J. Hay. 1985. Fine mapping and sequencing of a variable segment in the inverted repeat region of varicella-zoster virus DNA. J. Virol. 54:639-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalli-Sforza, L. L., P. Menozzi, and A. Piazza. 1994. The history and geography of human genes. Princeton University Press, Princeton, N.J.
- 7.Chou, S., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralizing epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 8.Cone, R., M. L. Huang, R. C. Ackman, and L. Corey. 1996. Coinfection with human herpesvirus 6 variants A and B in lung tissue. J. Clin. Microbiol. 34:877-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, C. L., D. Field, D. Metzgar, R. Saiz, P. A. Morin, I. L. Smith, S. A. Spector, and C. Wills. 1999. Numerous length polymorphisms at short tandem repeats in human cytomegalovirus. J. Virol. 73:6265-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 11.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329-341. [DOI] [PubMed] [Google Scholar]
- 12.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Varicella-zoster virus DNA from persistently infected cells contains novel tandem duplications. J. Gen. Virol. 69:2229-2249. [DOI] [PubMed] [Google Scholar]
- 13.Dumas, A. M., J. L. Geelen, M. W. Weststrate, P. Wertheim, and J. van der Noordaa. 1981. XbaI, PstI, and BglI restriction enzyme maps of the two orientations of the varicella-zoster virus genome. J. Virol. 39:390-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faga, B., W. Maury, D. A. Bruckner, and C. Grose. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280:1-6. [DOI] [PubMed] [Google Scholar]
- 15.Falk, K. I., J. Z. Zou, E. Lucht, A. Linde, and I. Ernberg. 1997. Direct identification by PCR of EBV types and variants in clinical samples. J. Med. Virol. 51:355-363. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1993. PHYLIP inference package, version 3.5. Department of Genetics, University of Washington, Seattle.
- 17.Franti, M., J.-T. Aubin, L. Poirel, A. Gautheret-Dejean, D. Candotti, J.-M. Huraux, and H. Agut. 1998. Definition and distribution analysis of glycoprotein B gene alleles of human herpes virus 7. J. Virol. 72:8725-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershon, A. A., S. P. Steinberg, and L. Gelb. 1984. Clinical reinfection with varicella-zoster virus. J. Infect. Dis. 149:137-142. [DOI] [PubMed] [Google Scholar]
- 19.Gessain, A., E. Boeri, R. Yanagihara, R. C. Gallo, and G. Franchini. 1993. Complete nucleotide sequence of a highly divergent human T-cell leukemia (lymphotropic) virus type-1 (HTLV-1) variant from Melanesia: genetic and phylogenetic relationship to HTLV-1 strains from other geographic regions. J. Virol. 67:1015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawrami, K., I. J. Hart, F. Pereira, S. Argent, B. Bannister, B. Bovill, D. Carrington, M. Ogilvie, S. Rawstorne, Y. Tryhorn, and J. Breuer. 1997. Molecular epidemiology of varicella-zoster virus in east London, England, between 1971 and 1995. J. Clin. Microbiol. 35:2807-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayakawa, Y., T. Yamamoto, K. Yamanishi, and M. Takahashi. 1986. Analysis of varicella-zoster virus DNAs of clinical isolates by endonuclease HpaI. J. Gen. Virol. 67:1817-1829. [DOI] [PubMed] [Google Scholar]
- 23.Hondo, R., and Y. Yogo. 1988. Strain variation of R5 direct repeats in the right-hand portion of the long unique segment of varicella-zoster virus DNA. J. Virol. 62:2916-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hondo, R., Y. Yogo, M. Yoshida, A. Fujima, and S. Itoh. 1989. Distribution of varicella-zoster virus strains carrying a PstI-site-less mutation in Japan and DNA change responsible for the mutation. Jpn. J. Exp. Med. 59:233-237. [PubMed] [Google Scholar]
- 25.Kinchington, P. R., J. Remenick, J. M. Ostrove, S. Straus, W. D. Ruyechan, and J. Hay. 1986. Putative glycoprotein gene of varicella-zoster virus with variable copy numbers of a 42-base-pair repeat sequence has homology to herpes simplex virus glycoprotein C. J. Virol. 59:660-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinchington, P. R., and S. E. Turse. 1995. Molecular basis for a geographic variation of varicella-zoster virus recognized by a peptide antibody. Neurology 45(Suppl. 8):S13-S14. [DOI] [PubMed] [Google Scholar]
- 27.Kinoshita, H., R. Hondo, F. Taguchi, and Y. Yogo. 1988. Variation of R1 repeated sequence present in open reading frame 11 of varicella-zoster virus strains. J. Virol. 62:1097-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Russa, P., O. Lungu, I. Hardy, A. Gershon, S. P. Steinberg, and S. Silverstein. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRussa, P., S. Steinberg, A. Arvin, D. Dwyer, M. Burgess, M. Menegus, K. Rekrut, K. Yamanishi, and A. Gershon. 1998. Polymerase chain reaction and restriction fragment length polymorphism analysis of varicella-zoster virus isolates from the United States and other parts of the world. J. Infect. Dis. 178(Suppl. 1): 64-66. [DOI] [PubMed] [Google Scholar]
- 30.Loparev, V. N., T. Argaw, P. R. Krause, M. Takayama, and D. S. Schmid. 2000. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loparev, V. N., K. McCaustland, B. P. Holloway, P. R. Krause, M. Takayama, and D. S. Schmid. 2000. Rapid genotyping of varicella-zoster virus vaccine and wild-type strains with fluorophore-labeled hybridization probes. J. Clin. Microbiol. 38:4315-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng, Y. X., T. J. Spira, G. J. Bhat, C. J. Birch, J. D. Druce, B. R. Edlin, R. Edwards, C. Gunthel, R. Newton, F. R. Stamey, C. Wood, and P. E. Pellett. 1999. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology 261:106-119. [DOI] [PubMed] [Google Scholar]
- 33.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlivan, M., K. Hawrami, W. Barrett-Muir, P. Aaby, A. Arvin, V. T. Chow, T. J. John, P. Matondo, M. Peiris, A. Poulson, M. Siqueira, M. Takahashi, Y. Talukder, K. Yamanishi, M. Leedham-Green, F. T. Scott, S. L. Thomas, and J. Breuer. 2002. The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J. Infect. Dis. 186:888-894. [DOI] [PubMed] [Google Scholar]
- 35.Santos, R. A., C. C. Hatfield, N. L. Cole, J. A. Padilla, J. F. Moffat, A. M. Arvin, W. T. Ruyechan, J. Hay, and C. Grose. 2000. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275:306-317. [DOI] [PubMed] [Google Scholar]
- 36.Straus, S. E., H. S. Aulakh, W. T. Ruyechan, J. Hay, T. A. Casey, G. F. Van de Woude, J. Owens, and H. A. Smith. 1981. Structure of varicella-zoster virus DNA. J. Virol. 40:516-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Straus, S. E., J. Hay, H. Smith, and J. Owens. 1983. Genome differences among varicella-zoster virus isolates. J. Gen. Virol. 64:1031-1041. [DOI] [PubMed] [Google Scholar]
- 38.Straus, S. E., W. Reinhold, H. A. Smith, W. T. Ruyechan, D. K. Henderson, R. M. Blaese, and J. Hay. 1984. Endonuclease analysis of viral DNA from varicella and subsequent zoster infections in the same patient. N. Engl. J. Med. 311:1362-1364. [DOI] [PubMed] [Google Scholar]
- 39.Takada, M., T. Suzutani, I. Yoshida, M. Matoba, and M. Azuma. 1995. Identification of varicella-zoster virus strains by PCR analysis of three repeat elements and a PstI-site-less region. J. Clin. Microbiol. 33:658-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayama, M., and N. Takayama. 2002. Long PCR amplification of varicella-zoster virus DNA in clinical specimens from the patients with varicella and herpes zoster. Kansenshogaku Zasshi. 76:347-354. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 41.Takayama, M., N. Takayama, N. Inoue, and Y. Kameoka. 1996. Application of long PCR method to identification of variations in nucleotide sequences among varicella-zoster virus isolates. J. Clin. Microbiol. 34:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thawaranantha, D., K. Balachandra, S. Jongtrakulsiri, W. Yamkunthong, K. Chimabutra, and J. Bhumiswasdi. 1995. Genome differences among varicella-zoster viruses isolated in Thailand. Southeast Asian J. Trop. Med. Public Health 26:677-683. [PubMed] [Google Scholar]
- 43.Umene, K., and M. Yoshida. 1993. Genomic characterization of two predominant genotypes of herpes simplex virus type 1. Arch. Virol. 131:29-46. [DOI] [PubMed] [Google Scholar]
- 44.Wagenaar, T., V. T. Chow, C. Buranathai, P. Thawasupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, S., H. Kabuto, and M. Shingu. 1991. Restriction endonuclease analysis of varicella-zoster virus DNAs. Kurume Med. J. 38:45-50. [DOI] [PubMed] [Google Scholar]
- 46.Zimber, U., H. K. Addinger, G. M. Lenoir, M. Vuillaume, M. V. Knebel-Doeberitz, G. Laux, C. Desgranges, P. Wittman, U. K. Freese, U. Schneider, and G. Bornkamm. 1986. Geographical prevalence of two types of Epstein-Barr virus. Virology 154:56-66. [DOI] [PubMed] [Google Scholar]
- 47.Zweerink, H. J., D. H. Morton, L. W. Stanton, and B. F. Neff. 1981. Restriction endonuclease analysis of the DNA from varicella-zoster virus: stability of the DNA after passage in vitro. J. Gen. Virol. 55:207-211. [DOI] [PubMed] [Google Scholar]