Abstract
MicroRNAs (miRNAs) are evolutionarily conserved, small, regulatory RNAs that negatively regulate gene expression. Extensive research in the last decade has implicated miRNAs as master regulators of cellular processes with essential role in cancer initiation, progression and metastasis, making them promising therapeutic tools for cancer management. In this review, we will briefly review the structure, biogenesis, functions and mechanism of action of these miRNAs, followed by a detailed analysis of the therapeutic potential of these miRNAs. We will focus on the strategies presently used for miRNA therapy; discuss their use and drawbacks, and the challenges and future directions for development of miRNA-based therapy for human cancers.
Keywords: microRNA, cancer, therapeutics, anti-miRNAs, miRNA mimics
Introduction
The discovery of microRNAs (miRNAs) in the last decade has been one of the most fascinating breakthroughs of recent times. The identification of these regulatory RNAs from the dark matter of the human genome initiated a shift in the paradigms of cancer biology, and facilitated a deeper understanding of the human biology. miRNAs are small (19-24nt), endogenous highly-conserved non-coding RNAs with imperative regulatory functions1. They negatively regulate gene expression by binding to the 3’-untranslated region (3’-UTR) of the target mRNAs, and causing mRNA degradation or translational repression. Thus, they are involved in post-transcriptional gene silencing and are predicted to target about 30% of the protein-coding genes2. Since its discovery, thousands of miRNAs have been identified in animals, plants and several viruses. miRNAs play an important role in multiple biological processes, including developmental timings, embryogenesis, cell differentiation, organogenesis, metabolism, apoptosis and various diseases, including cancers3. miRNA pathways have been implicated as a new layer of gene regulation important in both normal and diseased states, and thus further understanding of their molecular nature and functions would help identify novel tools for drug development.
miRNA biogenesis and function
miRNAs are usually transcribed from miRNA genes by RNA polymerase II (RNAPII), as autonomous transcription units, or as clusters from a polycistronic transcription unit, to give a long capped and polyadenylated primary miRNA transcript (pri-miRNA) with a stem-loop structure4. miRNA genes can be located in the exonic or intronic regions of non-protein coding regions, or in the intronic region of protein-coding transcription units. Pri-miRNAs are then cleaved by a Microprocessor complex (composed of Rnase III Drosha and double-stranded DNA binding domain protein DGCR8) to give hairpin-shaped precursors of microRNA (pre-miRNAs)5. The pre-miRNAs are then exported to the cytoplasm by the nuclear export factor Exportin-5/Ran GTP. Following their export from the nucleus, these pre-miRNAs are further processed to ~ 22 nucleotide miRNA:miRNA* duplexes by the cytoplasmic Rnase III enzyme Dicer, and its double-stranded RNA binding domain protein TRBP. Subsequently, the mature miRNA then gets incorporated into RISC (RNA-induced silencing complex), usually resulting in the repression of target gene expression. RISC usually binds to the 3’-UTR of target mRNAs and brings about cleavage or translational repression of the target mRNAs, depending on the degree of complementarity between miRNA and target mRNA. In humans, microRNAs usually bind to specific sequences with partial complementarity on target RNA transcripts, called ‘seed sequences’ or microRNA response elements (MREs), which then result in translational repression1.
Besides the canonical miRNA regulation pathway, several variations have also been reported for the miRNA-mediated regulation. Some miRNAs bind to the 5’-UTR or the open reading frame of the mRNA, resulting in activation rather than suppression of the target genes6. Few miRNAs also show decoy activity, by binding directly to the proteins, such as RNA-binding proteins, and thus inhibiting the interaction with their target RNA7. Moreover, in certain cases, miRNAs can also regulate gene expression at the transcriptional level8, by binding directly to the DNA regulatory elements. Thus miRNA-mediated regulation of gene expression is a complex science and is still an evolving concept.
miRNAs as preferred therapeutic targets for cancer
Since their discovery, miRNAs have been reported to have a variety of roles in cancer development and progression. Figure 1 depicts the widespread dysregulation and aberrant expression of miRNAs in human cancers. The seminal study linking miRNAs to cancer found that miRNA cluster miR-15-miR-16-1 were frequently deleted or downregulated in 69% of chronic lymphocytic leukemia patients (CLL)9. These miRNAs targeted anti-apoptotic proteins B cell lymphoma 2 (BCL2) and induced myeloid leukemia cell differentiation (MCL1) protein10. A knockout mouse model targeting the miR-15b/miR-16-1 cluster recapitulated the CLL-associated phenotype, validating the tumor suppressive functionality of these miRNAs in vivo11. Subsequent studies identified several other tumor suppressive miRNAs, such as the let-7 family members, miR-29, miR-34 family, miR-122 and miR-143/145 cluster, which suppress the expression of oncogenic protein-coding genes. Loss of tumor suppressive miRNA expression due to somatic alterations or germline mutations can initiate or enhance tumorigenecity.
Figure 1.
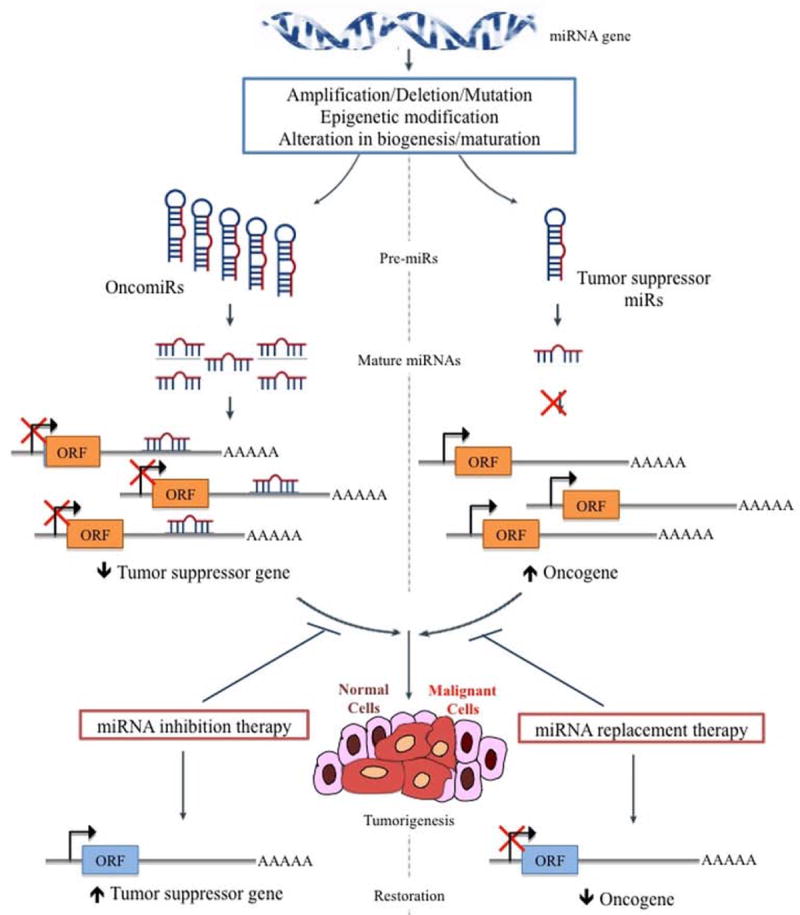
miRNA deregulation in human cancers.
miRNA genes frequently undergo chromosomal deletions or translocations, epigenetic regulation, alterations in miRNA promoter activity by oncogenes and tumor suppressor genes or alterations in the miRNA processing machinery resulting in widespread deregulation in their expression. Consequently, oncogenic miRNAs are upregulated, while tumor suppressive miRNAs are downregulated. This alteration at the miRNAome leads to aberrant changes in the transcriptome and proteome, causing cellular transformation and tumor progression.
In contrast, overexpressed miRNAs that promote tumorigenecity by targeting tumor suppressive genes are classified as oncomiRs12. One of the most prevalent oncomiR is miR-21, which is overexpressed in almost all human cancers and targets PTEN and PDCD4 (12). A recent study also reported the dependency of tumor cells on activated miR-21 for survival and proliferation advantage, and introduced the concept of OncomiR addiction13. Medina et al13 developed a conditional transgenic mouse overexpressing miR-21 and showed that these mice developed spontaneous pre-B malignant lymphoid-like phenotype. In the absence of miR-21, malignant cells undergo apoptosis and the tumors regress. Spontaneous tumorigenesis in mice models overexpressing miR-15514 and miR-22115 have established these miRNAs as bona fide oncomiRs. In addition, induction of lethal B cell malignancies in transgenic mice models of miR-17~92 cluster16 and miR-125b17 have highlighted miRNAs as causative agents in solid as well as hematological malignancies. These in vivo experimental findings strongly advocate the use for miRNAs as therapeutic intervention tools for cancer management.
Interestingly, it has been found that some miRNAs can behave as oncogenes in one cancer type and as tumor suppressive genes in others. For example, miR-221 is overexpressed in liver cancer and exerts an oncogenic function by targeting tumor suppressor PTEN, but it acts as a tumor suppressor in erythroblastic leukemia by silencing the KIT oncogene18, 19. Thus, identification of detailed biological functions and targets of miRNAs is an important aspect when considering miRNA therapeutics.
Using high throughput techniques such as microRNA microarrays and next-generation sequencing, widespread dysregulation and aberrant expression of miRNAs has been reported in almost all human cancers20, 21. The widespread dysregulation of miRNAs can be explained by their frequent location in cancer-associated hotspots of the human genome, including fragile sites, minimal regions of amplification, loss of heterozygosity sites, and common breakpoint regions22. Other mechanisms of this dysregulation include chromosomal deletions or translocations of regions with miRNA genes, epigenetic regulation of miRNA expression, alterations in miRNA promoter activity by oncogenes and tumor suppressor genes, alterations in the miRNA processing machinery (such as mutations in Dicer, TRBP2 or Exportin 5), and presence of mutated miRNA structural variants20. Aberrant miRNA expression has enabled clinicians to develop miRNA signature profiles that serve as important diagnostic, predictive, prognostic and theranostic biomarkers. In addition, miRNA expression patterns allow precise differentiation between different tumor types and identification of tissue of origin in poorly differentiated tumors23. The detection of stable circulating miRNAs in serum has opened up new avenues for non-invasive biomarkers in cancer prognosis24, 25. For example, high levels of miR-141 expression in the plasma have been associated with poor prognosis in patients with colorectal cancers26. Further, correlations between circulating miRNA levels and the response to a given anticancer agent have also been observed. For example, circulating miR-21 levels were higher in prostate cancer patients resistant to docetaxel-based chemotherapy, in comparison to those with chemosensitive disease27.
The recent discovery of competing endogenous RNAs (ceRNAs)28 lend further support to the potential use of miRNAs as therapeutic agents. The ceRNA hypothesis states that an extensive proportion of human transcriptome, including transcribed pseudogenes, mRNAs and long non-coding RNAs can interact or crosstalk with each other through miRNA responsive elements (MREs), establishing a comprehensive intricate regulatory network. For example, tumor suppressor PTEN is finely regulated by its ceRNAs, PTENP1 pseudogene and ZEB2 mRNA via common MREs29, 30. Placing this in the context of miRNA therapy, miRNA modulation may have more profound manifestations in an as-yet-uncharacterized RNA-dependent aspect. Thus, therapeutic modulation of miRNA levels might shift the balance and set up a cellular cascade enabling a more pronounced biological effect than previously anticipated. This discovery reveals a whole new realm of therapeutic possibilities for human cancers. More research to identify other cancer-associated ceRNA-miRNA networks needs to be done before the full therapeutic potential of such regulatory loops can be exploited.
miRNA-based therapeutic approaches
The major advances in the last decade have focused on designing novel targeted therapy capable of targeting specifically the malignant cells in a more rational way. The advent of miRNAs provide an additional layer of gene regulation on a broad spectrum of biological pathways by fine-tuning protein expression levels. The ability of miRNAs to regulate cellular processes, their inherent role in carcinogenesis as oncogenes or tumor suppressor genes, and the aberrant dysregulation of their expression levels in cancer illustrate the potential of miRNA modulation as a viable therapeutic strategy and a powerful intervention tool. By the virtue of their ability to target multiple protein-coding genes and their aberrant perturbations in widespread cancers, miRNAs have emerged to be promising novel therapeutic targets and intervention tools. Table 1 and Figure 2 describe the several approaches currently being explored for miRNA modulation.
Table 1.
miRNA-based therapeutic approaches
| Method | Definition | Mechanism of action | Clinical trial stage |
|---|---|---|---|
| AMOs73 | A single-stranded, RNA molecule designed to be complementary to a selected miRNA | Competitive inhibition of mature miRNA by Watson-Crick binding | Preclinical studies |
| 2’-O- modified AMOs | Modification of 2’-OH to 2’-O’methyl- and 2’-O’methoxyethyl- groups | Competitive inhibition of mature miRNA by Watson-Crick binding | Phase I clinical trial |
| Antagomirs43 | AMOs modified to have a phosphorothiolate backbone and conjugated with cholesterol | Competitive inhibition of mature miRNA; detailed mechanism still unclear | Preclinical studies |
| LNA anti-miRNAs45. 46 | Addition of an extra methylene bridge connecting the 2′-O atom and the 4′-C atom and ‘locks’ the ribose ring in a C3’-endo or C2’-endo conformation | High-affinity Watson-Crick hybridization with their RNA target molecules - Inhibition | Phase I and II clinical trials |
| miRNA sponges51 | RNA transcripts with multiple tandom repeats of miRNA binding sites for an endogenous miRNA | Stably interact with corresponding miRNA and prevent its interaction with its target mRNAs | Preclinical studies |
| miRNA masks53 | Single-stranded 2’-O’methyl-modified antisense oligonucleotides with entire complementary to the miRNA binding sites in the 3’-UTR of the target mRNA | ‘Mask’ the target mRNA from the endogenous miRNA and thus prevent its suppression | Preclinical studies |
| Small molecule inhibitors of miRNAs (SMIRs)39, 40, 54 | Small-molecule chemical compounds that interfere with miRNA biogenesis or matuation | Block specific miRNAs by structure-based docking onto the precursor or mature form of the miRNAs | Preclinical studies |
| miRNA expression vectors74 | Expression vectors that express a specific type of miRNA | Restoration of the expression and function of a specific miRNA | Preclinical studies |
| miRNA mimics74 | Small, chemically modified (2’-O’methoxy) RNA duplexes that can be loaded into RISC and achieve the downstream inhibition of the target mRNAs | Assist the miRNA function | Phase I clinical trials |
Figure 2.
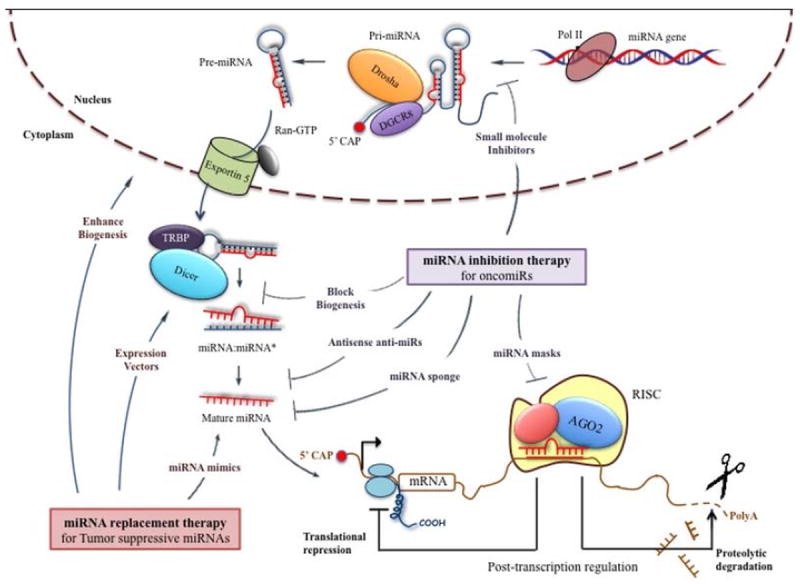
miRNA-based therapeutics for management of cancer.
miRNA inhibition therapy for OncomiRs include antisense anti-miRs (AMOs, modified AMOs, LNA-based AMOs, antagomirs), small molecule inhibitors, miRNA sponges and miRNA masks. miRNA replacement therapy for tumor suppressive miRs includes miRNA mimics, small molecule enhancers and expression vectors. These approaches act at different stages of miRNA biogenesis, including processing, maturation and strand selection.
miRNA restoration
Alterations or mutations in tumor suppressive miRNAs impede their inhibitory effect and contribute to oncogenic activation in cancer cells. An effective solution is to restore the normal function of these miRNAs by replacing or substituting the lost miRNA using synthetic miRNA-like molecules called miRNA mimics or with miRNAs encoded in expression vectors. miRNA mimics are small, chemically modified (2’-O’methoxy) RNA duplexes that are processed into single-strand form inside the cells and loaded into RISC to achieve the downstream inhibition of the target mRNAs. For example, introduction of miRNA mimics for miR-15a in prostate cancer cell lines induced marked apoptosis and blocked proliferation31. However, an effective in vivo delivery system greatly influences the stability and uptake of miRNA mimics. Systemic delivery of neutral lipid-based vehicle for miR-34a and let-7 mimics reduced tumor burden in KRAS-activated non-small cell lung cancer mouse model32. Additionally, systemic delivery of atelocollagen-conjugated miR-34a and miR-16 in a mouse xenograft model effectively inhibited progression of colon cancer and prostate cancer, respectively33.
An alternative approach is engineering longer pre-miRNA-like shRNAs from plasmid or viral vectors driven by strong promoters to induce tissue- or tumor-specific expression34. For example, intranasal administration of an adenovirus expressing let-7a RNA hairpin in a KRAS mutant mouse effectively restrained the growth of the lung cancer xenograft mouse model35, 36. Further, systemic administration of miR-26a using adeno-associated virus in hepatocellular carcinoma resulted in tumor-specific inhibition of cell proliferation, induction of apoptosis and blocked tumor progression37. Another example includes a RNA polymerase II driven expression vector for miR-155 that has been shown to effectively increase miR-155 expression levels in vitro and in in vivo xenograft models38.
The primary causes of reduced miRNA expression in human cancers are genetic deletion of miRNA loci, epigenetic silencing via CpG island hypermethylation in the promoter region of miRNA genes or decreased biogenesis and/or processing of miRNAs22. Reversal of these processes using small molecules can help restore the global miRNA expression levels. Hypomethylating agents such as decitabine or 5-azacytidine has been shown to reverse epigenetic silencing of miRNAs as well as mRNAs, and are approved for treatment of myelodysplastic syndromes39. Another example is the small molecule enoxacin, which has been shown to promote miRNA biogenesis and processing by binding to TARBP2 (TAR RNA-binding protein 2)40. Enoxacin treatment augmented the production of a subset of miRNAs and reduced tumor growth in mouse models of colon cancer (40). These studies highlight the effectiveness of restoring downregulated miRNAs in cancer cells. However, these strategies are non-specific and induce re-expression of a spectrum of miRNAs.
The first miRNA replacement therapy – MRX34, an intravenously injected liposome-based miR-34 mimic – is currently in Phase I clinical trials for patients with advanced hepatocellular carcinoma. MRX34 will be administered intravenously twice a week for three weeks with one week off to investigate its safety, pharmacokinetics and pharmacodynamics profile (on clinicaltrials.gov, Identifier: NCT01829971).
miRNA inhibition
OncomiRs are frequently overexpressed in cancers, and inhibition of these miRNAs would help restore the normal function of its target tumor suppressive genes. Several miRNA inhibitory agents have been recently tested in preclinical and clinical studies and include antisense anti-miR oligonucleotides, locked nucleic acid (LNA-) anti-miRNAs, miRNA sponges, miRNA molecule and small molecule inhibitors of miRNAs.
Synthetic anti-miRNA oligonucleotides are single-stranded RNA molecules complementary to the target miRNA41, 42 that function as competitive inhibitors by obstructing their interaction with target mRNAs. Thus targeted inhibition of a specific miRNA and subsequent upregulation of its target mRNAs can be achieved. Addition of different 2’-ribose modifications to AMOs (2’-O-methyl AMOs and 2’-O-methoxyethyl) contributes to nuclease resistance and improved binding affinity, stability and specificity, and further addition of a phosphorothioate moiety along with the 2’ modification further enhances the resistance to endonucleases, and better serum stability and cellular uptake42. Another optimal modification is the addition of cholesterol functionality at the 3’-position of the nucleic acid to generate ‘antagomirs’43. The cholesterol modification further enhances the cellular uptake of these AMOs. Intravenous injection of antagomirs against miR-122 afforded a marked and specific inhibition of endogenous miR-122 in mice, with an upregulation of target mRNAs43. The effects were long-lasting (up to 23 days) with no noticeable immune response or toxicity. Targeted suppression of the mature miR-122 was observed in liver, lung, kidney, heart, intestine, fat, skin, bone marrow, muscle, ovaries and adrenals, except brain. In another instance, systemic treatment of a metastatic breast cancer mouse model with miR-10b antagomirs did not reduce primary tumor but effectively suppressed breast cancer metastasis to lung44. miR-10b antagomir significantly reduced miR-10b levels and increased the levels of Hoxd10, an important miR-10b target.
Another example of modified AMOs are the Locked Nucleic Acid anti-miRs, in which an extra methylene bridge connecting the 2′-O atom and the 4′-C atom ‘locks’ the ribose ring in a C3’-endo or C2’-endo conformation45. LNA-modified oligonucleotides create high-affinity Watson-Crick binding to target mRNAs, and thus exhibit higher thermal stability and superior hybridization with their RNA target molecules. Furthermore, they display higher aqueous solubility and increased metabolic stability for in vivo delivery. For instance, intravenous injection of LNA anti-miR against miR-122 showed distinctly superior efficiency in antagonizing miR-122 compared to cholesterol-conjugated antagomirs46, and was completely safe and efficacious in primate models, such as African green monkeys46 and chimpanzees47. An efficient depletion of miR-122 in the liver was reported in each case without any evidence of LNA-associated toxicities or histopathological changes in the animals. Miravirsen (SPC3649), an LNA-anti-miR-122 by Santaris Pharma, was recently evaluated in Phase I and Phase IIa clinical trials for treatment of hepatitis C virus, it was found that miravirsen successfully reduced HCV RNA in a dose-dependent manner in patients with chronic HCV genotype 1 infection48.
Modified LNA anti-miRs that are 8 nucleotides long and specifically bind to the 5’-seed sequence (called tiny LNAs) have also been developed49. Treatment with tiny LNAs resulted in inhibition of several miRNAs families that share the same seed region, and a consequent upregulation of their direct targets. Systemic intravenous delivery of unconjugated tiny LNAs showed long-term miRNA silencing with high specificity in orthotopic breast cancer model. It was also shown that an 8-mer antimiR-155-LNA inhibits Waldenstrom Macroglobulinemia (WM) and CLL cell proliferation in vitro, while systemically delivered antimiR-155-LNA showed significantly decreased tumor growth in vivo50.
Ebert and colleagues51 reported a novel system for miRNA antagonism called ‘miRNA sponges’ or ‘miRNA decoys’. These are RNA transcripts with multiple tandem repeats of miRNA binding sites for an endogenous miRNA. These sponges stably interact with or ‘soak up’ the corresponding miRNA and prevent its interaction with its target mRNAs. A remarkable attribute of this approach is the ability of miRNA sponge to affect all closely related miRNAs within a family that share overlapping targets. In order to imitate the miRNA-mRNA binding like in the natural setting, a bulge was introduced at position 9-12, which achieved better activity due to increased miRNA retention. For example, inhibiting miR-9 expression using a miRNA sponge effectively inhibited pulmonary micrometastasis in orthotopic mouse models of breast cancer52.
Inhibition of a specific miRNA using the aforementioned strategies causes de-repression of the entire array of target genes that show miRNA binding sites for that particular miRNA. A gene-specific anti-miRNA therapy that can reverse the repressive action of miRNAs in an mRNA-selective manner has also been developed. These ‘miRNA-masks’ consists of single-stranded 2’-O’methyl-modified antisense oligonucleotides with locked 5’ and 3’ ends that are entirely complementary to the miRNA binding sites in the 3’-UTR of the target mRNA. In effect, they ‘mask’ the target mRNA from the endogenous miRNA and thus prevent its suppression. This principle has been successfully applied to abrogate the activity of miR-430 on TGFβ (transforming growth factor β) signaling pathway in a zebrafish model53.
Potential miRNA-specific small molecule inhibitors (SMIRs) were identified from a large-scale drug screening using a luciferase-based reporter assay54. Diazobenzene and its derivatives were found to reduce the transcription of the miR-21 gene into pri-miR-21 in vitro. However, no cytotoxic or non-specific effects on other processes were tested. Identification of other such small molecule inhibitors of miRNAs might be a useful supplement to conventional chemotherapeutic agents.
Recent studies have reported that miRNAs can also be packaged into vesicles and secreted into the extracellular environment as hormones by donor cells55. These then trigger a receptor-mediated response in different recipient cells. For instance, Yang et al56 reported that macrophages regulate the invasiveness of breast cancer cells by exosome-mediated transport of miR-223. Thus, targeting these miRNAs in the donor cells can have an impact on the functionality of a different recipient cell. Furthermore, extracellular miRNAs (for example, let-7) can also directly interact with Toll-like receptors (TLRs) and trigger activation of downstream pathway57. Targeting these extracellular miRNAs provides novel therapeutic options for cancer management, and further studies will be needed to determine the effectiveness of such approaches.
Strategies for miRNA therapeutics to treat human cancers
Cancer represents a complex genetic disease, and treatment with any individual therapy confines them to the ‘one-drug-one-target’ paradigm and renders them susceptible to resistance in due course. The current knowledge available from gene therapy and RNAi technologies can be easily adapted to miRNA therapy. Exploiting the current understanding of the genetic nature of cancer and the molecular pathways involved in malignant transformation, and the wide spectrum of new molecular drugs available, several novel therapeutic combination regimens can be evaluated. One strategy is to target a major molecular alteration clearly linked to disease pathogenesis by using multiple therapeutic agents, called the ‘Sandwich RNA inhibition’ approach. A recent study exemplified this strategy by targeting EphA2, an important ovarian cancer oncogene, using a combination of EphA2-targeting siRNAs and miR-520d-3p mimics58. Dual targeting of EphA2 prominently decreased EphA2 protein levels, suppressed tumor growth, migration and invasion, and exhibited synergistic anti-tumor efficiency than either monotherapy alone, both in vitro and in vivo. Thus regimens using a cocktail of RNAi (RNA interference) therapeutics to target dominant oncogenes might achieve better therapeutic outcomes in human cancers.
An alternative strategy is to target various molecular defects in the multistep pathways of specific cancers, called the ‘Multiplex RNA inhibition’ approach. For instance, for CLL, strategies to restore expression of miR-15/16 family may induce better therapeutic response by suppressing multiple members of the anti-apoptotic BCL2 family10. Finally, combining these strategies (miRNAs/anti-miRs combination ‘cocktails’) with conventional chemotherapy might afford synergistic effects and improved therapeutic efficiency for clinical cancer management.
Clinical considerations for miRNA-based therapy
miRNA-based therapies offer several advantages over the conventional chemotherapy. The most attractive quality is their ability to regulate multiple genes, often in the perspective of a common network, achieving a more cumulative effect on a set of related target proteins at multiple levels in the same pathway. Considering that cancer is a heterogeneous disease wherein distinct biological processes often get deregulated and hence cannot be treated by a single-agent therapy, this ability of miRNAs make them particularly valuable.
The small size and low molecular weight of miRNAs render them as attractive options for clinical drug development. Moreover, the discovery of more than 1500 human miRNA adds significantly to the pool of novel druggable targets amenable to pharmacological intervention in cancer management. In addition, since these occur naturally in human body, their intercellular presence does not evoke an immune response and reduces adverse side effects. Thus, restoring proteomic homeostasis by modifying miRNA expression in cancer cells may rewire the cell and reverse the cancer phenotype with minimal toxicity.
The major challenge to miRNA-based therapeutics is an effective mode of delivery. The biological instability of unmodified naked RNAs and AMOs due to rapid degradation by cellular and serum nucleases makes their direct delivery an unfeasible option59, 60. Additionally, their small size and negative charge could interfere with their ability to cross the cell membranes. Thus, they require some form of targeted delivery vehicle for sufficient tissue specificity and cellular uptake. Both viral and non-viral vehicles have been considered for RNA delivery (Figure 3).
Figure 3.
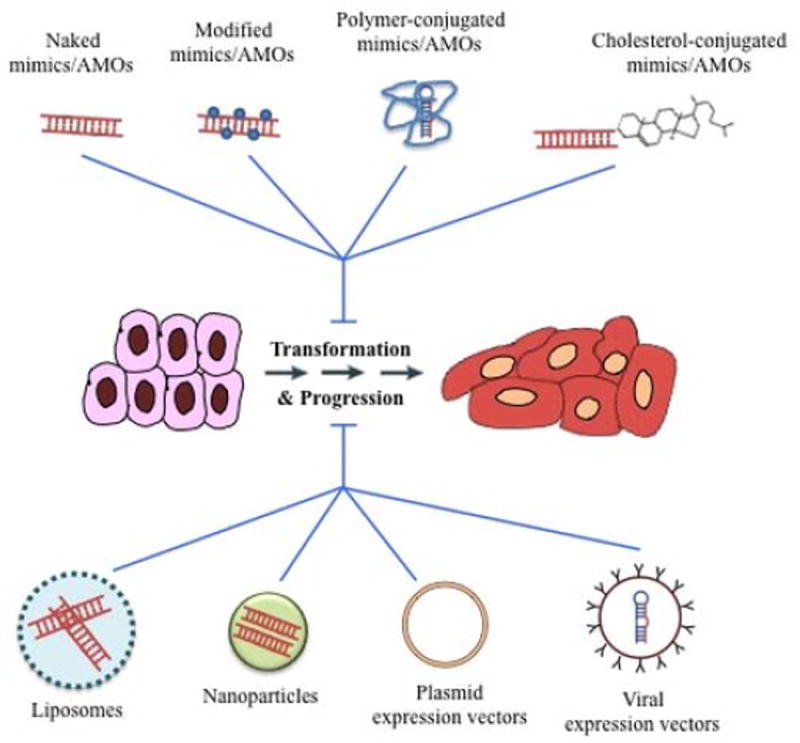
miRNA delivery options into the cells.
Schematic representation of different delivery approaches currently investigated for delivery of miRNA-based therapeutics in cancer cells. Synthetic miRNAs can be delivered naked with different chemical modifications, conjugated to natural or synthetic polymers, conjugated with cholesterol molecules, complexed with liposomes, encapsulated in nanoparticles, using DNA-plasmid expression vectors or via viral expression vectors.
The non-viral delivery options include chemical modifications of the oligonucleotides, lipid complexes with small molecules, liposomes, polymers, antibodies and nanoparticles. Chemical modifications to the primary structure of the RNA such as phosphorothioate backbone, cholesterol conjugation, peptide nucleic acid peptides or fluorine derivatives show improved stability and availability, but they are frequently associated with impaired function and severe toxicities61. Delivery of miRNA using a carrier agent has shown increased efficacy and stability with reduced toxicity. Use of cationic lipid nanoparticles to deliver pre-miR-107 in a preclinical model of head and neck squamous cell carcinoma showed 45.2% reduction in tumor burden compared to lipoplexes containing pre-miR controls62. Another study reported that co-administration of siRNAs against c-Myc/MDM2/VEGF and and miR-34a using liposome- polycation-hyaluronic acid (LPH) nanoparticle formulation modified with tumor targeting single chain antibody fragment successfully reduced lung metastasis load of B16F10 melanoma model63. To overcome the toxicity caused by the cationic nature of the liposomes, a neutral lipid emulsion was developed, and delivery of miR-34a and let-7a using this emulsion reduced lung tumor burden in non-small cell lung cancer64. However, neutral liposomes preferentially accumulate in lungs, hindering their application to treatment of other tumors64. Other research groups used a polyurethane-short branch polythylenimine (PU-PEI) as a carrier of miR-145 and demonstrated significant tumor inhibition in xenograft models of lung adenocarcinoma65 and glioblastomas66. Use of these synthetic carriers has become popular models of delivery due to improved stability, cellular uptake and biodistribution profile; however their toxicity and immunogenicity still remain a concern. Recently, another study reported that intravenously injection of modified exosomes with GE11 peptide on their surfaces delivered let-7a specifically to EGFR-expressing tissues in xenograft breast cancer models67. Since exosomes are natural vesicles secreted by several cells and tissues, they might not be immunogenic and thus present reduced toxicity. Another approach to targeted delivery is by tagging the modified oligonucleotides or carriers with antibodies. An additional challenge with this approach is their large molecular size which hampers the release of miRNA from the blood into the target tissue through the capillary endothelium68. In addition to synthetic polymeric materials, naturally occurring polymers such as chitosan, protamine and attelocollagen have also been explored; however, their utility is limited mainly by their immunogenicity69.
Viral options include the use of adeno-associated virus vectors or lentiviral vectors. Long-lasting and stable knockdown of target transcription can be achieved by expression of shRNAs from these vectors with either Pol II or Pol III promoters. However, toxicity and immunogenicity limit the clinical application of viral vectors. Furthermore, continuous expression of miRNAs can lead to saturation of the endogenous processing machinery and lead to toxic side effects70.
Local administration is advantageous since it needs lower RNA doses, causes minor toxic reactions and achieves better bioavailability of the drug to the target tissue71, 72. However, local administration can be useful only for solid tumors, since local distribution will be of no use in hematological malignancies like leukemia. Local administration also does not facilitate exposure of metastasizing cancer cells in circulation to the RNA drugs. Systemic delivery is thus a better route of administration since it provides a more effective biodistribution to the target tissues. Nonetheless, barriers to in vivo systemic delivery of miRNAs include renal clearance, failure to cross the capillary endothelium (for RNAs conjugated with large molecules), limited passage through extracellular matrix, destruction by scavenging macrophages, etc72.
Conclusion
Current times represent an important stage in the development of novel RNA-based therapeutic modalities for cancer treatment. The entry of two miRNA-based therapeutic agents in clinical trials should promote development of new miRNA-targeting anticancer drugs with improved specificity and efficacy for therapeutic application. Research efforts on improving their chemical designs and developing better delivery options to achieve superior sensitivity and specificity will be of crucial importance. Insights into the non-classical functions of miRNAs will help further expand their clinical application spectrum. miRNAs that upregulate protein translation can be targeted to control the translation of specific oncogenes or tumour suppressor genes. Furthermore, regulating the production, transportation and release of exosome miRNAs may have beneficial effects in controlling cancer development. Finally, more mechanistic studies to identify the interactions of miRNAs with human genome, transcriptome and proteome will enable us to comprehend the underlying intricate network of molecular regulation. miRNA-based therapy has the potential to bring an exciting new facet to personalized medicine for cancer treatment; however, a deeper and clearer understanding of its biology is warranted.
Acknowledgments
G.A.C. is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is also supported as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar and by the CLL Global Research Foundation. Work in G.A.C.’s laboratory is supported in part by the US National Institutes of Health (NIH)/US National Cancer Institute (NCI) grant CA135444; a US Department of Defense Breast Cancer Idea Award; Developmental Research Awards in breast cancer, ovarian cancer, brain cancer, prostate cancer, multiple myeloma and leukaemia (P50 CA100632) as well as head and neck cancer (P50 CA097007) from the Specialized Programs of Research Excellence (SPOREs); a Sister Institution Network Fund (SINF) grant from the MD Anderson Cancer Center and the German Cancer Research Center (DKFZ) in chronic lymphocytic leukaemia; a SINF grant in colorectal cancer, the Laura and John Arnold Foundation; the RGK Foundation; and The Estate of C. G. Johnson, Jr. M. Y. S. is a Rosalie B. Hite fellow.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 6.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 7.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–5. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 14.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callegari E, Elamin BK, Giannone F, Milazzo M, Altavilla G, Fornari F, Giacomelli L, D’Abundo L, Ferracin M, Bassi C, et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;6(3):1025–33. doi: 10.1002/hep.25747. [DOI] [PubMed] [Google Scholar]
- 16.Sandhu SK, Fassan M, Volinia S, Lovat F, Balatti V, Pekarsky Y, Croce CM. B-cell malignancies in microRNA Eμ-miR-17~92 transgenic mice. Proc Natl Acad Sci USA. 2013;110(45):18208–13. doi: 10.1073/pnas.1315365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enomoto Y, Kitaura J, Hatakeyama K, Watanuki J, Akasaka T, Kato N, Shimanuki M, Nishimura K, Takahashi M, Taniwaki M, et al. Eμ/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia. 2011;25(12):1849–56. doi: 10.1038/leu.2011.166. [DOI] [PubMed] [Google Scholar]
- 18.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 25.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–331. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 28.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 32.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 36.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 40.Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, Rossi S, Setien F, Casanovas O, Simo-Riudalbas L, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci USA. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 44.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotech. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–41. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 46.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 47.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. NEJM. 2013;368(18):1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 49.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nature Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, Sacco A, Liu Y, Ngo H, Quang P, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- 51.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–4. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 54.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah MY, Calin GA. The mix of two worlds: non-coding RNAs and hormones. Nucleic Acid Ther. 2013;23(1):2–8. doi: 10.1089/nat.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nature Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 58.Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, et al. Therapeutic Synergy between microRNA and siRNA in Ovarian Cancer Treatment. Cancer Discov. 2013;3(11):1302–15. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao X, Pan F, Holt CM, Lewis AL, Lu JR. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert Opin Drug Deliv. 2009;6:673–86. doi: 10.1517/17425240902992894. [DOI] [PubMed] [Google Scholar]
- 61.Vorhies JS, Nemunaitis J. Nonviral delivery vehicles for use in short hairpin RNA-based cancer therapies. Expert Rev Anticancer Ther. 2007;7:373–82. doi: 10.1586/14737140.7.3.373. [DOI] [PubMed] [Google Scholar]
- 62.Piao L, Zhang M, Datta J, Xie X, Su T, Li H, Teknos TN, Pan Q. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol Ther. 2012;20(6):1261–9. doi: 10.1038/mt.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18(9):1650–6. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–22. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiou GY, Cherng JY, Hsu HS, Wang ML, Tsai CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, et al. Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J Control Release. 2012;159(2):240–50. doi: 10.1016/j.jconrel.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Yang YP, Chien Y, Chiou GY, Cherng JY, Wang ML, Lo WL, Chang YL, Huang PI, Chen YW, et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 2012;33(5):1462–76. doi: 10.1016/j.biomaterials.2011.10.071. [DOI] [PubMed] [Google Scholar]
- 67.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–91. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18(5-6):282–9. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 70.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 71.Spizzo R, Rushworth D, Guerrero M, Calin GA. RNA inhibition, microRNAs, and new therapeutic agents for cancer treatment. Clin Lymphoma Myeloma. 2009;9(Suppl 3):S313–8. doi: 10.3816/CLM.2009.s.030. [DOI] [PubMed] [Google Scholar]
- 72.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–6. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 74.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]


