Abstract
Human parainfluenza virus type 3 (HPIV-3) is an airborne pathogen that infects human lung epithelial cells from the apical (luminal) plasma membrane domain. In the present study, we have identified cell surface-expressed nucleolin as a cellular cofactor required for the efficient cellular entry of HPIV-3 into human lung epithelial A549 cells. Nucleolin was enriched on the apical cell surface domain of A549 cells, and HPIV-3 interacted with nucleolin during entry. The importance of nucleolin during HPIV-3 replication was borne out by the observation that HPIV-3 replication was significantly inhibited following (i) pretreatment of cells with antinucleolin antibodies and (ii) preincubation of HPIV-3 with purified nucleolin prior to its addition to the cells. Moreover, HPIV-3 cellular internalization and attachment assays performed in the presence of antinucleolin antibodies and purified nucleolin revealed the requirement of nucleolin during HPIV-3 internalization but not during attachment. Thus, these results suggest that nucleolin expressed on the surfaces of human lung epithelial A549 cells plays an important role during HPIV-3 cellular entry.
Human parainfluenza virus type 3 (HPIV-3), belonging to the Paramyxoviridae family, is an enveloped, single-stranded, negative-sense virus that primarily infects lung epithelial cells of the airway (13, 42). Airborne infection by HPIV-3 not only manifests in disease states including pneumonia and bronchiolitis in infants but also causes high morbidity among immunocompromised adults (13, 42). HPIV-3 initiates infection following the engagement of its two envelope proteins, the hemagglutinin-neuraminadase (HN) and fusion (F) proteins, with the cell surface receptor(s) present on the plasma membrane of airway epithelia. It is evident that HN promotes the attachment function following its interaction with a cell surface sialic acid-containing receptor(s) (SAR). These initial interactions promote F-mediated fusion of the viral membrane with the cellular plasma membrane, leading to the penetration of the virus into the cells (1, 13, 42). Although F and HN proteins are critically required during the initial phases of virus entry, additional functions of these proteins during the life cycle of the virus have been reported. For example, HN, possessing neuraminadase activity, is also required for the efficient cell surface budding of HPIV-3 following cleavage of SAR (13, 31, 42). Moreover, homotypic coexpression of both HPIV-3 HN and HPIV-3 F proteins is required for cell-cell fusion and syncytium formation (32, 34). These findings suggest that the cellular receptor specificity of HPIV-3 envelope proteins may vary depending on the specific function of these proteins during the virus life cycle, i.e., entry, budding, and cell-cell fusion.
Although the envelope proteins of HPIV-3 are capable of performing various functions during the viral life cycle, HN and F are primarily required during cellular entry of HPIV-3. It is well documented that cell surface SAR serves as the initial attachment receptor for HPIV-3 following its interaction with HN. It was recently demonstrated that, apart from the SAR, cell surface heparan sulfate (HS) is also required for the efficient cellular entry of HPIV-3 in human lung epithelial A549 cells (9). Moreover, it is speculated that an additional non-SAR and/or non-HS cell surface molecule(s) may also serve as a secondary receptor(s) for HPIV-3, since (i) HN of HPIV-3 uses specific SAR and does not indiscriminately bind to all sialic acid-containing molecules on the cell surface (49), (ii) HPIV-3 cellular entry was not completely abolished in the absence of cell surface sialic acid molecules (46, 47, 48), (iii) complete inhibition of HPIV-3 entry did not occur in cells lacking HS (9), (iv) a recombinant HPIV-3 lacking the neuramindase activity was capable of entering the cells (56, 57), and (v) previous studies (8) on the mechanism of HPIV-3 entry and budding in polarized human lung epithelial A549 cells have revealed preferential utilization of the apical plasma membrane domain by HPIV-3 for these processes, thus demonstrating that the apical plasma membrane domain of lung epithelial cells preferentially expresses the cell surface molecule(s) utilized by HPIV-3 to gain entry into the cells. Thus, these studies have suggested that specific sialyated/nonsialyated and/or nonproteoglycan cell surface molecule(s) may act as the primary and/or secondary entry receptor(s) for HPIV-3. Moreover, the majority of studies (1, 47, 48, 49, 56, 67) dealing with the mechanism of HPIV-3 cellular entry and fusion were performed with nonepithelial cells such as HeLa, LLC-MK2 and CV-1 cells, cells that are not of lung origin. Since viruses are capable of utilizing different sets of molecules depending on the cell type during the entry process, we investigated whether any nonproteoglycan and/or non-SAR molecule is involved in the entry of HPIV-3 into human lung epithelial cells, the cells that are the primary target of HPIV-3 during the normal course of infection via the airway.
In the present study, we have identified cell surface nucleolin as an additional cell surface molecule that is required for the efficient entry of HPIV-3 into human lung epithelial A549 cells. Nucleolin is a multifunctional protein (65) that shuttles between nucleus and cytoplasm and has been reported to be expressed on the surfaces of various cells (38, 41, 62), acting as a receptor for various ligands, including lipoproteins (61), cytokines, growth factors (11, 60, 68), the extracelullar matrix (18, 29, 39), bacteria (63), and viruses (17, 33, 51, 52, 53, 54). We demonstrate that, similar to these functions of cell surface nucleolin, nucleolin is expressed on the surfaces of A549 cells with preferential enrichment on the apical plasma membrane domain and that its interaction with HPIV-3 envelope protein(s) leads to the efficient cellular entry of HPIV-3.
MATERIALS AND METHODS
Virus and cells.
HPIV-3 and vesicular stomatitis virus (VSV) (Indiana serotype, Mudd-Summers strain) were propagated in CV-1 and BHK-21 cells, respectively, as described previously (9, 10). Human lung epithelial A549, BHK-21, and CV-1 cells were maintained in Dulbecco modified Eagle medium (GIBCO-BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum, penicillin, streptomycin, and glutamine. The viral titer was monitored by plaque assay analysis with CV-1 (for HPIV-3) or L929 (for VSV) cells as described previously (9, 10, 55). [35S]methionine (Perkin-Elmer, Wellesley, Mass.) labeling of HPIV-3 was essentially performed as described previously (8). HPIV-3 and VSV were also biotinylated as described previously (35, 50). Briefly, 1 mg of purified HPIV-3 and VSV pellet was dissolved in sodium bicarbonate buffer (pH 8.6) containing 150 mM NaCl and 150 μg of N-hydroxysuccinimidobiotin (NHS-Biotin; Pierce, Rockford, Ill.)/ml. The mixture was incubated for 2 h on a rotor at 4°C. Following incubation, the biotinylation reaction was quenched by adding Tris-HCl (pH 8.8) at a final concentration of 50 mM. The biotinylated virus pellet was obtained following centrifugation (110,000 × g) at 36 K for 2 h, and 1× phosphate-buffered saline (PBS) was added to the virus pellet.
VOPBA.
Total cell lysates obtained from A549 cells and fractionated on an anion exchange column were used for a virus overlay protein binding assay (VOPBA) (5, 23, 25, 37, 45, 71). The A549 cells were lysed as described previously (14). The lysate (15 mg of protein) was subjected to fast protein liquid chromatography with an anion exchange column (Hi trap-Q column; Pharmacia-Amersham, Uppsala, Sweden) on an AKTA purifier (Pharmacia-Amersham). The proteins were eluted with a linear gradient from 20 mM to 1 M NaCl as 500-μl fractions. The eluted fractions were concentrated with Centricon (Millipore, Billerica, Mass.), and a portion of each fraction was used for VOPBA. For VOPBA (5), the fractions were subjected to sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-7.5% PAGE) analysis and the separated proteins were transferred to the nitrocellulose membrane. Following transfer, the membrane was washed once with 1× PBS (10 min, room temperature [RT]), and 4% bovine serum albumin (BSA) in 1× PBS was added to the blot for overnight incubation at 4°C. The membrane was then incubated with 5% nonfat dry milk in 1× PBS (1 h, RT) and washed three times with 1× PBS (10 min each, RT). 35S-HPIV-3 (5 million cpm) was added to the blot in the presence of minimal essential medium containing 10% fetal bovine serum. Following incubation for 12 h at 4°C, the blot was washed four times with 2% BSA in 1× PBS (15 min each, RT) and twice with 1× PBS containing 0.1% NP-40 (15 min each, RT). The dried blot was then subjected to fluorography. In addition, VOPBA was also carried out by adding 35S-HPIV-3 following preincubation of the blot with 20-fold excess nonradioactive (cold) HPIV-3.
Protein sequencing.
The eluted fractions containing the 35S-HPIV-3 interacting 110-kDa protein band (as deduced by VOPBA) were used for amino-terminal sequence analysis. These fractions were subjected to SDS-7.5% PAGE, and the separated proteins were transferred to the polyvinylidene difluoride (PVDF) membrane. Following transfer, the PVDF membrane was stained with Coomassie blue and the 110-kDa band was excised from the membrane for amino-terminal sequencing by the Edman degradation method with an Applied Biosystems Procise automated sequencer (model 492).
Cell surface and domain-specific biotinylation.
For cell surface biotinylation (43), A549 cells chilled at 4°C for 2 h were washed once with chilled 1× PBS, and NHS-Biotin (Pierce) (250 μg/ml) was added to the washed cells in the presence of cold 1× PBS. The cells were incubated with biotin at 4°C for 90 min with the addition of fresh biotin after the first 45 min. The biotinylation reaction was quenched following the washing of the cells (four times, 10 min each, 4°C) with 1× PBS (cold) containing 100 mM glycine. The cells were then washed twice with 1× PBS and lysed. Avidin conjugated to agarose beads was added to the cell lysates, and, following 8 h of incubation at 4°C, the beads were washed extensively with 1× PBS and SDS-PAGE sample buffer was directly added to the washed beads for Western blot analysis (see below) with antinucleolin or anti-β-catenin antibodies. For the domain-specific biotinylation assay (7), A549 cells were grown on filter inserts (0.45-μm-pore-size MCE membrane; Millipore) for the formation of a polarized monolayer (8). The polarized cells were chilled, biotin was added either from the upper (apical) or the lower (basolateral) chamber, and the reaction was performed as described above. The cell lysates prepared from cells treated with biotin from either the apical or the basolateral side were subjected to precipitation with avidin-agarose and to Western blot analysis as described above.
Purification of nucleolin and β-catenin.
Human nucleolin was purified from Hep-2 cells as described previously (63). Nucleolin was purified to homogeneity as deduced by Coomassie blue staining (data not shown) (63). The recombinant histidine (His)-tagged β-catenin (a gift from Pierre D. McCrea, University of Texas M. D. Anderson Cancer Center, Houston, Tex.) cloned into the baculovirus vector was used to infect Hi-5 cells. The infected cell extract was subjected to nickel-agarose affinity chromatography, and the bound His-β-catenin was eluted by imidazole as described previously (70). The eluted β-catenin was homogeneous, as determined by Coomassie blue staining (data not shown).
Interaction of biotinylated HPIV-3 with nucleolin.
Biotinylated HPIV-3 (2 multiplicities of infection [MOI]) was added to A549 cells, and following 2 h of adsorption at 37°C the cells were washed extensively with 1× PBS and the cell lysate was subjected to immunoprecipitation with antinucleolin or anti-β-catenin antibody in the presence of protein A-Sepharose beads (Amersham-Pharmacia) as described previously (10). Following 12 h of incubation at 4°C, the beads were washed, the bound proteins were subjected to SDS-7.5% PAGE, and the separated proteins were transferred to the nitrocellulose membrane. The membrane was blotted with avidin conjugated to horseradish peroxidase (avidin-HRP) (Pierce) to visualize the biotinylated envelope proteins of HPIV-3. Similarly, the cell lysates obtained from the biotinylated HPIV-3-treated (incubated at 37 or 4°C) or VSV-treated (incubated at 37°C) cells were precipitated with avidin-agarose as described above and the bound proteins were subjected to Western blot analysis with antinucleolin antibody.
Western blot analysis.
Western blot analysis was performed essentially as described previously (8, 9, 10). Following SDS-7.5% PAGE, the proteins transferred to the nitrocellulose membrane were blocked with 1× PBS containing 10% nonfat dry milk (blocking buffer) (2 h, RT). The various primary antibodies, including antinucleolin (monoclonal) (Santa Cruz, Santa Cruz, Calif.), anti-β-catenin (monoclonal) (Sigma-Aldrich, St. Louis, Mo.), anti-HPIV-3 RNP (polyclonal, recognizes HPIV-3 N protein), antihemagglutinin (anti-HA; monoclonal) (Sigma-Aldrich), and anti-VSV P protein antibodies (polyclonal) were added to the blot in the presence of the blocking buffer and incubated for 12 h at 4°C. The blots were then washed with 1× PBS containing 0.05% NP-40 and incubated with the secondary antibodies (anti-rabbit or -mouse HRP [Santa Cruz]) for 2 h at RT. The washed blots were processed for enhanced chemiluminescence according to the manufacturer's protocol (Amersham-Pharmacia).
Transfection of HA-nucleolin and F and HN proteins.
HA-tagged human nucleolin (HA-nucleolin) subcloned into pcDNA3 (Invitrogen, Carlsbad, Calif.) (a gift from Nancy Meizel, University of Washington, Seattle, Wash.) and empty pcDNA3 vectors were used for transfecting A549 cells grown to 70 to 80% confluence. The cDNAs (1.5 μg/ml) were transfected by using Lipofectin (GIBCO-BRL) as described previously (10, 16). Forty-eight hours posttransfection, cells were lysed and the lysate was subjected to Western blot analysis with anti-HA antibody. The lysates were also immunoprecipitated with anti-HA antibody conjugated to the agarose beads (Sigma-Aldrich). The bound proteins were subjected to VOPBA with 35S-HPIV-3 in the absence or presence of excess cold HPIV-3 as described above. FLAG-tagged HPIV-3 F and HN proteins (FLAG-F and FLAG-HN, respectively) subcloned into the pGEM4 vector were used to express these proteins in A549 cells. Since the pGEM4 vector possesses the T7 polymerase promoter site, T7 polymerase was expressed in trans prior to transfection by infecting A549 cells (10 MOI) with a recombinant vaccinia virus (MVA-T7) expressing T7 polymerase. Following 2 h of infection with MVA-T7, cells were transfected with either F or HN cDNA (1 μg/ml) by using Lipofectin (15, 16). An empty vector was also transfected as a control. Thirty-six hours posttransfection, the cells were incubated with methionine-free medium (GIBCO-BRL) for 3 h and the cells were labeled with [35S]methionine (300 μCi/ml) for 4 h at 37°C. The cell lysates were immunoprecipitated with anti-FLAG antibody conjugated to the agarose beads (Sigma-Aldrich). The proteins bound to the washed beads were subjected to SDS-7.5% PAGE and fluorography as described previously (15, 16).
Coimmunoprecipitation of nucleolin with HPIV-3 envelope proteins.
For the coimmunoprecipitation analysis, cells were transfected with FLAG-F and FLAG-HN individually or together as detailed above. An empty vector was also transfected as a control. Thirty-six hours posttransfection, the cells were lysed and the lysates were immunoprecipitated with anti-FLAG-agarose. Following 12 h of incubation at 4°C in a rotor, the beads were washed extensively and the bound proteins were subjected to SDS-7.5% PAGE and Western blot analysis with antinucleolin antibody.
Virus infection in the presence of antinucleolin antibodies and purified nucleolin.
The effects of nucleolin antibodies and purified nucleolin on HPIV-3 and VSV infection were studied by using A549 cells. For antibody-blocking experiments, A549 cells were incubated with polyclonal (Santa Cruz) or monoclonal (MBL, Watertown, Mass.) nucleolin antibodies (25 and 50 μg/ml) or control polyclonal β-catenin (Sigma-Aldrich) antibody (70 μg/ml) for 1 h at 37°C. Following antibody incubation, HPIV-3 or VSV (0.2 MOI) was added to the cells and the adsorption was continued for 2 h at 37°C. The cells were then washed to remove unbound viruses, and the infection was continued for an additional 36 h. Thirty-six hours postinfection, the cell lysates were used for Western blot analysis with either anti-HPIV-3 RNP or anti-VSV P protein antibodies to monitor the intracellular HPIV-3 N or VSV P protein levels. In addition, the culture supernatants were collected to measure virus yield by plaque assay of CV-1 or L929 cells. The effect of purified nucleolin on virus infection was investigated following incubation of VSV or HPIV-3 (0.2 MOI) with either purified nucleolin (5, 15, and 30 nM) or purified β-catenin (50 nM) for 3 h at room temperature. The preincubated virus was then added to the A549 cells, and adsorption was carried out for 2 h at 37°C. Following adsorption, the cells were washed to remove unbound viruses and infection was continued for an additional 36 h. The infectivity was monitored by Western blot analysis and the plaque assay as described above. The efficiency of virus infection (based on the plaque assay results), expressed as percentage of infection, was calculated based on a ratio that is detailed in the figure legends. This method of calculating the percentage of HPIV-3 infection efficiency was previously utilized in several studies (8, 9, 26, 55).
Attachment and internalization of 35S-HPIV-3 in the presence of nucleolin antibodies and purified nucleolin.
The 35S-HPIV-3 attachment and internalization assays were performed as described previously (8). The kinetics of 35S-HPIV-3 attachment was studied by adding 0.25 to 2 MOI of 35S-HPIV-3 (1 × 105 to 8 × 105 cpm) to chilled A549 cells. After 2 h of incubation at 4°C (the temperature that supports attachment but not internalization), the cells were washed extensively with chilled 1× PBS, the washed cells were lysed, and the lysate radioactivity representing the attached 35S-HPIV-3 was counted with a scintillation counter. For the attachment assay in the presence of nucleolin antibodies, A549 cells were chilled at 4°C for 2 h, followed by the addition of nucleolin and control antibodies. The cells were incubated with the antibodies for 2 h at 4°C, and 35S-HPIV-3 (1 MOI) (4 × 105 cpm) was added to these cells for attachment at 4°C. After 2 h of incubation, the cells were washed extensively with chilled 1× PBS, the washed cells were lysed, and the lysate radioactivity representing the attached 35S-HPIV-3 was counted with a scintillation counter. Similarly, 35S-HPIV-3 preincubated with purified nucleolin or β-catenin was added to chilled A549 cells for the attachment assay.
For the internalization assay (8, 9), chilled A549 cells were incubated with the antibodies for 2 h at 4°C and 35S-HPIV-3 (1 MOI) (4 × 105 cpm) was added to the cells for attachment at 4°C. Following attachment for 2 h, the cells were washed extensively with 1× PBS and the temperature was shifted to 37°C to allow internalization of the attached virus. At 0.5, 1, and 2 h postinternalization, cells were washed extensively with 1× PBS and trypsinized for 15 min at 37°C to remove cell surface-attached viruses as described previously (8). The protease activity was neutralized with complete Dulbecco modified Eagle medium, and the cells were washed twice with 1× PBS. The washed cells were lysed, and the lysates representing the internalized 35S-HPIV-3 were counted (counts per minute) with a scintillation counter. A similar internalization assay was performed with 35S-HPIV-3 preincubated with either purified nucleolin or β-catenin.
For the assays described above, the background counts were determined by counting cell lysates obtained from cells not incubated with 35S-HPIV-3 and the background was subtracted from all of the counts derived from cells incubated with the radiolabeled virus. The efficiency of internalization or attachment, expressed as a percentage of internalization or attachment, was calculated based on a ratio that is detailed in the applicable figure legend.
RESULTS
Identification of nucleolin as an HPIV-3 binding protein.
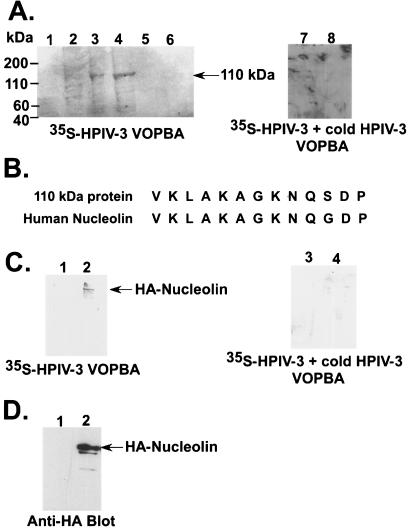
In order to identify proteins that interact with the envelope proteins of HPIV-3, VOPBA was performed with total protein extracts from A549 cells. This technique was utilized previously to identify receptor proteins for several viruses, including encephalomyocarditis virus, mouse hepatitis virus, and cytomegalovirus (23, 25, 37, 45, 71). Prior to VOPBA, total protein extracts obtained from A549 cells were subjected to anion exchange chromatography. The column-eluted fractions were subjected to VOPBA with 35S-HPIV-3. For VOPBA (5), the fractions were subjected to SDS-PAGE and the separated proteins were transferred to the nitrocellulose membrane. The proteins transferred to the membrane were partially renatured overnight at 4°C in the presence of BSA. The blot was then incubated with 35S-HPIV-3 for 12 h at 4°C, and, finally, the washed blot was subjected to fluorography. As shown in Fig. 1A, VOPBA revealed a prominent protein band of 110 kDa in two fractions (lanes 3 and 4), suggesting the interaction of HPIV-3 with a 110-kDa protein from A549 cells. The specificity of this interaction was borne out by the observation that 35S-HPIV-3 failed to interact with the 110-kDa band (Fig. 1A, lanes 7 and 8) in the presence of 20-fold excess cold (nonradiolabeled) HPIV-3 following VOPBA analysis of the active fractions (Fig. 1A, lanes 3 and 4) containing the 35S-HPIV-3 interacting 110-kDa band. In order to identify the 110-kDa protein, the relevant fractions were subjected to SDS-PAGE and the separated proteins were transferred to the PVDF membrane for amino-terminal sequencing. It is important to note that the 110-kDa protein fractions also contained an additional three to four proteins of lower molecular weights, as visualized by Coomassie blue staining (data not shown). However, HPIV-3 specifically interacted with the 110-kDa protein and the 110-kDa protein band was well separated from the other proteins. Moreover, the amino-terminal sequencing of the 110-kDa band revealed that a single protein is represented. The amino-terminal sequencing identified the 110-kDa band as human nucleolin (Fig. 1B).
FIG. 1.
Identification of nucleolin as an HPIV-3 envelope binding protein. (A) VOPBA of A549 protein fractions eluted from the anion exchange column with 35S-HPIV-3 in the absence (lanes 1 to 6) and presence (lanes 7 and 8) of excess nonradioactive (cold) HPIV-3. The 35S-HPIV-3 interacting 110-kDa protein band is marked. (B) Comparison of the amino-terminal primary sequence of human nucleolin with the sequence of the 110-kDa protein. (C) 35S-HPIV-3 VOPBA in the absence (lanes 1 and 2) and presence (lanes 3 and 4) of excess nonradioactive (cold) HPIV-3 was performed by using anti-HA immunoprecipitated cell lysates obtained following transfection with HA-nucleolin (lanes 2 and 4) or an empty vector (lanes 1 and 3). (D) Western blot analysis of cell lysates (10 μg of protein) obtained from cells transfected with HA-nucleolin (lane 2) or an empty vector (lane 1) with anti-HA antibody.
To further confirm our VOPBA results, we next transfected HA-nucleolin in A549 cells, and the expressed HA-nucleolin was immunoprecipitated by anti-HA antibody conjugated to agarose beads and the bound proteins were subjected to VOPBA analysis with 35S-HPIV-3. As shown in Fig. 1C, while HPIV-3 interacted with HA-nucleolin (lane 2), no interaction was observed following the immunoprecipitation of control (transfected with an empty plasmid) cell lysates with anti-HA-agarose (lane 1). The specificity of the interaction of HPIV-3 with HA-nucleolin was borne out by the observation that incubation of the VOPBA blot with 20-fold excess cold (nonradioactive) HPIV-3 prior to the addition of 35S-HPIV-3 resulted in a loss of interaction of 35S-HPIV-3 with HA-nucleolin (Fig. 1C, lane 4). The intracellular expression of HA-nucleolin under the above-mentioned experimental condition was tested by Western blot analysis of transfected cell lysates with anti-HA antibody. As shown in Fig. 1D, Western blot analysis revealed abundant expression of transfected HA-nucleolin (lane 2), while no protein was detected in control cells (lane 1). These results suggest that HPIV-3 envelope proteins interact with nucleolin expressed in A549 cells.
Nucleolin is expressed on the apical plasma membrane domain of human lung epithelial A549 cells.
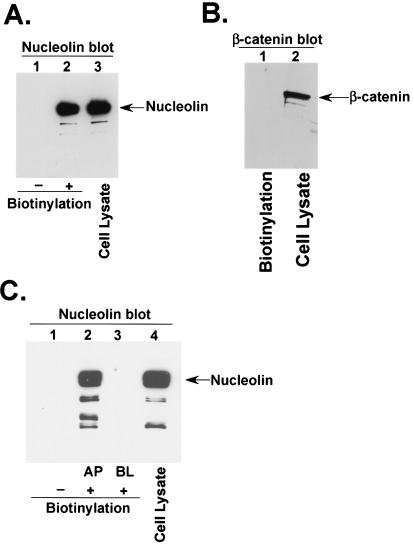
Based on the ability of HPIV-3 virion particles to interact with nucleolin, we hypothesized that nucleolin may play a role during HPIV-3 cellular entry. If indeed nucleolin serves as an entry cofactor for HPIV-3 in A549 cells, it should be expressed on the cell surface. Therefore, to investigate the plasma membrane expression levels of nucleolin in A549 cells, a cell surface biotinylation assay (43) was performed as described in Materials and Methods. Following biotinylation, the biotinylated and nonbiotinylated A549 cell lysates were precipitated with avidin-agarose and the bound proteins were subjected to Western blot analysis with antinucleolin or anti-β-catenin (control) antibodies. As shown in Fig. 2A, a significant amount of nucleolin was expressed on the A549 plasma membrane (lane 2), while no protein was observed in nonbiotinylated lysates (lane 1). In contrast to the results for nucleolin, control experiments revealed the absence of β-catenin, an intracellular protein (70), in biotinylated A549 lysates (Fig. 2B, lane 1). It is important to note that approximately 5% of the total cellular nucleolin was expressed on the cell surface, since 100 μg of biotinylated cell lysate (Fig. 2A, lane 2) possessed amounts of nucleolin protein similar to those possessed by 4 μg of total cellular lysate (Fig. 2A, compare lanes 2 and 3).
FIG. 2.
Cell surface expression of nucleolin. (A) Nonbiotinylated (lane 1) and biotinylated (lane 2) A549 cell lysates (100 μg of protein) were subjected to precipitation with avidin-agarose and Western blot analysis with antinucleolin antibody. A549 cell lysates (lane 3) (4 μg of protein) served as a control. (B) Biotinylated A549 cell lysates (lane 1) (200 μg protein) were subjected to precipitation with avidin-agarose and Western blot analysis with anti-β-catenin antibody. A549 cell lysates (lane 2) (5 μg of protein) served as a control. (C) Nonbiotinylated (lane 1) and biotinylated cell lysates (250 μg protein) obtained following biotinylation from either the apical (AP) (lane 2) or the basolateral (BL) (lane 3) side of a filter-grown polarized monolayer of A549 cells were subjected to precipitation with avidin-agarose and Western blot analysis with antinucleolin antibody. Cell lysates (lane 4) (12 μg of protein) from polarized A549 cells served as a control.
Once it was established that nucleolin is expressed on the surfaces of A549 cells, we studied the polarized expression (the apical versus the basolateral plasma membrane domains) of nucleolin in A549 cells, since earlier studies demonstrated that the apical pole of A549 cells is preferentially utilized by HPIV-3 during entry (8). Domain-specific biotinylation (apical or basolateral) (7) of A549 cells grown on filter inserts was essentially performed as described previously (7). Briefly, biotin was added to either the upper (apical) or the lower (basolateral) chamber of A549 filter inserts. The cell lysates obtained from these cells were precipitated with avidin-agarose, and the avidin-bound proteins were subjected to Western blot analysis with antinucleolin antibody. As shown in Fig. 2C, a significantly high level of cell surface nucleolin was observed in the apical (lane 2) plasma membrane domain of A549 cells compared to that observed in the basolateral pole (lane 3), which failed to express nucleolin. The control experiments revealed the absence of protein bands in nonbiotinylated cell lysates (Fig. 2C, lane 1). These results demonstrate not only the presence of nucleolin on the A549 cell surface but also its preferential enrichment on the apical plasma membrane domain, the domain utilized by HPIV-3 during entry into the polarized epithelial cells. Thus, these observations, coupled with the results obtained following VOPBA (Fig. 2A and C), suggest that the envelope proteins of HPIV-3 virion particles may interact with cell surface-expressed nucleolin, probably during viral entry.
Interaction of HPIV-3 with nucleolin during cellular entry.
In order to establish a possible role of nucleolin during HPIV-3 entry, we next examined whether HPIV-3 is capable of interacting with cell surface-expressed nucleolin during the adsorption stage. For these studies, purified HPIV-3 was biotinylated as described in Materials and Methods and the plaque assay analysis revealed that biotinylated HPIV-3 was as infectious as the untreated virus, indicating that the biotinylation reaction had no adverse affect on virus infectivity (data not shown). Biotinylated HPIV-3 was added to A549 cells, and the virus was allowed to adsorb for 2 h at 37°C. Following adsorption, the cells were washed and the cell lysates were immunoprecipitated with antinucleolin or β-catenin (control) antibodies and the proteins bound to the Sepharose-antibody beads were subjected to Western blot analysis with avidin-HRP (to detect biotinylated HPIV-3). As shown in Fig. 3A, antinucleolin (lane 2) but not the control β-catenin antibody (lane 1) specifically precipitated biotinylated envelope proteins of HPIV-3. These results suggest that HPIV-3 interacts with nucleolin, possibly during the early stages of the virus life cycle. The biotinylation efficiency of the HPIV-3 envelope proteins was clearly visible, since avidin-HRP blot analysis (Fig. 3A, lane 3) of biotinylated HPIV-3 detected two major proteins, namely, the F and HN proteins. These results are consistent with the biotinylation reaction being specifically restricted to only the extracellular domain of the plasma membrane and virus envelope proteins.
FIG. 3.
Interaction of nucleolin with biotinylated HPIV-3. (A) Lysates obtained from A549 cells incubated with biotinylated HPIV-3 (Biotin-HPIV-3) (2 MOI) at 37°C were immunoprecipitated with either β-catenin (lane 1) or nucleolin (lane 2) antibodies. The bound proteins were then subjected to SDS-7.5% PAGE and blotting with avidin-HRP. Biotinylated HPIV-3 (lane 3) served as a control to demonstrate the biotinylated envelope proteins F and HN. (B) Lysates obtained from A549 cells incubated in the absence (lane 1) or presence (lane 2) of biotinylated HPIV-3 (Biotin-HPIV-3) (2 MOI) at 37°C were precipitated with avidin-agarose. The bound proteins were then subjected to Western blotting with antinucleolin antibody. A549 cell lysate (lane 3) served as a control. (C) Lysates obtained from A549 cells incubated in the absence (lane 1) or presence (lane 2) of biotinylated VSV (Biotin-VSV) (2 MOI) at 37°C were precipitated with avidin-agarose. The bound proteins were then subjected to Western blotting with antinucleolin antibody. A549 cell lysate (lane 3) served as a control. (D) Lysates obtained from A549 cells incubated in the absence (lane 1) or presence (lane 2) of biotinylated HPIV-3 (Biotin-HPIV-3) (2 MOI) at 4°C were precipitated with avidin-agarose. The bound proteins were then subjected to Western blotting with antinucleolin antibody. A549 cell lysate (lane 3) served as a control.
The results presented in Fig. 3A were further confirmed by precipitating the cell lysates obtained from A549 cells adsorbed with biotinylated HPIV-3 for 2 h at 37°C with avidin-agarose. The avidin-bound proteins were subjected to Western blot analysis with antinucleolin antibody. As shown in Fig. 3B, nucleolin was specifically precipitated by avidin (lane 2), suggesting an interaction of nucleolin with the biotinylated HPIV-3 envelope proteins early during infection. Nucleolin was not detected in cell lysates obtained from uninfected cells (Fig. 3B, lane 1), and the A549 cell lysate served as a control (Fig. 3B, lane 3). The specificity of interaction between nucleolin and the HPIV-3 envelope proteins was further borne out by the observation that biotinylated VSV (an enveloped RNA virus possessing G protein as its envelope protein) (59) failed to bind with nucleolin under experimental conditions similar to those utilized for HPIV-3 (Fig. 3C). Moreover, we failed to detect interaction of biotinylated HPIV-3 with nucleolin during the attachment stage of virus entry, since the incubation of biotinylated HPIV-3 with A549 cells at 4°C (the temperature that restricts virus fusion and internalization) resulted in a lack of interaction of nucleolin with the envelope proteins (Fig. 3D). These results suggest that cell surface nucleolin interacts with the HPIV-3 envelope proteins during cellular entry of the virus. Moreover, this interaction seems to occur at a postattachment stage, probably during the fusion and/or internalization process.
HPIV-3 F protein, but not HN, interacts with nucleolin.
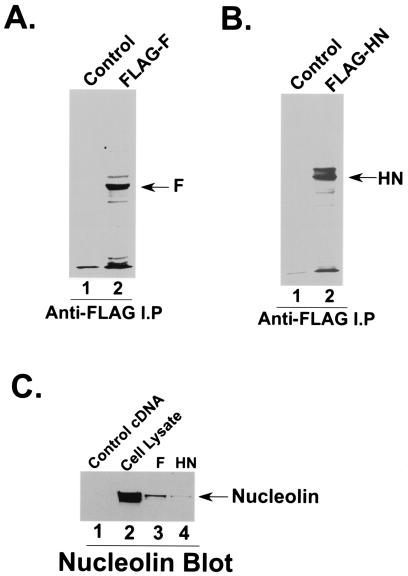
Since nucleolin interacts with the HPIV-3 envelope proteins, we next investigated whether F and/or HN binds to nucleolin in A549 cells. In order to study such interactions, A549 cells were transfected individually with FLAG-F or FLAG-HN or an empty vector (control). Following transfection, cells were labeled with [35S]methionine and the radiolabeled cell lysates were immunoprecipitated with anti-FLAG-agarose beads. The radiolabeled proteins bound to the washed beads were then subjected to SDS-PAGE analysis and fluorography. As shown in Fig. 4A and B, comparable amounts of FLAG-F (Fig. 4A, lane 2) and FLAG-HN (Fig. 4B, lane 2) were detected in A549 cells, while no proteins were detected in cells transfected with a control vector (Fig. 4A and B, lane 1).
FIG. 4.
Interaction of nucleolin with HPIV-3 F and HN proteins. (A) A549 cells transfected with either an empty vector (lane 1) or HPIV-3 FLAG-F cDNA (lane 2) were pulse labeled with [35S]methionine, and the radioactive lysate was immunoprecipitated with anti-FLAG antibody prior to SDS-7.5% PAGE and fluorography. (B) A549 cells transfected with either an empty vector (lane 1) or HPIV-3 FLAG-HN cDNA (lane 2) were pulse labeled with [35S]methionine, and the radioactive lysate was immunoprecipitated with anti-FLAG antibody prior to SDS-7.5% PAGE and fluorography. (C) Lysates (100 μg of protein) obtained from A549 cells transfected with either an empty vector (lane 1), HPIV-3 FLAG-F (lane 3) or HPIV-3 FLAG-HN (lane 4) cDNA were immunoprecipitated with anti-FLAG antibody. The proteins bound to the washed anti-FLAG-agarose beads were subjected to Western blot analysis with antinucleolin antibody. An A549 cell lysate (lane 2) (20 μg of protein) served as a control.
Once the expression of F and HN in A549 cells had been confirmed, we examined the possible interaction of these proteins with nucleolin. Cell lysates obtained from A549 cells transfected with either an empty vector, FLAG-F, or FLAG-HN were immunoprecipitated with anti-FLAG-agarose, and the bound proteins were subjected to Western blot analysis with antinucleolin antibody. As shown in Fig. 4C, the HPIV-3 F protein specifically interacted with nucleolin (lane 3), while the interaction of HN with nucleolin was barely detectable (lane 4). No detectable nucleolin protein was present in cells transfected with an empty vector (Fig. 4C, lane 1), and A549 cell lysate served as a control (Fig. 4C, lane 2). In addition, coexpression of both F and HN resulted in the detection of nucleolin at amounts comparable to what was observed following the expression of F alone (data not shown). The specificity of the interaction of nucleolin with HPIV-3 F protein was borne out by the observation that nucleolin failed to interact with HPIV-3 nucleocapsid protein (N protein, a nonenvelope protein of HPIV-3 involved in genome RNA encapsidation) (13) when it was expressed in A549 cells (data not shown). These results demonstrate that nucleolin is capable of interacting specifically with the F protein of HPIV-3, suggesting a possible role of nucleolin during HPIV-3 fusion and internalization.
Inhibition of HPIV-3 infection by antinucleolin antibodies.
In order to obtain evidence for a functional role of nucleolin during the entry and replication process of HPIV-3, the infection of A549 cells was performed following incubation of these cells with antinucleolin antibodies. A549 cells were incubated with nucleolin polyclonal and monoclonal antibodies (25 and 50 μg/ml) or control (β-catenin polyclonal) antibodies (70 μg/ml) for 1 h at 37°C. Following 1 h of incubation, HPIV-3 was added to the cells; following 2 h of adsorption at 37°C, the cells were washed extensively to remove unbound viruses. The infection was then continued for 36 h at 37°C, and the medium supernatants were subjected to plaque assay analysis. As shown in Fig. 5A, both polyclonal and monoclonal nucleolin antibodies at a concentration of 50 μg/ml inhibited HPIV-3 replication significantly, while control antibody at a concentration of 70 μg/ml failed to inhibit virus replication. It is important to note that using higher amounts of nucleolin antibodies (a concentration of more than 50 μg/ml) did not result in significantly increased inhibition in virus replication (data not shown). The value obtained from the plaque assay result (from Fig. 5A) was utilized to demonstrate the extent of inhibition of HPIV-3 infection (the percentage of infection inhibition) by nucleolin antibodies (Fig. 5B). The plaque assay results were further confirmed by Western blot analysis of A549 cell lysates with anti-HPIV-3 RNP antibody. As shown in Fig. 5C, significant reductions in HPIV-3 N protein amounts were visualized for cells treated with nucleolin antibody (lane 4) compared to the untreated (lane 2) or control antibody-treated (lane 3) cells. It is important to note that although nucleolin antibodies inhibited HPIV-3 replication significantly, they failed to completely abolish HPIV-3 infection, thus suggesting a role of nucleolin in efficient HPIV-3 infection.
FIG. 5.
Effect of nucleolin antibodies (Ab) on virus replication. (A) Culture supernatants collected from A549 cells mock infected or infected with HPIV-3 (0.2 MOI) in the absence or presence of nucleolin (Nuc) polyclonal (Poly) and monoclonal (Mono) antibodies or β-catenin polyclonal antibody (control) were added to CV-1 cells for a plaque assay. The plaque assay results, reflecting the viral titers, are expressed in PFU per milliliter. Each value represents the mean ± standard deviation for three determinations. (B) The average plaque assay values (in PFU per milliliter) from panel A were used to show the percentages of inhibition of infection in the presence of nucleolin antibodies. The percentage of infection, reflecting the percentage of virus release, was calculated as a ratio of the PFU-per-milliliter value obtained for cells infected with HPIV-3 in the presence of the antibodies to the value obtained for cells infected with HPIV-3 in the absence of the antibodies. The 100% level of infection represents the value (20 × 105 PFU/ml) obtained for untreated cells. (C) A549 cell lysates (10 μg of protein) obtained from mock-infected (lane 1) and HPIV-3-infected cells (36 h postinfection) in the absence (lane 2) or in the presence of β-catenin (control) (lane 3) or nucleolin (Nuc) (lane 4) antibodies were subjected to Western blot analysis with HPIV-3 anti-RNP antibody. (D) Culture supernatants collected from A549 cells mock infected or infected with VSV (0.2 MOI) in the absence or presence of nucleolin (Nuc) polyclonal (Poly) and monoclonal (Mono) antibodies were added to L929 cells for a plaque assay. The percentage of infection, reflecting the percentage of virus release, was calculated as a ratio of the PFU-per-milliliter value obtained for cells infected with VSV in the presence of the antibodies to the value obtained for cells infected with VSV in the absence of the antibodies. The 100% infection level represents the value (in PFU per milliliter) obtained for untreated cells. (E) A549 cell lysates (10 μg of protein) obtained from mock-infected (lane 1) and VSV-infected cells (36 h postinfection) in the absence (lane 2) or in the presence of β-catenin (control) (lane 3) or nucleolin (Nuc) (lane 4) antibodies were subjected to Western blot analysis with VSV anti-P antibody.
The specificity of nucleolin antibodies for the inhibition of HPIV-3 replication was borne out by the observation that nucleolin antibody treatment under the same experimental conditions used for HPIV-3 had no effect on VSV replication. Plaque assay analysis revealed no changes in VSV replication even in the presence of 70 μg of nucleolin antibody/ml (Fig. 5D). In addition, nucleolin antibody (Fig. 5E, lane 4) had no effect on VSV protein levels compared to the untreated (lane 2) and control antibody-treated (lane 3) cells, as confirmed by Western blot analysis with VSV antiphosphoprotein (P protein) antibody. Similar to the anti-HPIV-3 effect of nucleolin antibodies, preincubation of cells with the cell surface nucleolin-blocking peptide HB-19 (the peptide that specifically binds the cell surface nucleolin) (51, 52, 54), but not preincubation with the control peptide TW (blocking peptide for the CXCR receptor), inhibited HPIV-3 but not VSV infection (data not shown). These results suggest that cell surface nucleolin serves an essential function during the infection process of HPIV-3.
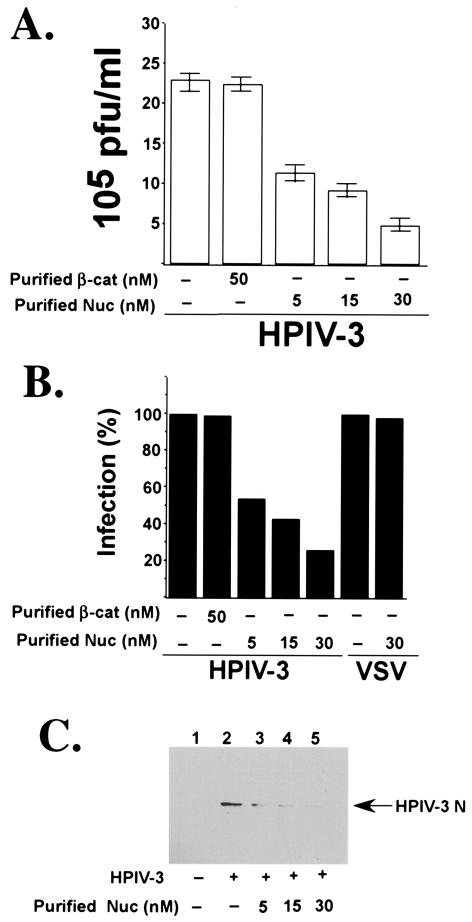
HPIV-3 preincubated with purified nucleolin fails to infect cells efficiently.
To further examine the involvement of nucleolin, specifically during HPIV-3 entry, human nucleolin purified to homogeneity (63) was utilized. HPIV-3 was preincubated with either purified nucleolin (5, 15, or 30 nM) or purified β-catenin (control, 50 nM) for 3 h at room temperature, and the preincubated virus was added to the A549 cells. Following adsorption at 37°C for 2 h, the cells were washed extensively to remove unbound viruses. The infection was then extended for an additional 36 h at 37°C, and the medium supernatants were subjected to plaque assay analysis. As shown in Fig. 6A, preincubation of HPIV-3 with 30 nM nucleolin significantly reduced virus infection, while preincubation of virus with β-catenin had no effect on HPIV-3 replication. The value obtained from the plaque assay result (Fig. 6A) and VSV-infected cells (data not shown) was utilized to demonstrate the extent of inhibition of HPIV-3 and VSV infection (the percentage of infection inhibition) by purified nucleolin (Fig. 6B). Concomitantly, Western blot analysis (Fig. 6C) of A549 cell lysates with anti-HPIV-3 RNP antibody revealed a significant reduction in HPIV-3 N protein amounts from cells infected with HPIV-3 preincubated with either 5 nM (lane 3), 15 nM (lane 4), or 30 nM (lane 5) purified nucleolin compared to the cells infected with untreated HPIV-3 (lane 2). In contrast to the results for HPIV-3, preincubation of VSV with 30 nM nucleolin had no effect on the VSV titer (Fig. 6B) and viral protein levels (data not shown). Once again, similar to the observed antiviral effect of nucleolin antibodies, purified nucleolin failed to completely abolish HPIV-3 infection. Thus, these results suggest that an additional cell surface molecule(s) is also required for HPIV-3 entry, and nucleolin's role may be limited to the augmentation of entry efficiency.
FIG. 6.
Effect of purified nucleolin on virus replication. (A) Culture supernatants collected from A549 cells mock infected or infected with HPIV-3 (0.2 MOI) preincubated with either purified nucleolin (Nuc) or β-catenin (β-cat) were added to CV-1 cells for a plaque assay. The plaque assay results, reflecting the viral titers, are expressed in PFU per milliliter. Each value represents the mean ± standard deviation for three determinations. (B) The average plaque assay values (in PFU per milliliter) from panel A were used to show the percentages of inhibition of infection of HPIV-3 in the presence of purified nucleolin. The percentage of infection, reflecting the percentage of virus release, was calculated as a ratio of the PFU-per-milliliter value obtained for cells infected with HPIV-3 in the presence of purified proteins to the value obtained for cells infected with HPIV-3 in the absence of purified proteins. The 100% level of infection represents the value (23 × 105 PFU/ml) obtained for untreated cells. Similarly, culture supernatants collected from A549 cells mock infected or infected with VSV (0.2 MOI) in the absence or presence of purified nucleolin were added to L929 cells for a plaque assay. The percentage of infection, reflecting the percentage of virus release, was calculated as a ratio of the PFU-per-milliliter value obtained for cells infected with VSV in the presence of the purified protein to the value obtained for cells infected with VSV in the absence of the purified protein. The 100% level of infection represents the value (in PFU per milliliter) obtained from un-treated cells. (C) A549 cell lysates (5 μg of protein) obtained from mock-infected (lane 1) and HPIV-3-infected (36 h postinfection) cells following preincubation in the absence (lane 2) or presence of 5 nM (lane 3), 15 nM (lane 4), and 30 nM (lane 5) purified nucleolin (Nuc) were subjected to Western blot analysis with HPIV-3 anti-RNP antibody.
Nucleolin is required for postattachment internalization of HPIV-3.
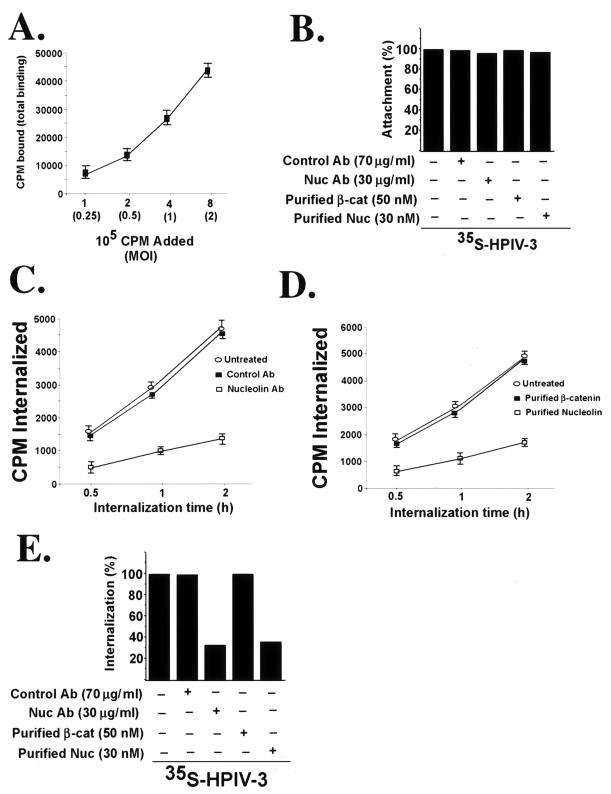
Finally, we examined the precise entry stage (attachment versus internalization) at which nucleolin serves its essential function during HPIV-3 entry. The attachment and internalization assays were performed with 35S-HPIV-3 as described previously (8, 9). Initially, the kinetics of 35S-HPIV-3 binding to A549 cells was examined by adding increasing amounts of 35S-HPIV-3 (0.25 to 2 MOI) (1 × 105 to 8 × 105 cpm) to chilled A549 cells. The virus was incubated at 4°C (the temperature that supports attachment but not internalization) for 2 h, and following virus attachment the cells were washed extensively and the lysed cells were measured for bound radioactivity (expressed in counts per minute) with a scintillation counter. As shown in Fig. 7A, the binding of 35S-HPIV-3 occurred in a dose-dependent manner, since virus attachment increased linearly with increasing amounts (0.25 to 2 MOI) of 35S-HPIV-3. The results from the attachment assay also revealed that approximately 5 to 7% of the input virus bound to the A549 cell surface during the time frame of our experiment and that saturable binding was not achieved at a concentration of at least 1 MOI. Based on these results, we chose to use 1 MOI (representing the nonsaturable concentration for virus attachment) of 35S-HPIV-3 (4 × 105 cpm) for our subsequent attachment and internalization assays. The rationale for using 1 MOI of virus is based on previous studies in which a nonsaturable amount of virus was used to perform attachment and internalization assays (24, 69). Moreover, a nonsaturable amount of virus closely mimics the physiological concentration of virus that binds and enters the cells during productive infection.
FIG. 7.
Effect of nucleolin antibodies (Ab) and purified nucleolin (Nuc) on cellular attachment and internalization of HPIV-3. (A) The kinetics of 35S-HPIV-3 attachment to A549 cells was examined by adding different amounts of virus (0.25 to 2 MOI, or 1 × 105 to 8 × 105 cpm) to chilled A549 cells. Following attachment at 4°C for 2 h, the cells were washed extensively and the cell-associated radioactivity (in counts per minute) representing the attached virus was measured by counting the cell lysate with a liquid scintillation counter. Each value represents the mean ± standard deviation for three determinations. (B) Attachment of 35S-HPIV-3 (1 MOI, or 4 × 105 cpm) to chilled A549 cells pretreated with antibodies or the virus preincubated with the purified proteins was determined following the attachment of the virus at 4°C for 2 h. Following adsorption, the cells were washed extensively and the cell-associated radioactivity (in counts per minute) representing the attached virus was measured by counting the cell lysate with a liquid scintillation counter. The percentage of attachment was calculated as a ratio of the amount of radioactivity present in cells incubated with 35S-HPIV-3 in the presence of the antibodies or purified proteins to the amount of radioactivity present in cells incubated with 35S-HPIV-3 alone. (C) Internalization of 35S-HPIV-3 (1 MOI, or 4 × 105 cpm) into A549 cells pretreated with antibodies was determined following incubation of attached (2 h, 4°C) virus at 37°C for 0.5, 1, and 2 h. The cell-associated radioactivities (in counts per minute) representing the internalized virus at different time points were measured by counting the cell pellet with a liquid scintillation counter. Each value represents the mean ± standard deviation for three determinations. (D) Internalization of 35S-HPIV-3 (1 MOI, or 4 × 105 cpm) into A549 cells following preincubation of the virus with the purified proteins was determined following incubation of attached (2 h, 4°C) virus at 37°C for 0.5, 1, and 2 h. The cell-associated radioactivities (in counts per minute) representing the internalized virus at different time points were measured by counting the cell pellet with a liquid scintillation counter. Each value represents the mean ± standard deviation for three determinations. (E) The average internalization values (counts per minute internalized) at 2 h postinternalization at 37°C from panels C and D were used to show the percentages of inhibition of internalization in the presence of nucleolin antibody and purified nucleolin. The percentage of internalization was calculated as a ratio of the amount of radioactivity present in cells infected with 35S-HPIV-3 in the presence of the antibodies or purified proteins to the amount of radioactivity present in cells infected with 35S-HPIV-3 alone. β-cat, β-catenin.
For the attachment assay in the presence of antibodies, chilled A549 cells were preincubated with either control (β-catenin) or nucleolin antibodies for 2 h at 4°C, followed by the addition of 35S-HPIV-3 (1 MOI, or 4 × 105 cpm). Similarly, 35S-HPIV-3 preincubated with purified nucleolin or β-catenin (control) was added to chilled cells. The virus was incubated at 4°C for 2 h, and following virus attachment the cells were washed extensively and the lysed cells were measured for bound radioactivity with a scintillation counter. As shown in Fig. 7B, the attachment assay revealed no change in HPIV-3 cellular binding for cells in the presence of either nucleolin antibody or purified nucleolin compared to the untreated cells. These results suggest that nucleolin is not involved during the initial cellular attachment of HPIV-3.
We next investigated whether nucleolin is required during HPIV-3 internalization, since nucleolin was not involved during HPIV-3 attachment. The internalization assay was performed in a manner similar to that used for the attachment assay, but following the incubation of 35S-virus (1 MOI, 4 × 105 cpm) at 4°C, the cells were washed extensively and the temperature was shifted to 37°C to promote the internalization of the attached virus particles. Following 0.5, 1, and 2 h of incubation at 37°C, the cells were washed extensively and trypsinized. The trypsinized cell pellet was further washed with PBS, and the cell pellet was counted for cell-associated radioactivity (representing the internalized virus) with a scintillation counter. As shown in Fig. 7C and D, both nucleolin antibody (Fig. 7C) and purified nucleolin (Fig. 7D) inhibited the internalization of 35S-HPIV-3 at different postinternalization time points, while control antibody and purified β-catenin had no effect on virus internalization. The internalized counts-per-minute values shown in Fig. 7C and D were used to generate a bar graph demonstrating the percentage of inhibition of internalization of 35S-HPIV-3 in the presence of purified nucleolin and nucleolin antibody (Fig. 7E). Similar inhibition of 35S-HPIV-3 internalization was observed in the presence of HB-19 but not in the presence of the control TW peptide (data not shown). While nucleolin-blocking agents inhibited 35S-HPIV-3 internalization, similar treatments had no effect on 35S-VSV internalization (data not shown). Interestingly, the nucleolin inhibitory agents failed to completely abolish virus internalization, a finding similar to what was observed during the HPIV-3 infection process in the presence of these agents.
DISCUSSION
In the present study, we have identified cell surface nucleolin as a cofactor required for the efficient cellular entry of HPIV-3. The requirement of nucleolin during HPIV-3 infection and entry was evident from several observations, including (i) the binding of 35S-HPIV-3 with nucleolin following VOPBA; (ii) the interaction of biotinylated HPIV-3 with nucleolin during cellular entry (adsorption stage); (iii) the specific interaction of HPIV-3 F, but not HN, with nucleolin; (iv) the inhibition of HPIV-3 infection and cell surface internalization by nucleolin antibodies; and (v) that preincubation of HPIV-3 with purified nucleolin reduced HPIV-3 infection and cell surface internalization. Moreover, the expression of nucleolin on the apical plasma membrane domain of polarized human lung epithelial cells, the domain preferentially utilized by HPIV-3 during entry (8, 13, 42), additionally suggests the involvement of nucleolin during HPIV-3 entry.
Nucleolin is a major RNA binding protein of the nucleolus that shuttles between the nucleus and the cytoplasm (65). Although nucleolin functions in the nucleus, it is now well established that a portion of intracellular nucleolin is also expressed on the surfaces of a variety of cells (11, 18, 29, 38, 39, 41, 60, 61, 62, 68). In addition, the present study has demonstrated that in human lung epithelial A549 cells, nucleolin is preferentially expressed on the apical plasma membrane domain. Cell surface nucleolin has been demonstrated to act as an adherence receptor for a bacterium, enterohemorrhagic Escherichia coli O157:H7 (63), and as an entry cofactor for viruses like HIV-1 (33, 51, 52, 53, 54) and coxsackie B virus (17). For HIV-1, cell surface nucleolin was found to act as one of the coreceptors during virus entry. It is interesting that a recent study has reported that nucleolin may also be involved during the assembly of another retrovirus, murine leukemia virus (3). In accordance with the previous studies on the role of cell surface nucleolin during cellular adherence and the entry of human pathogens, the present study has suggested similar involvement of nucleolin during HPIV-3 entry.
Although nucleolin is expressed on the cell surface, it does not possess a typical transmembrane domain. This observation led to the speculation that nucleolin may be localized on the cell surface as a peripheral membrane protein bound to an integral protein(s). Interestingly, a recent study has demonstrated that, like several secretory proteins (e.g., IL-1β, thioredoxin, and FGF), the cell surface nucleolin utilizes a nonclassical endoplasmic reticulum-Golgi pathway during its plasma membrane trafficking (53). The physiological significance of cell surface nucleolin is borne out by previous observations that it functions as a receptor for several ligands. These ligands include cytokine midkine, lipoproteins, laminin, fructosyllysine, factor J, T-cell receptor, cell surface ectoprotein kinases, urokinase and its receptor, L-selectin, and heparan binding protein syndecans (11, 18, 29, 38, 39, 41, 53, 60, 61, 62, 63, 68). The interactions of cell surface nucleolin with these extracellular ligands were demonstrated to play an important role in cell proliferation, mitogenesis, differentiation, and immunogenic responses. Interestingly, a protein named NRP (nucleolin-related protein) bearing high (95%) homology to nucleolin was also reported to be expressed on the apical pole of polarized kidney epithelial cells (64). In that context, a recent study has demonstrated that a bacterium, enterohemorrhagic E. coli O157:H7, that infects the epithelial cells from the luminal (apical) domain also utilizes nucleolin as one of its adherence receptors (63).
Cellular entry of enveloped viruses constitutes a complex and orchestrated process whereby virus envelope proteins interact with an array of host proteins to facilitate efficient entry. The complexity of the viral entry mechanism is also evident from the observation that primarily two stages, attachment and fusion (internalization), comprise the entry process, whereby different cellular proteins are utilized during either of these steps. Furthermore, the interaction of envelope proteins with cell surface molecules at each step results in conformational changes, leading to their subsequent interaction with an additional primary and/or secondary receptor(s). These interactions have been well documented during the entry of HIV-1, which utilizes several attachment and fusion receptors and coreceptors (6, 58). For HPIV-3, it was demonstrated previously that SAR serves as the initial attachment receptor for the virus via its interaction with HN. It is postulated that this interaction induces conformational changes in the F protein (complexed with HN on the virus envelope), leading to the exposure of its fusogenic peptide and subsequent fusion of the host target membrane with the envelope. Interestingly, like HPIV-3, HIV-1 also initially attaches to the cell surface following the interaction of its envelope protein gp-160 with HS (58). This event not only sequesters virus on the cell surface but also renders conformational changes in gp-160 that lead to its interaction with additional coreceptors that are required for efficient fusion. In that context, it is noteworthy that HIV-1 gp-160 shares a high degree of structural homology with paramyxovirus F protein (19, 20, 42). Thus, it is possible that although HPIV-3 requires SAR during attachment, an additional cell surface cofactor(s) may be required for efficient internalization and/or fusion. Indeed, Sendai virus, an HPIV-3-related mouse parainfluenza virus that possesses both HN and F as its envelope proteins (42) and requires SAR during entry (13, 42), also utilizes additional entry receptors, including glycophorins (72), ganglioside GD1α (21), and asialoglycoprotein receptor (44), in a cell-specific manner. In that context, previous studies (9) have demonstrated that both HS and SAR are required for efficient HPIV-3 entry into A549 cells, and in the present study we have shown that while cell surface nucleolin is not required for HPIV-3 attachment, it plays a role during postattachment internalization of the virus. Thus, we speculate that HPIV-3 utilizes SAR along with HS and nucleolin for efficient entry into human lung epithelial A549 cells. This hypothesis is further strengthened by our observation that nucleolin-blocking agents failed to completely abolish HPIV-3 infection and internalization. Similar utilization of SAR and/or HS and additional cell surface proteins during entry has been documented for several viruses (4, 12, 27, 30, 36, 45).
For example, rotavirus was shown to utilize both SAR and cell surface-expressed heat shock cognate protein 70 (hsc 70) during entry (27). It is interesting that hsc 70, like nucleolin, is also a nucleocytoplasmic protein that does not possess a transmembrane domain and an endoplasmic reticulum-specific signal sequence. Moreover, like nucleolin during HPIV-3 entry, hsc 70 is not required for the attachment of rotaviruses but is required during postattachment events. The role of nucleolin during cellular entry of HPIV-3 could possibly be mediated by the chaperonin function of nucleolin, since nucleolin acts as a chaperone during the nuclear import or export of nuclear or cytoplasmic proteins (2). It is well established that viral envelope proteins undergo major conformational changes during entry and the uncoating process as a result of their multiple contacts with various cell surface molecules. In that scenario, it could be speculated that chaperonin activity is required to obtain the conformationally favorable folding required for efficient entry. This phenomenon may also be true for rotaviruses, as suggested by a recent study (27), since rotaviruses also utilize a nucleocytoplasmic chaperone protein hsc 70 for cellular entry.
Although, the present study has suggested a role of nucleolin in the entry of HPIV-3 into human lung epithelial cells, an additional cell surface molecule(s) is also required for HPIV-3 entry. This speculation is based on the observation that nucleolin-blocking agents failed to completely abolish HPIV-3 infection and internalization in A549 cells. In that scenario, nucleolin may act as a secondary cofactor during infection, since earlier studies have reported that the inhibition of primary receptors usually results in complete (95 to 100%) inhibition of virus infectivity (22, 28, 40, 66). Thus, the role of nucleolin during HPIV-3 entry may be limited to the augmentation of the efficiency of HPIV-3 entry into human lung epithelial A549 cells. Currently, studies are in progress to dissect the sequential and/or cooperative utilization of HS, nucleolin, and SAR by HPIV-3 envelope proteins during virus entry.
In summary, the present study has demonstrated that cell surface nucleolin serves as one of the cofactors required for the efficient entry of HPIV-3 into human lung epithelial cells. The role of nucleolin during this process is at the virus internalization stage but not at the attachment stage. However, based on the present study and previous observations, we speculate that an additional cell surface molecule(s) also plays an important role during HPIV-3 entry. Therefore, the identification and characterization of the function of this molecule(s) will provide novel insights into the mechanism of host-virus interactions and lead to the development of novel antiviral therapies.
Acknowledgments
We thank Nancy Meizel (University of Washington, Seattle, Wash.) and Pierre D. McCrea (University of Texas M. D. Anderson Cancer Center, Houston, Tex.) for the HA-nucleolin and His-β-catenin constructs. We also thank Ara Hovanessian (Institut Pasteur, Paris, France) for the HB-19 peptide. We thank the Cleveland Clinic Molecular Biotechnology Core laboratory for the amino terminal sequence analysis and Sanker Subramaniam for help with the fast protein liquid chromatography analysis. We are also grateful to the Cleveland Clinic Virus Core laboratory for their help in preparing recombinant proteins from baculoviruses. We also thank Ratan Maitra and Satya P. Yadav for helpful discussions during the project.
S.B. was supported by a fellowship from the Morgenthaler Foundation. This work was supported by U.S Public Health Service grant AI32027 (to A.K.B.).
REFERENCES
- 1.Ah-Type, C., K. Schwartz, E. Huberman, E. Carlin, and A. Moscona. 1999. Virus-receptor interactions of human parainfluenza viruses type 1, 2, and 3. Microb. Pathog. 27:329-336. [DOI] [PubMed] [Google Scholar]
- 2.Allain, F. H., P. Bouvet, T. Dieckmann, and J. Feigon. 2000. Molecular basis of sequence recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19:6870-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral gag proteins and inhibits virion assembly. J. Virol. 74:11027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, E., C. Forrest, J. Connolly, J. Chappel, Y. Liu, F. Schnell, A. Nusrat, C. Parkos, and T. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 5.Benito-Salas, J. S., and R. M. del Angel. 1997. Identification of two surface proteins from C6/36 cells that bind dengue type 4 virus. J. Virol. 71:7246-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 7.Bose, S., S. Seetharam, N. M. Dahms, and B. Seetharam. 1997. Bipolar functional expression of transcobalamin II receptor in human intestinal epithelial Caco-2 cells. J. Biol. Chem. 272:3538-3543. [DOI] [PubMed] [Google Scholar]
- 8.Bose, S., A. Malur, and A. Banerjee. 2001. Polarity of human parainfluenza virus type 3 infection in polarized human lung epithelial A549 cells: role of microfilament and microtubule. J. Virol. 75:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bose, S., and A. Banerjee. 2002. Role of heparan sulfate in human parainfluenza virus type 3 infection. Virology 298:73-89. [DOI] [PubMed] [Google Scholar]
- 10.Bose, S., M. Mathur, P. Bates, N. Joshi, and A. K. Banerjee. 2003. Requirement of cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 84:1687-1699. [DOI] [PubMed] [Google Scholar]
- 11.Callebaut, C., S. Nisole, J. Briand, B. Krust, and A. Hovanessian. 2001. Inhibition of HIV infect ion by the cytokine midkine. Virology 281:248-264. [DOI] [PubMed] [Google Scholar]
- 12.Caruso, M., L. Belloni, O. Sthandier, P. Amati, and M. Garcia. 2003. α4β1 integrin acts as a cell receptor for murine polyomavirus at the postattachment level. J. Virol. 77:3913-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanock, R. M., B. R. Murphy, and P. L. Collins. 2001. Parainfluenza viruses, p. 1341. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 14.Choudary, S., J. Gao, D. Leaman, and B. De. 2001. Interferon action against human parainfluenza virus type 3: involvement of a novel antiviral pathway in the inhibition of transcription. J. Virol. 75:4823-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhary, S. K., A. G. Malur, Y. Huo, B. P. De, and A. K. Banerjee. 2002. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 302:373-382. [DOI] [PubMed] [Google Scholar]
- 16.De, B., M. Hoffman, S. Choudhary, C. Huntley, and A. Banerjee. 2000. Role of NH2 and COOH terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J. Virol. 74:5886-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Verdugo, U. R., H. C. Selinka, M. Huber, B. Kramer, J. Kellermann, P. H. Hofschneider, and R. Kandolf. 1995. Characterization of a 100-kilodalton binding protein for the six serotypes of coxsackie B viruses. J. Virol. 69:6751-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumler, I., V. Stepanova, U. Jerke, O. Mayboroda, F. Vogel, P. Bouvet, V. Tkachuk, H. Haller, and D. Gulba. 1999. Urokinase-induced mitogenesis is mediated by casein kinase 2 and nucleolin. Curr. Biol. 9:1468-1476. [DOI] [PubMed] [Google Scholar]
- 19.Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597-612. [DOI] [PubMed] [Google Scholar]
- 20.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epand, R., S. Nir, M. Parolin, and T. Flanagan. 1995. The role of the ganglioside GD1α as a receptor for Sendai virus. Biochemistry 34:1084-1090. [DOI] [PubMed] [Google Scholar]
- 22.Feldman, S., S. Audet, and J. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox, J. M., and M. E. Bloom. 1999. Identification of a cell surface protein from Crandell feline kidney cells that specifically binds Aleutian mink disease parvovirus. J. Virol. 73:3835-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentsch, J. R., and A. F. Pacitti. 1985. Effect of neuraminidase treatment of cells and effect of soluble glycoproteins on type 3 reovirus attachment to murine L cells. J. Virol. 56:356-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gershoni, J., M. Lapidot, N. Zakai, and A. Loyter. 1986. Protein blot analysis of virus receptors: identification and characterization of the Sendai virus receptor. Biochem. Biophys. Acta 856:19-26. [DOI] [PubMed] [Google Scholar]
- 26.Greengard, O., N. Poltoratskaia, E. Leikina, J. Zimmerberg, and A. Moscona. 2000. The anti-influenza virus agent 4-GU-DANA (Zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J. Virol. 74:11108-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerrero, C., D. Bouyssounade, S. Zárate, P. Isa, T. López, R. Espinosa, P. Romero, E. Méndez, S. López, and C. Arias. 2002. Heat shock cognate protein 70 is involved in rotavirus cell entry. J. Virol. 76:4096-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallak, L., P. Collins, W. Knudson, and M. Peeples. 2000. Iduronic acid containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 29.Harms, G., R. Kraft, G. Grelle, B. Volz, J. Dernedde, and R. Tauber. 2001. Identification of nucleolin as a new L-selectin ligand. Biochem. J. 360:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewish, M., Y. Takada, and B. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horga, M. A., G. L. Gusella, O. Greengard, N. Poltoratskaia, M. Porotto, and A. Moscona. 2000. Mechanism of interference mediated by human parainfluenza virus type 3 infection. J. Virol. 74:11792-11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horvath, C., R. Paterson, M. Shaughnessy, et al. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66:4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hovanessian, A., F. Puvion-Dutilleul, S. Nisole, J. Svab, E. Perret, J. Deng, and B. Krust. 2000. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp. Cell Res. 261:312-328. [DOI] [PubMed] [Google Scholar]
- 34.Hu, X., R. Ray, and R. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ighirami, G., M. Nakamura, J. Balow, A. Notkins, and P. Casali. 1988. Model for studying virus attachment: identification and quantitation of Epstein-Barr virus-binding cells by using biotinylated virus in flow cytometry. J. Virol. 62:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson, T., D. Sheppard, M. Denyer, W. Blakemore, and A. King. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin, Y., I. Pardoe, A. Burness, and T. Michalak. 1994. Identification and characterization of the cell surface 70-kilodalton sialoglycoprotein(s) as a candidate receptor for encephalomyocarditis virus on human nucleated cells. J. Virol. 68:7308-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan, P., H. Heid, V. Kinzel, and D. Kubler. 1994. Major cell surface-located protein substrates of an ecto-protein kinase are homologs of known nuclear proteins. Biochemistry 33:14696-14706. [DOI] [PubMed] [Google Scholar]
- 39.Kibbey, M., B. Johnson, R. Petryshyn, M. Jucker, and H. Kleinman. 1995. A 110-kD nuclear shuttling protein, nucleolin, binds to the neurite-promoting IKVAV site of laminin-1. J. Neurosci. Res. 42:314-322. [DOI] [PubMed] [Google Scholar]
- 40.Klimstra, W., K. Ryman, and R. Johnston. 1998. Adaption of Sindbis virus to BHK cells selects for use of heparin sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krantz, S., R. Salazar, R. Brandt, J. Kellermann, and F. Lottspeich. 1995. Purification and partial amino acid sequencing of a fructosyllysine-specific binding protein from cell membrane of the monocyte-like cell line U937. Biochim. Biophys. Acta 1266:109-112. [DOI] [PubMed] [Google Scholar]
- 42.Lamb, R., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Luo, Y., P. M. Vassilev, X. Li, Y. Kawanabe, and J. Zhou. 2003. Native polycystin 2 functions as a plasma membrane Ca2+-permeable cation channel in renal epithelia. Mol. Cell. Biol. 23:2600-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markwell, M., A. Porter, and A. Schwartz. 1985. An alternative route of infection for viruses: entry by means of the asialoglycoprotein receptor of a Sendai virus mutant lacking its attachment protein. Proc. Natl. Acad. Sci. USA 82:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martínez-Barragan, J., and R. Angel. 2001. Identification of a putative coreceptor on Vero cells that participates in dengue 4 virus infection. J. Virol. 75:7818-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moscona, A., and R. W. Peluso. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 65:2773-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moscona, A., and R. W. Peluso. 1992. Fusion properties of cells infected with human parainfluenza virus type 3: receptor requirements for viral spread and virus-mediated membrane fusion. J. Virol. 66:6280-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moscona, A., and R. W. Peluso. 1993. Relative affinity of the human parainfluenza virus type 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J. Virol. 67:6463-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moscona, A., and R. W. Peluso. 1996. Analysis of human parainfluenza virus type 3 receptor binding variants: evidence for the use of a specific sialic-acid containing receptor. Microb. Pathog. 20:179-184. [DOI] [PubMed] [Google Scholar]
- 50.Nauwynck, H., X. Duan, H. Favoreel, P. Van Oostveldt, and M. Pensaert. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor mediated endocytosis. J. Gen. Virol. 80:297-305. [DOI] [PubMed] [Google Scholar]
- 51.Nisole, S., B. Krust, C. Callebaut, G. Guichard, S. Muller, J. Briand, and A. Hovanessian. 1999. The anti-HIV psuedopeptide HB-19 forms a complex with the cell-surface-expressed nucleolin independent of heparan sulfate proteoglycans. J. Biol. Chem. 274:27875-27884. [DOI] [PubMed] [Google Scholar]
- 52.Nisole, S., B. Krust, E. Dam, A. Bianco, N. Seddiki, S. Loaec, C. Callebaut, G. Guichard, S. Muller, J. Riand, and A. Hovanessian. 2000. The HB-19 pseudopeptide 5[Kpsi(CH2N)PR]-TASP inhibits attachment of T lymophocyte- and macrophage-tropic HIV to permissive cells. AIDS Res. Hum. Retrovir. 16:237-249. [DOI] [PubMed] [Google Scholar]
- 53.Nisole, S., B. Krust, and A. Hovanessian. 2002. Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp. Cell Res. 276:155-173. [DOI] [PubMed] [Google Scholar]
- 54.Nisole, S., E. Said, C. Mische, M. Prevost, B. Krust, P. Bouvet, A. Bianco, J. Briand, and A. Hovanessian. 2002. The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J. Biol. Chem. 277:20877-20886. [DOI] [PubMed] [Google Scholar]
- 55.Perlman, S., M. Jordan, R. Brossmer, O. Gerrngard, and A. Moscana. 1999. The use of a quantitative fusion assay to evaluate HN-receptor interaction for human parainfluenza virus type 3. Virology 265:57-65. [DOI] [PubMed] [Google Scholar]
- 56.Porotto, M., O. Greengard, N. Poltoratskaia, M. A. Horga, and A. Moscona. 2001. Human parainfluenza virus type 3 HN-receptor interaction: effect of 4-guanidino-Neu5Ac2en on a neuraminidase-deficient variant. J. Virol. 75:7481-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prince, G. A., M. G. Ottolini, and A. Moscona. 2001. Contribution of the human parainfluenza virus type 3 HN-receptor interaction to pathogenesis in vivo. J. Virol. 75:12446-12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rider, C. C. 1997. The potential for heparin and its derivatives in the therapy and prevention of HIV-1 infection. Glycoconj. J. 14:639-642. [DOI] [PubMed] [Google Scholar]
- 59.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 60.Said, E., B. Krust, S. Nisole, J., Svab, J. Briand, and A. Hovanessian. 2002. The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J. Biol. Chem. 277:37492-37502. [DOI] [PubMed] [Google Scholar]
- 61.Semenkovich, C. F., R. E. Ostlund, M. O. Olson, and J. W. Yang. 1990. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry 29:9708-9713. [DOI] [PubMed] [Google Scholar]
- 62.Shibata, Y., T. Muramatsu, M. Hirai, T. Inui, T. Kimura, H. Saito, L. M. McCormick, G. Bu, and K. Kadomatsu. 2002. Nuclear targeting by the growth factor midkine. Mol. Cell. Biol. 22:6788-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinclair, J., and A. O'Brien. 2002. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-γ of enterohemorrhagic Escherichia coli. J. Biol. Chem. 277:2876-2885. [DOI] [PubMed] [Google Scholar]
- 64.Sorokina, E., and J. Kleinman. 1999. Cloning and preliminary characterization of a calcium-binding protein closely related to nucleolin on the apical surface of inner medullary collecting duct cells. J. Biol. Chem. 274:27491-27496. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava, and, M. H. Pollard. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 13:1911-1922. [PubMed] [Google Scholar]
- 66.Su, C., C. Liao, Y. Lee, and Y. Lin. 2001. Highly sulfated forms of heparin sulfate are involved in Japanese encephalitis virus infection. Virology 286:206-215. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki, T., A. Portner, R. A. Scroggs, M. Uchikawa, N. Koyama, K. Matsuo, Y. Suzuki, and T. Takimoto. 2001. Receptor specificities of human respiroviruses. J. Virol. 75:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Take, M., J. Tsutsui, H. Obama, M. Ozawa, T. Nakayama, I. Maruyama, T. Arima, and T. Muramatsu. 1994. Identification of nucleolin as a binding protein for midkine (MK) and heparin-binding growth associated molecule (HB-GAM). J. Biochem. 116:1063-1068. [DOI] [PubMed] [Google Scholar]
- 69.Tamura, M., K. Natori, M. Kobayahi, T. Miyamura, and N. Takeda. 2000. Interaction of recombinant Norwalk virus particles with the 105-kilodalton cellular binding protein, a candidate receptor molecule for virus attachment. J. Virol. 74:11589-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tao, Y. S., R. A. Edwards, B. Tubb, S. Wang, J. Bryan, and P. D. McCrea. 1996. Beta-catenin associates with the actin-bundling protein fascin in a noncadherin complex. J. Cell Biol. 134:1271-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor, H., and N. Cooper. 1990. The human cytomegalovirus receptor on fibroblasts is a 30-kilodalton membrane protein. J. Virol. 64:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wybenga, L., R. Epand, S. Nir, J. Chu, F. Sharom, T. Flanagan, and R. Epand. 1996. Glycophorin as a receptor for Sendai virus. Biochemistry 35:9513-9518. [DOI] [PubMed] [Google Scholar]