Abstract
Aquaporins (AQPs) are major intrinsic proteins (MIPs) that mediate bidirectional flux of water and other substrates across cell membranes, and play critical roles in plant-water relations, dehydration stress responses and crop productivity. However, limited data are available as yet on the contributions of these proteins to the physiology of the major crop barley (Hordeum vulgare). The present work reports the identification and expression analysis of the barley MIP family. A comprehensive search of publicly available leaf mRNA-seq data, draft barley genome data, GenBank transcripts and sixteen new annotations together revealed that the barley MIP family is comprised of at least forty AQPs. Alternative splicing events were likely in two plasma membrane intrinsic protein (PIP) AQPs. Analyses of the AQP signature sequences and specificity determining positions indicated a potential of several putative AQP isoforms to transport non-aqua substrates including physiological important substrates, and respond to abiotic stresses. Analysis of our publicly available leaf mRNA-seq data identified notable differential expression of HvPIP1;2 and HvTIP4;1 under salt stress. Analyses of other gene expression resources also confirmed isoform-specific responses in different tissues and/or in response to salinity, as well as some potentially inter-cultivar differences. The work reports systematic and comprehensive analysis of most, if not all, barley AQP genes, their sequences, expression patterns in different tissues, potential transport and stress response functions, and a strong framework for selection and/or development of stress tolerant barley varieties. In addition, the barley data would be highly valuable for genetic studies of the evolutionarily closely related wheat (Triticum aestivum L.).
Introduction
Aquaporins (AQPs) belong to the superfamily of membrane channels called the major intrinsic proteins (MIPs). The plant MIPs (often generically called ‘aquaporins’) are typically divided into seven subfamilies: the plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin-26-like intrinsic proteins (NIPs), small, basic intrinsic proteins (SIPs) [1] and the novel, less common subfamilies of GlpF-like intrinsic protein (GIP1;1) [2], hybrid intrinsic proteins (HIPs) and the uncategorized X intrinsic proteins (XIPs) [3]. Based on the ar/R (aromatic/Arginine) selectivity filter residues, NIPs are typically divided into group I (WVAR; e.g., AtNIP1;1, OsNIP1;1), group II [A(V/I/A)(G/A)R; e.g., AtNIP5;1, OsNIP3;1;] and group III (GSGR; e.g., OsNIP2;1) [4]. Four main subfamilies (PIPs, TIPs, NIPs, SIPs) have been identified in both primitive and higher plants, while HIPs and GIPs have only been identified in mosses [3] and XIPs identified in mosses and several dicotyledonous plants, e.g., tobacco, potato and tomato [5] and soybean [6], but not monocots. The plant MIP families show a high gene multiplicity, with 36 isoforms reported in maize [7], 38 in rice [8], and 66 in soybean [6]. All MIPs exhibit a number of characteristic features (reviewed in [1]), including (i) six trans-membrane helices (TM1-TM6) and five inter-helical loops (LA-LE); (ii) two short helices (HB, HE) that contain the highly conserved Asn-Pro-Ala (NPA) motifs that form the pore that allows a single-file passage of water molecules; (iii) the ar/R selectivity filter, comprised of four residues (one from TM2, one from TM5, two from LE), and shown to significantly influence the nature of transport substrates; (iv) five positions (P1-P5) likely important for discriminating between AQPs and glycerol transporters and (v) a conserved motif Ala-Glu-Phe (AEF or AEFXXT) in TM1, of unclear function.
Plant MIPs have been shown to have significant roles in water homeostasis and response to salinity and drought, and additionally, many isoforms also transport other substrates such as ammonia, silicon, CO2 and boron (reviewed in [1, 8]). Reduced expression of certain PIP aquaporins and reduced water uptake under nitrogen supply provided as ammonium rather than nitrate [9] further suggests a mechanism of direct control to reduce ammonium toxicity (sole ammonium being toxic to many plants compared to a mixed-feed with nitrate), although how the nutrient/antinutrient is distinguished and the stimulus is linked to modulations of aquaporin expression and water transport remains unclear. The diverse aquaporins thus play important roles in many life processes of plants such as photosynthesis, nitrogen fixation, nutrient acquisition, reduced uptake and/or detoxification of toxic compounds, and other environmental stress responses, and hold immense genetic potential for crop improvement via varietal selection or transgenic strategies. Drought, salinity and nutrient stresses impede the productivity of barley and/or wheat significantly. However, despite the nutritional and economic importance of these crops, the roles of AQPs as genetic factors that may affect the growth and yield of these crops remain rather poorly understood. In case of wheat, the presence of at least 35 AQPs (PIPs and TIPs only) has been reported [10], with some likely discrepancies in distinction of some genes versus homeologues due to its polyploidy, and its NIP and SIPs are little studied. Similarly, studies on barley have been limited to some individual genes, especially PIPs [11– 15]. This study focusses on analysing the MIP superfamily in barley.
Barley is an important food crop which belongs to grass family Poaceae, subfamily Pooideae and tribe Triticeae, which also includes the larger cereal crop, wheat. However, barley is more adaptable and resilient than wheat [16]. It is more tolerant to drought, salinity and cold, and can be cultivated at higher altitudes and latitudes and farther into deserts than other cereal crops [17]. Barley seems to be able to maintain the root hydraulic conductivity (important for maintenance of transpiration), at least under moderate salinity stress [13] as compared to wheat [18], possibly explaining its better tolerance. Aquaporins such as PIPs have crucial roles in water uptake, its transmembrane transport and osmotic mechanisms. Hence a deeper understanding of these genes in barley will be essential for crop improvement through selective breeding and transgenics, both in barley and wheat. Its close evolutionary relatedness to wheat and its smaller, diploid, now-sequenced genome make it an excellent candidate for this purpose. The findings of the present study have led to possibly the entire AQP family of barley, including the gene sequences, expression patterns and the potential transport substrates of the encoded proteins.
Materials and Methods
Identification of barley aquaporin sequences
Barley AQP sequences were identified from the leaf mRNA-seq dataset obtained earlier, the NCBI UniGene database, and the barley draft genome sequence, as follows. For mining the leaf mRNA-seq data (publicly available, deposited as Sequence Read Archive accession number SRA062960 ([19]; including supplemental data) to identify the transcripts representing AQPs, keyword searches were carried out for putative protein encoding sequences annotated as 'aquaporin', 'PIP', 'TIP', 'NIP', 'SIP', 'intrinsic', 'channel', 'transmembrane' and 'nodulin-like' based on their similarity to rice proteins. For accessing the NCBI UniGenes, the accession numbers of cDNAs corresponding to each barley UniGene were retrieved from their profile in NCBI (http://www.ncbi.nlm.nih.gov/unigene/; last accessed December 2014). The cDNAs were translated in Gene Runner (http://www.generunner.net/) and inspected for conserved features such as NPA motifs. The nearest barley relatives of rice AQP genes identified earlier [8] were mined from the NCBI UniGene database. All thus-identified UniGenes were also translated and the barley cDNAs representing these identified from NCBI as above. For searching the barley draft genome, the cDNAs identified in mRNA-seq and the rice AQPs (http://rice.plantbiology.msu.edu/; last accessed December 2014) were subjected to BLASTn against the International Barley Genome Sequencing Consortium (IBGSC) [20] IPK Barley Blast Server (http://webblast.ipk-gatersleben.de/barley/; last accessed December 2014) high confidence CDS (HC_genes_CDS_Seq) and full length cDNA databases (ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/genes/; last accessed December 2014) (e-value threshold <0.01). All data were used to compile a comprehensive list of all identified barley AQP cDNAs. These were then subjected to BLASTn against the IBSC assembly_WGSMorex and assembly_WGSBowman databases (http://webblast.ipk-gatersleben.de/barley/; last accessed December 2014) (e-value <0.01) to obtain their genomic sequences and physical locations. Additionally, the low confidence gene set (LC_genes_CDS_Seq) was also mined to note any other potential AQPs.
Sequence translations, analyses, alignments, construction of phylogenetic trees
All cDNAs were translated in Gene Runner and the putative protein sequences analysed. The subcellular locations of putative proteins were predicted with WoLF PSORT (http://wolfpsort.org/), PSORT (http://psort.hgc.jp/form.html) and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html). The DNA and protein sequences were aligned in Bioedit v7.1.3 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) using CLUSTALW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Phylogenetic trees were constructed based on the nucleotide alignments shown in S1 Fig using MEGA5 (http://www.megasoftaware.net/) and maximum likelihood method, with bootstrapping set at 1000 replications. Alignments of the genomic sequences and cDNAs were conducted using the gene structure display server (GSDS; http://gsds.cbi.pku.edu.cn/) to determine the intron-exon structures.
Analysis of expression of barley aquaporins
The mRNA-seq data were analysed for relative expression of AQPs in the control and salt-stressed leaf RNAs. Details of each barley EST corresponding to a UniGene (cultivar, tissue, treatment, development stage) were sourced from the NCBI EST database (http://www.ncbi.nlm.nih.gov/nucest/16322814) (data not shown). Also, the probeset IDs corresponding to AQPs were obtained from HarvEST:Barley v1.83 (http://harvest.ucr.edu/) using the ‘search by GenBank number, EST name or unigene number’ tool, and some also from Besse et al. [21]. The IDs were used for analysis of their expression profiles in Genevestigator (https://www.genevestigator.com/gv/). The RNA-seq data of the eight tissues related to the IBGSC high confidence gene predictions (ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/expression/README) were used to obtain expression scores for genes with MLOC numbers and generate a heat map of log2 normalised expression.
Results
Identification of barley aquaporin genes
The search for AQPs in the SUT barley leaf transcriptome developed earlier [19] [GenBank SRA062960] by keyword and rice locus identified thirty-one UniGenes. The corresponding cDNA numbers to these (e.g., http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?UGID=2919645&TAXID=4513&SEARCH=Hv.23281) showed the cDNAs represented twenty-two annotated (eleven PIPs, seven TIPs, four NIPs) and eight unannotated genes (Table 1; S1 Table). By mining the IBGSC barley genome project CDS and genomic databases with the cDNAs identified from the MSU rice cDNA database and the barley leaf mRNA-seq data, 36 CDSs and 40 genomic sequences could be identified altogether. By comparing these to the rice and barley cDNAs, 22 genomic sequences could be annotated as above, while sixteen were unannotated, comprised of eight as above and eight additional ones. Additionally, a genomic sequence identified in Bowman and Morex genomes (contig_222714; contig_401367) was not present in the CDS database, and a BLASTn against NCBI revealed that both contigs corresponded to HvTIP5;1 cDNA (AB540227). Based on % identities to other reported barley sequences (Table 1) or to orthologs in rice, maize or sorghum (data not shown), it appears that some annotations in Genbank may need to be revised, i.e., (i) AB540229, currently annotated as HvNIP2;1, seems better annotated as HvNIP2;2; (ii) AB710142 annotated as HvNIP2;2 should be HvNIP2;1; (iii) GU584119 annotated as HvTIP1;3 should be HvTIP1;2; (iv) GU584121 annotated as HvTIP2;1 should be HvTIP2;3 (Table 1). The BLASTn of the unannotated sequences against the Rice MSU cDNA database revealed they represented seven PIP2s (tentatively called HvPIP2a-g), three TIPs (HvTIP3, HvTIP4a, HvTIP4b), four NIPs (HvNIP2, HvNIP3a, HvNIP3b, HvNIP4) and two SIPs (HvSIP1, HvSIP2). BLAST queries of PpGIP1;1 [2], PpHIP1;1 [3] and GmXIP1;1 [6] against the barley genome database led to no hits, indicating these subfamilies are absent or highly divergent in barley, consistent with reports of their absence/loss in monocots [3, 5]. Additionally, searches of the low confidence (LC) gene set led to nine sequence, of which one (MLOC_43388.1) was the longest (S2 Table). However, their putative translation products indicated they were partial and lacked NPA motifs, except two sequences (MLOC_37440.1 and MLOC_43388.1) that exhibited one NPA. The LC genes need confirmation and were not included in further analyses. In summary, the methods collectively led to the barley AQP superfamily comprising of at least forty members, twenty four being annotated and sixteen unannotated (Table 1).
Table 1. Summary of the identified barley aquaporins.
| Barley AQP | GenBank accession numbers | Corresponding barley CDS from genome database # | Corresponding barley gDNA contigs from genome database | Chromosomal location* | Method used to identify aquaporins | |||
|---|---|---|---|---|---|---|---|---|
| Barley genome database | mRNA-seq | |||||||
| Bowman | Morex | gDNA | CDS | |||||
| PIPs | ||||||||
| HvPIP1;1 | AB286964; AK249573; X76911 | MLOC_80094 | contig_863807 | contig_88585 | 2HL | ✓ | ✓ | ✓ |
| HvPIP1;2 | AB275278 | - | contig_72237 | contig_95629 | 5HL | ✓ | x | ✓ |
| HvPIP1;3 | AB009308; AK251251 | - | - | contig_1569089 | 6HL | ✓ | ✓ | ✓ |
| HvPIP1;4 | AB275279 | - | contig_2001921 | contig_280723 | 6HL | ✓ | x | ✓ |
| HvPIP1;5 | AB009309; AK360427; AK359326 | MLOC_10855 | contig_879787 | contig_1559936 | 6HL | ✓ | ✓ | ✓ |
| HvPIP2;1 | AB219366; AB009307; AK250654 | MLOC_13871 | contig_64927 | contig_1566694 | 6HL | ✓ | ✓ | ✓ |
| HvPIP2;2 | AB377269; AK250563; AK253017 | - | contig_1993266 | contig_40687 | 2HS | ✓ | ✓ | ✓ |
| HvPIP2;2a | - | MLOC_56278 | contig_1993266 | contig_40687 | 2HS | ✓ | ✓ | x |
| HvPIP2;3 | AB275280; AK376080; AK353861; AK249631 | - | contig_9266 | contig_140919 | 2HL | ✓ | ✓ | ✓ |
| HvPIP2;4 | AB219525; AK252600 | - | contig_869787 | contig_1616200 | 2HL | ✓ | ✓ | ✓ |
| HvPIP2;5 | AB377270; AK370379; AK370703 | MLOC_54419 | contig_65610 | contig_39125 | 2HS | ✓ | ✓ | ✓ |
| HvPIP2;6 | - | MLOC_62649 | contig_164695 | contig_46809 | 5HS | ✓ | ✓ | ✓ |
| HvPIP2;7 | GU584120; AK359099; AK359187; AK248491 | MLOC_552 | contig_396978 | contig_103970 | 5HL | ✓ | ✓ | ✓ |
| HvPIP2;7a | AK359996 | - | contig_396978 | contig_103970 | 5HL | ✓ | ✓ | ✓ |
| HvPIP2;8 | AK359199; AK356299; AB808658 | MLOC_44991 | contig_16921 | contig_275925 | 5HL | ✓ | ✓ | ✓ |
| HvPIP2;9 | AK361545; AK361542 | MLOC_61081 | contig_1576537 | contig_45084 | 6HS | ✓ | ✓ | ✓ |
| HvPIP2;10 | AK373720 | MLOC_72670 | contig_869109 | contig_62088 | 7HS | ✓ | ✓ | ✓ |
| HvPIP2;11 | - | MLOC_17384 | contig_282313 | contig_1576537 | 4HL | ✓ | ✓ | x |
| HvPIP2;12 | - | MLOC_18325 | contig_72035 | contig_1580129 | 5HL | ✓ | ✓ | x |
| TIPs | ||||||||
| HvTIP1;1 | AB540221; X80266; AK359670; AK367756 | MLOC_73301 | contig_2001586 | contig_63334 | 4HL | ✓ | ✓ | ✓ |
| HvTIP1;2 | AB540226; GU584119 a ; AK355942; AK372282; AK253104; AK367251 | MLOC_58872 | contig_844644 | contig_43071 | 3HL | ✓ | ✓ | ✓ |
| HvTIP2;1 | AB540222; AK250814; AK251090 | MLOC_66094 | contig_9681 | contig_51093 | 6HL | ✓ | ✓ | ✓ |
| HvTIP2;2 | AB540223; AK363660 | - | contig_222714 | contig_401367 | 2HL | ✓ | ✓ | ✓ |
| HvTIP2;3 | AB540224; EU872296; GU584121 b ; AB261102; AK248215; AK249965 | MLOC_22808 | contig_1985016 | contig_161234 | 7HL | ✓ | ✓ | ✓ |
| HvTIP3;1 | AB540228; AK376769 | MLOC_51183 | contig_16911 | contig_368665 | 1H | ✓ | ✓ | ✓ |
| HvTIP3;2 | AK373620 | MLOC_72436 | contig_46720 | contig_61640 | - | ✓ | ✓ | ✓ |
| HvTIP4;1 | AB540225; AK364960; AK368258; AK358374 | MLOC_71237 | contig_268752 | contig_59399 | 4HL | ✓ | ✓ | ✓ |
| HvTIP4;2 | - | MLOC_71267 | contig_12177 | contig_5946 | 3HS | ✓ | ✓ | x |
| HvTIP4;3 | - | MLOC_69640 | contig_872426 | contig_56741 | 3HS | ✓ | ✓ | x |
| HvTIP5;1 | AB540227 | - | contig_222714 | contig_401367 | 2HL | ✓ | x | x |
| NIPs | ||||||||
| HvNIP1;1 | AB540230; AK356027 | - | contig_848627; contig_135880 | contig_2551848; contig_95435 | 7HS | ✓ | ✓ | ✓ |
| HvNIP1;2 | AB540231; AK365010 | MLOC_36500 | contig_1982025; contig_75289 | contig_2554420; contig_2546891 | 5HL | ✓ | ✓ | ✓ |
| HvNIP2;1 | GQ496520; AK363953; GQ496519; AB447482; c AB710142 | MLOC_67894 | contig_1985750 | contig_53853 | 6HL | ✓ | ✓ | ✓ |
| HvNIP2;2 | AB447484; AB540229 d | - | contig_62067 | contig_45067 | 7HS | ✓ | x | ✓ |
| HvNIP2;3 | AK360552; AK357908 | - | contig_202862 | contig_1633799 | - | ✓ | ✓ | ✓ |
| HvNIP3;1 | - | MLOC_64918 | contig_71289 | contig_49507 | 1H | ✓ | ✓ | ✓ |
| HvNIP3;2 | - | MLOC_14646 | contig_221593 | contig_1568583 | 3HL | ✓ | ✓ | x |
| HvNIP4;1 | AK373249 | MLOC_62234 | contig_15550 | contig_46327 | 3HS | ✓ | ✓ | x |
| SIPs | ||||||||
| HvSIP1;1 | AK355004; AK252830 | - | contig_422844; contig_2006162; contig_889326 | contig_157697; contig_27; contig_53238 | 4HS | ✓ | ✓ | ✓ |
| HvSIP2;1 | AK364835; AK364572 | - | contig_1983253 | contig_48750 | 4HL | ✓ | ✓ | ✓ |
Barley AQPs annotated in this study are underlined;
‘✓’ indicates present and
‘x’ indicates absent;
‘-’: no information available.
# The corresponding barley CDS was obtained through Blast searches against the IPK Barley Blast Server (http://webblast.ipk-gatersleben.de/barley/) high confidence CDS (HC_genes_CDS_Seq) and full length cDNA databases.
*Chromosomal location data was obtained from the IBGSC barley genome database. The Genbank accession numbers likely to have been currently annotated incorrectly are shown in bold.
aannotated as HvTIP1;3 instead of HvTIP1;2;
bannotated as HvTIP2;1 instead of HvTIP2;3;
cannotated as HvNIP2;2 instead of HvNIP2;1;
dannotated as HvNIP2;1 instead of HvNIP2;2.
Phylogenetic analyses, new annotations and predicted intron-exon structures
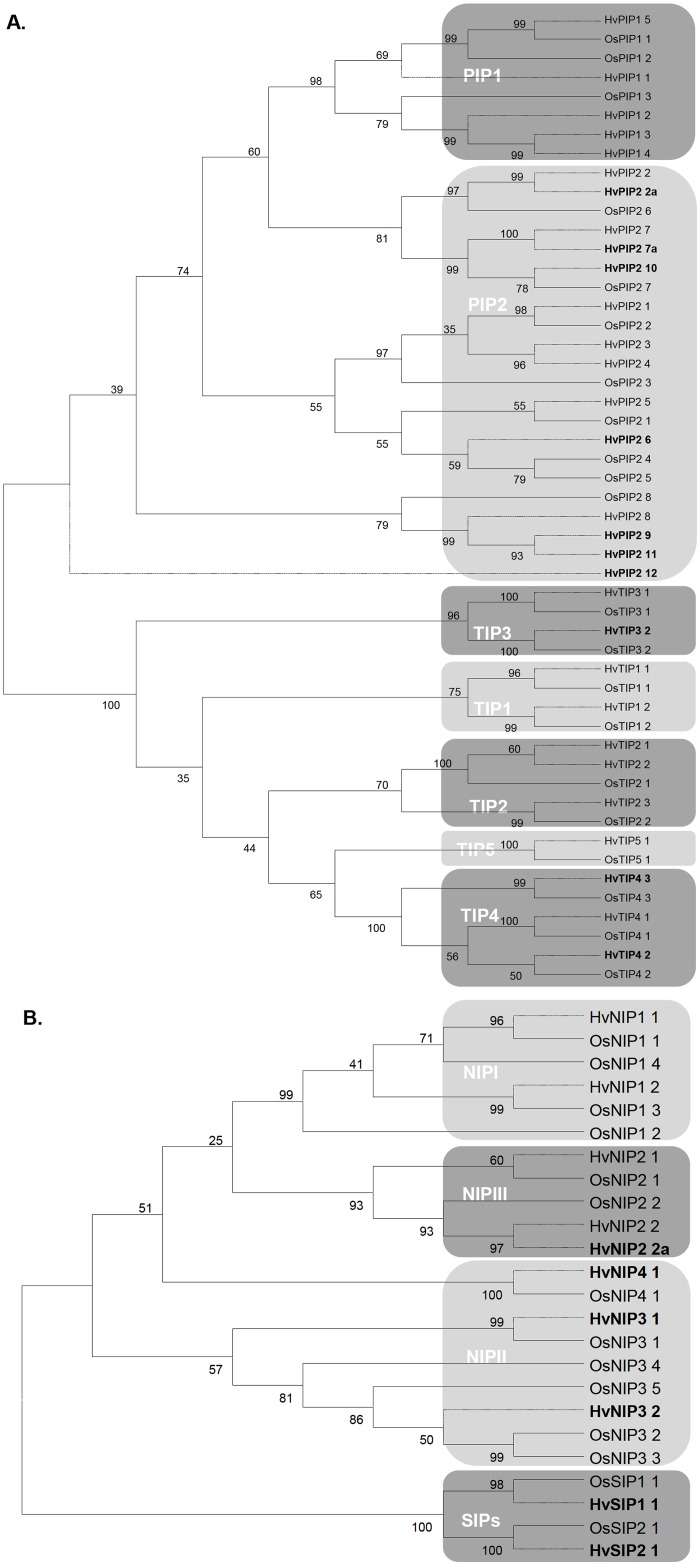
The phylogenetic analysis based on the nucleotide sequence alignments (S1 Fig) showed that the forty AQPs clustered into the four major sub-families (PIPs, TIPs, NIPs, SIPs) in relation to rice orthologs, and that all unannotated PIPs clustered with OsPIP2s (Fig 1). HvPIP2c clustered with OsPIP2;8 and the previously annotated HvPIP2;8 [22] and was annotated as HvPIP2;9. A BLASTx of HvPIP2b against NCBI identified Sorghum bicolor SbPIP2;6 (XM_002461888/XM_002461891) as the closest ortholog, hence it was annotated as HvPIP2;6, instead of the GU989200 currently annotated so in NCBI. HvPIP2d possessed two insertions (115 bp and 116 bp) identical to two introns in HvPIP2;7 genomic sequence. Likewise, HvPIP2e displayed an 86 bp insertion identical to an HvPIP2;2 intron; hence these were annotated as HvPIP2;7a and HvPIP2;2a, respectively. The respective translations revealed a frame-shift at amino acid 246, and premature stop codons. Thus these two may be either mis-annotated as cDNA in NCBI and barley genome database, or may undergo alternative splicing, or may be pseudogenes. Based on the closest rice orthologs, other unannotated barley genes were renamed HvPIP2;10 (HvPIP2a), HvPIP2;11 (HvPIP2f), HvPIP2;12 (HvPIP2g), HvTIP3;2 (HvTIP3); HvTIP4;2 (HvTIP4b), HvTIP4;3 (HvTIP4a), HvNIP3;1 (HvNIP3a), HvNIP3;2 (HvNIP3b) and HvNIP4;1 (HvNIP4). HvNIP2 clustered with HvNIP2;2 (Fig 1) and the sequences differed at both DNA and amino acid levels, hence HvNIP2 was annotated as HvNIP2;3. HvSIP1 and HvSIP2 represent HvSIP1;1 and HvSIP2;1, respectively, of Besse et al. [21] (Table 1). Pairwise identity scores were then obtained by alignments of the rice and barley cDNAs and putative protein sequences to identify the closest orthologs (S3 Table).
Fig 1. Phylogenetic trees of putative barley and rice aquaporin sequences.
A. PIPs and TIPs; B. NIPs and SIPs. The phylogenetic trees were constructed in MEGA4.1 (maximum likelihood with bootstraps) based on the sequence alignments shown in S1 Fig. The different sub-families and groups are highlighted in different shades of grey. The AQPs annotated in this study are shown in bold. The branches corresponding to barley AQPs are shown as dashed lines while those for rice are solid lines.
Comparison of the cDNAs (from leaf mRNA-seq data [19] and IBGSC barley genome project) and the available complete genomic sequences (from IBGSC barley genome project) using GSDS indicated that the HvPIPs had zero to three introns, as noted in rice OsPIPs [8] but unlike the typically three introns in dicot PIPs, e.g., soybean GmPIPs [6] (Table 2; AQPs which lack complete gDNA sequences are not included). The HvPIPs exhibited a diversity in intron sizes (71 to 1,245 bp), while most introns of TIPs were <200 bp (Table 2). Similar to soybean [6], two introns were common for HvTIPs, except HvTIP1s and HvTIP4;3. The NIP introns varied greatly in length (79 to 4,765 bp) and NIPIIIs showed the highest intron number (four). All introns displayed the standard GT/AG splice junctions except for the first intron of HvPIP2;7 and HvPIP2;10 which exhibited GC/AG.
Table 2. Intron-exon structures of the identified barley aquaporin genes.
| Barley AQP | Gene length (bp) | Gene Structure | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of | Intron and exon sizes (bp) | |||||||||||
| Exon | Intron | E1 | I1 | E2 | I2 | E3 | I3 | E4 | I4 | E5 | ||
| PIPs | ||||||||||||
| HvPIP1;1 | 1,644 | 4 | 3 | 334 | 101 | 296 | 122 | 141 | 554 | 96 | ||
| HvPIP1;2 | 1,082 | 3 | 2 | 642 | 91 | 141 | 112 | 96 | ||||
| HvPIP1;4 | 1,088 | 3 | 2 | 642 | 95 | 141 | 114 | 96 | ||||
| HvPIP1;5 | 2,790 | 4 | 3 | 337 | 990 | 296 | 292 | 141 | 635 | 99 | ||
| HvPIP2;1 | 2,974 | 4 | 3 | 318 | 116 | 301 | 1,245 | 142 | 741 | 111 | ||
| HvPIP2;2 | 950 | 2 | 1 | 735 | 86 | 129 | ||||||
| HvPIP2;2a | 954 | 1 | 954 | |||||||||
| HvPIP2;3 | 2,214 | 3 | 2 | 615 | 895 | 141 | 446 | 117 | ||||
| HvPIP2;5 | 2,179 | 3 | 2 | 618 | 1,212 | 141 | 94 | 114 | ||||
| HvPIP2;6 | 1,076 | 3 | 2 | 609 | 92 | 141 | 123 | 111 | ||||
| HvPIP2;7 | 1,107 | 3 | 2 | 307 | 115 | 437 | 116 | 132 | ||||
| HvPIP2;7a | 1,107 | 1 | 1,107 | |||||||||
| HvPIP2;8 | 876 | 1 | 876 | |||||||||
| HvPIP2;9 | 879 | 1 | 879 | |||||||||
| HvPIP2;10 | 1,219 | 4 | 3 | 337 | 71 | 299 | 105 | 141 | 128 | 138 | ||
| HvPIP2;11 | 879 | 1 | 879 | |||||||||
| HvPIP2;12 | 840 | 1 | 840 | |||||||||
| TIPs | ||||||||||||
| E1 | I1 | E2 | I2 | E3 | I3 | E4 | I4 | E5 | ||||
| HvTIP1;1 | 1,146 | 2 | 1 | 133 | 393 | 620 | ||||||
| HvTIP1;2 | 880 | 2 | 1 | 133 | 121 | 626 | ||||||
| HvTIP2;1 | 951 | 3 | 2 | 127 | 119 | 251 | 82 | 372 | ||||
| HvTIP2;2 | 960 | 3 | 2 | 127 | 86 | 251 | 124 | 372 | ||||
| HvTIP2;3 | 937 | 3 | 2 | 130 | 98 | 248 | 92 | 369 | ||||
| HvTIP3;1 | 1,036 | 3 | 2 | 154 | 145 | 248 | 99 | 390 | ||||
| HvTIP3;2 | 1,160 | 3 | 2 | 139 | 105 | 248 | 266 | 402 | ||||
| HvTIP4;1 | 1,378 | 3 | 2 | 142 | 164 | 254 | 440 | 378 | ||||
| HvTIP4;2 | 1,668 | 3 | 2 | 127 | 791 | 251 | 130 | 369 | ||||
| HvTIP4;3 | 851 | 2 | 1 | 127 | 95 | 629 | ||||||
| HvTIP5;1 | 984 | 3 | 2 | 127 | 108 | 251 | 87 | 411 | ||||
| NIPs | ||||||||||||
| E1 | I1 | E2 | I2 | E3 | I3 | E4 | I4 | E5 | ||||
| HvNIP1;2 | 2,300 | 4 | 3 | 306 | 512 | 423 | 95 | 62 | 691 | 211 | ||
| HvNIP2;1 | 2,983 | 5 | 4 | 150 | 110 | 225 | 1,427 | 195 | 122 | 62 | 436 | 256 |
| HvNIP2;2 | 3,025 | 5 | 4 | 168 | 91 | 225 | 1,365 | 195 | 129 | 62 | 537 | 253 |
| HvNIP3;1 | 5,881 | 4 | 3 | 225 | 4,765 | 426 | 104 | 62 | 103 | 196 | ||
| HvNIP3;2 | 827 | 3 | 2 | 429 | 79 | 62 | 109 | 148 | ||||
| HvNIP4;1 | 1,410 | 3 | 2 | 147 | 154 | 416 | 405 | 288 | ||||
| SIPs | ||||||||||||
| HvSIP2;1 | 3,529 | 3 | 2 | 318 | 2,150 | 258 | 629 | 174 | ||||
AQPs where the intron-exon structures could not be determined due to the lack of complete gDNA sequences are not included in the table.
Characteristic features and potential transport abilities of the putative aquaporins
The putative protein sequences of the barley AQPs showed conservation of both NPA motifs in all isoforms except for HvNIP3;1 which exhibited NPS and NPV, and HvSIP1;1 and HvSIP2;1 exhibited Leu or Thr at the Ala of first NPA (Table 3; S2 Fig). The ar/R substrate selectivity filter residues for PIPs were F-H-T-R, similar to those in maize [7], wheat and rice [8, 10]. The product of HvPIP2;7a lacked both NPAs and had unusual ar/R residues due to the frameshift (see above). The HvTIPs exhibited six different combinations, i.e., H-I-A-V (TIP1s), H-I-G-R (TIP2s), H-I-A-R (TIP3s), Q-T-A-R (HvTIP4;1), H-V-A-R (HvTIP4;3) and Q-V-A-R (HvTIP5;1), confirming the variability in TIPs in other plants [6, 8]. HvNIP2;1 and HvNIP2;2 showed G-S-G-R, similar to other NIPIIIs such as OsNIP2;1 and OsNIP2;2 [8]. The HvNIPI residues were W-V-A-R, characteristic of NIPIs in rice [8] and soybean [6]. The HvNIP3;2 presented a new combination (V-I-A-R). The P1-P5 positions deemed important for discriminating between authentic ‘aquaporins’ and glyceroporins [23], were more conserved at P2-P4 in all subfamilies (Table 3).
Table 3. Key structural features and predicted non-aqua transport substrates of putative barley aquaporin proteins.
| Barley AQP | NPA motif | Ar/R selectivity filter | P1—P5 | Subcellular location | Predicted transport substrate a | Rice ortholog (% identity)* |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LB/LE | TM2 | TM5 | LE1 | LE2 | P1 | P2 | P3 | P4 | P5 | cDNA | Protein | |||
| PIPs | ||||||||||||||
| HvPIP1;1 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO2, urea | OsPIP1;2 (89.6) | OsPIP1;1 (92.3) |
| HvPIP1;2 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO2, urea, water 8 | OsPIP1;3 (85.7) | OsPIP1;3 (89.3) |
| HvPIP1;3 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron 1, CO2, urea | OsPIP1;3 (82.8) | OsPIP1;3 (88.0) |
| HvPIP1;4 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron 1, CO2, urea | OsPIP1;3 (83.2) | OsPIP1;3 (88.6) |
| HvPIP1;5 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO2, urea | OsPIP1;1 (92.3) | OsPIP1;1 (94.1) |
| HvPIP2;1 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO 2 2; 5, urea, water 7 | OsPIP2;2 (87.5) | OsPIP2;2 (88.2) |
| HvPIP2;2 | NPA/ NPA | F | H | T | R | M | S | A | F | W | PM | CO 2 5, glycerol, urea, water 7; 8 | OsPIP2;1 (78.0) | OsPIP2;1 (72.2) |
| HvPIP2;2a | NPA/ NPA | F | H | T | R | M | S | A | Y | L | PM | glycerol, urea | OsPIP2;6 (78.3) | OsPIP2;6 (75.5) |
| HvPIP2;3 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO 2 5, urea, water 7 | OsPIP2;3 (90.2) | OsPIP2;3 (89.3) |
| HvPIP2;4 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, urea, water 7 | OsPIP2;3 (90.1) | OsPIP2;3 (88.3) |
| HvPIP2;5 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO2 5, urea, water7; 8 | OsPIP2;1 (90.1) | OsPIP2;1 (90.0) |
| HvPIP2;6 | NPA/ NPA | F | H | T | R | Q | S | A | F | W | PM | boron, CO2, urea | OsPIP2;4 (51.6) | OsPIP2;4 (51.0) |
| HvPIP2;7 | NPA/ NPA | F | H | T | R | A | S | A | F | W | PM | CO2, water 8 | OsPIP2;7 (75.0) | OsPIP2;7 (77.4) |
| HvPIP2;7a | - | F | - | - | - | - | - | - | - | - | - | - | OsPIP2;7 (48.5) | OsPIP2;7 (44.1) |
| HvPIP2;8 | NPA/ NPA | F | H | T | R | H | S | A | F | W | PM | - | OsPIP2;5 (71.0) | OsPIP2;5 (68.0) |
| HvPIP2;9 | NPA/ NPA | F | H | T | R | H | S | A | F | W | PM | - | OsPIP2;5 (68.3) | OsPIP2;5 (63.0) |
| HvPIP2;10 | NPA/ NPA | F | H | T | R | M | S | A | F | W | PM | CO2, glycerol, urea | OsPIP2;7 (76.9) | OsPIP2;7 (63.8) |
| HvPIP2;11 | NPA/ NPA | F | H | T | R | H | S | A | F | W | PM | - | OsPIP2;1 (68.3) | OsPIP2;3 (66.4) |
| HvPIP2;12 | NPA/ NPA | F | H | T | R | H | S | A | F | W | PM | - | OsPIP2;1 (74.2) | OsPIP2;1 (70.0) |
| TIPs | ||||||||||||||
| HvTIP1;1 | NPA/ NPA | H | I | A | V | T | S | A | Y | W | PM/V | H2O2, urea, water 8 | OsTIP1;1 (89.3) | OsTIP1;1 (89.6) |
| HvTIP1;2 | NPA/ NPA | H | I | A | V | T | S | A | Y | W | V | H2O2, urea, water 8 | OsTIP1;2 (87.3) | OsTIP1;2 (90.8) |
| HvTIP2;1 | NPA/ NPA | H | I | G | R | T | S | A | Y | W | V/PM | ammonia, formamide, H2O2 | OsTIP2;1 (89.3) | OsTIP2;1 (90.7) |
| HvTIP2;2 | NPA/ NPA | H | I | G | R | T | S | A | Y | W | PM | ammonia, formamide, H2O2 | OsTIP2;1 (68.6) | OsTIP2;1 (88.3) |
| HvTIP2;3 | NPA/ NPA | H | I | G | R | T | S | A | Y | W | V/PM | ammonia, formamide, H2O2, water 8 | OsTIP2;2 (91.2) | OsTIP2;2 (90.7) |
| HvTIP3;1 | NPA/ NPA | H | I | A | R | T | V | A | Y | W | PM/M | H2O2 | OsTIP3;1 (88.0) | OsTIP3;1 (88.3) |
| HvTIP3;2 | NPA/ NPA | H | I | A | R | S | A | A | Y | W | Chl | H2O2 | OsTIP3;2 (84.1) | OsTIP3;2 (81.3) |
| HvTIP4;1 | NPA/ NPA | Q | T | A | R | T | S | A | Y | W | V/PM | - | OsTIP4;1 (81.1) | OsTIP4;1 (80.9) |
| HvTIP4;2 | NPA/ NPA | H | T | A | R | T | S | A | Y | W | V/PM | glycerol, urea | OsTIP4;2 (75.0) | OsTIP4;2 (70.2) |
| HvTIP4;3 | NPA/ NPA | H | T | A | R | A | S | A | Y | W | PM | - | OsTIP4;3 (63.7) | OsTIP4;3 (52.8) |
| HvTIP5;1 | NPA/ NPA | Q | V | A | R | S | S | A | Y | W | Chl | - | OsTIP5;1 (86.1) | OsTIP5;1 (79.2) |
| NIPs | ||||||||||||||
| HvNIP1;1 | NPA/ NPA | W | V | A | R | F | T | A | Y | V | PM | Glycerol, water 6 | OsNIP1;1 (83.1) | OsNIP1;1 (84.9) |
| HvNIP1;2 | NPA/ NPA | W | V | A | R | F | T | A | Y | I | PM | glycerol, arsenite 6 , water 6 | OsNIP1;3 (69.9) | OsNIP1;3 (67.8) |
| HvNIP2;1 | NPA/ NPA | G | S | G | R | L | T | A | Y | F | PM/ER | antimony, arsenite, boron 3, silicon 4, urea, water 6 | OsNIP2;1 (84.0) | OsNIP2;1 (78.9) |
| HvNIP2;2 | NPA/ NPA | G | S | G | R | L | T | A | Y | F | PM/Chl | antimony, arsenite, boron, silicon, urea, water 6 | OsNIP2;2 (88.5) | OsNIP2;2 (87.0) |
| HvNIP2;3 | NPA/ NPA | G | S | G | R | L | T | A | Y | F | PM/Chl | antimony, arsenite, boron, silicon, urea | OsNIP2;2 (87.6) | OsNIP2;2 (87.7) |
| HvNIP3;1 | NPS/ NPV | A | I | G | R | F | T | A | Y | L | V | antimony, arsenite, boron | OsNIP3;1 (71.6) | OsNIP3;1 (88.5) |
| HvNIP3;2 | NPA/ NPA | V | I | A | R | Y | T | A | Y | L | PM/ER | - | OsNIP3;3 (55.7) | OsNIP3;3 (58.9) |
| HvNIP4;1 SIPs | NPA/ NPA | C | G | G | R | M | S | A | Y | V | PM | - | OsNIP4;1 (72.2) | OsNIP4;1 (59.8) |
| HvSIP1;1 | NPT/ NPA | L | V | P | N | M | A | A | Y | W | ER | - | OsSIP1;1 (83.0) | OsSIP1;1 (85.0) |
| HvSIP2;1 | NPL/ NPA | S | H | G | S | F | A | A | Y | W | PM/V | - | OsSIP2;1 (83.7) | OsSIP2;1 (74.5) |
‘-’ Indicates no information available.
aPotential substrate transported predicted using the signature sequences developed earlier [31]. The underlined substrates have been experimentally proven for the particular barley AQPs: 1[48], 2[50], 3[49], 4[51], 5[40], 6[26], 7[13], 8[21].
*Only the highest % identity score of each sequence is listed. The identity scores were obtained based on alignments of the cDNAs and putative protein sequences by pairwise identities in BioEdit (S3 Table). The AQPs annotated in this study are shown in bold. Sub-cellular locations: Chl (chloroplast), ER (endoplasmic reticulum), M (mitochondria), PM (plasma membrane) and V (vacuole).
The radish RsPIP1s have lower water permeability than RsPIP2s, and a loop E residue in RsPIP1;3 (Ile244) compared to RsPIP2;2 (Val235) is suggested to be relevant [24]. All HvPIP1s possessed Ile while the HvPIP2s had Val (S2 Fig), suggesting the HvPIP1s may have lower water permeability. Further, the difference between OsPIP1s (lower water permeability) and OsPIP2s (higher water permeability) was attributed to a residue in TM2, where OsPIP1s have Ala and OsPIP2s have Ile/Val [25]. At this position, HvPIP1;1 and HvPIP1;5 had Ala, but all HvPIP2s as well as HvPIP1;2, HvPIP1;3, HvPIP1;4 had Val (S2 Fig), suggesting the latter group may be permeable to water and supporting the observation of Besse et al. [21] that HvPIP1;2 is permeable to water. Some of the HvTIPs and HvNIPs also possessed a Val here (e.g. HvTIP2;1, HvTIP2;3, HvNIP1;1, HvNIP1;2), suggesting this residue may be functionally important for water transport of TIPs and NIPs. In line with this suggestion, a recent study observed that HvNIP1;1, HvNIP1;2, HvNIP2;1 and HvNIP2;2 all of which possessed the Val residue in TM2 (S2 Fig), were water transporters [26].
AQPs are known to function as tetramers and a highly conserved Cys (Cys80 in ZmPIP2;1) in loop A of PIP1s and PIP2s is shown to be essential for formation of disulphide bonds between PIP monomers, increasing the oligomer stability [27]. Alignments showed conservation of this Cys in all HvPIP1s and PIP2s (S2 Fig), suggesting a similar role for it in these, but its absence in TIPs, NIPs and SIPs suggests other residue(s) may be involved in this role. All PIPs were predicted to localise to the plasma membrane, as the subfamily name suggests and consistent with localisation of HvPIP2;1 [12]. However, an N-terminal diacidic motif Asp-Ile/Val-Glu [28, 29] found important for exit of newly synthesised PIPs from the ER to plasma membrane was noted only in HvPIP2;1, HvPIP2;3 and HvPIP2;4. Further, the subfamily names do not always indicate location (reviewed in [8]). In congruence with this, the TIPs were predicted to localise to the plasma membrane, mitochondria and chloroplast, in addition to the expected tonoplast (Table 3). Some NIPs were predicted to localise to the plasma membrane and HvSIP1;1 was predicted in the endoplasmic reticulum. All HvPIPs except HvPIP2;7 and HvPIP2;10 possessed a His corresponding to His193 in SoPIP2;1 considered important for pH-dependent gating [30] (S2 Fig), but the TIPs, NIPs and SIPs did not show this residue. Gating of SoPIP2;1 by phosphorylation of Ser115 and Ser274 has been shown, with blocking by Leu197 [30]. All three equivalent residues were observed for most HvPIPs. However, most TIPs possessed a Thr at Ser115, lacked Ser274, and only in HvTIP4;3 and HvTIP5;1 showed Leu197. All NIPs (except HvNIP3;2) had a Ser at Ser262 in GmNOD26 involved in phosphorylation. Alignments with AtPIP2;1 showed a conserved Ser in most HvPIP2s corresponding to S280 and S283, which are phosphorylated under salt stress [31] (S2 Fig).
Our earlier analysis of plant AQPs with non-aqua transport substrates had revealed distinct signature sequences and ‘specificity determining positions’ (SDPs) for each substrate group [32]; hence the putative barley AQPs were analysed for these. A number of isoforms were predicted to have the potential to transport micronutrients and molecules involved in plant growth and vigour (boron also being toxic in high concentrations) (Table 3). The potential to transport urea (many PIPs, TIPs and NIPs) and boron (many HvPIPs but no TIPs) was widespread, but that for ammonia was restricted to some TIPs (HvTIP2;1 HvTIP2;2; HvTIP2;3), CO2 to HvPIPs (all except HvPIP2;7), and silicon to NIPs (HvNIP2;1, HvNIP2;2; HvNIP2;2). The potential to transport the signalling molecule H2O2 seemed restricted to the TIPs (Table 3). NIPs had the most diverse predicted substrates, as found in other studies, including the potentially toxic arsenite.
Modulation of expression of barley aquaporin genes
In mRNA-seq, gene expression level is generally measured as the number of sequence reads mapping to a gene [33]. In the present study, the normalised sequence reads (base mean expression) were used. Most PIPs and some TIPs exhibited higher expression levels than NIPs and SIPs (S3 Fig), in line with Arabidopsis [34]. HvPIP1;4 had the most abundant transcripts in control plants, which nearly doubled upon exposure to salt stress (fold change (FC) +1.93; S1 Table). FC of ≥ +1.5 or ≤ -1.5 is considered notable [35], hence in the present context, such genes may have roles in osmotic regulation under salinity stress. Several other PIPs also showed notable changes, the largest being noted for HvPIP1;3 (FC +2.93) but that in HvPIP1;2 (FC +1.97) being statistically significant. Among the TIPs, HvTIP1;2 and HvTIP4;1 were the most abundant AQP transcripts in control plants, but only HvTIP4;1 displayed a statistically significant decline (FC -15.89) under salinity, suggesting a critical role for it. HvTIP1;1 and HvTIP2;3 showed moderate expression and up-regulation (FC +1.62, +2.60, respectively). HvTIP2;1, HvTIP2;2, HvTIP3;1 and HvTIP3;2 exhibited very low expression levels, but HvTIP3;2 was significantly up-regulated (FC +2.40). HvNIP2;2 was the most abundant NIP, but expressed at much lower levels than many PIPs and TIPs, and showed little change. The five other NIPs exhibited low-moderate expression and only HvNIP2;1 showed notable down-regulation. HvSIP2;1 had higher and more differential expression than HvSIP1;1. In summary, thirteen genes showed up-regulation and five showed a decrease under salinity, of which the changes in HvTIP4;1 and HvPIP1;2 were statistically significant (p-value <0.05).
The ESTs corresponding to the forty genes above were analysed and grouped as follows; apex, callus, epidermis, flower (anther, carpel, inflorescence and pistil), leaf, maternal, root, seed (caryopsis, coleoptile, embryo, endosperm, pericarp, testa), shoot, and spike (rachis) (S4 Table). Based on the total number of ESTs, the PIPs (2,043) and TIPs (1,260) appeared to have much higher expression than the NIPs (51) and SIPs (40), confirming other reports [34]. The PIPs and TIPs also typically showed higher expression in the leaf (PIPs: 285; TIPs: 112), root (546; 305), seed (215; 417) and shoot (798; 281) compared to other tissues (S4 Fig). Some ESTs were noted in response to stresses such as cold, drought, salinity or waterlogging (S5 Table). The heat-map generated for the IBGSC RNA-seq data of eight tissues related to the high-confidence gene predictions supported high expression levels of PIPs and TIPs in shoots (including tillers), inflorescences and developing grain, but not roots and germinating seed (S5 Fig). HvTIP1;1, HvTIP2;1, HvTIP2;3 and HvPIP1;1 were highly expressed in a number of tissues. In many tissues the expression levels of NIPs were lower than or the same as the TIPs and PIPs, except for high levels of HvNIP3;1 (developing grain) and HvNIP4;1 (inflorescence). A number of isoform-specific patterns were also noted.
Further, microarrays have been employed to monitor gene expression under salt stress in barley cv. Maythorpe and Golden Promise, the latter being more effective in its Na+ exclusion ability [36]. Analysis of this data in Genevestigator demonstrated that ten sequences (HvPIP1;1, HvPIP1;5, HvPIP2;2, HvPIP2;5, HvTIP1;1, HvTIP2;3, HvNIP1;1, HvNIP1;2, HvNIP2;3, HvNIP4;1) showed similar trends (up- or down-regulation) in both cultivars, but others differed (Table 4; S1 Table). Differences were also noted in the expression of some isoforms between the root and shoot of a variety (e.g., HvPIP2;1, HvPIP2;4 of Golden Promise). Further, comparison of the microarray data (cv. Maythorpe and Golden Promise) to leaf mRNA-seq (cv. Hindmarsh) indicated inter-cultivar differences in expression of many isoforms, while eight genes showed a common trend (e.g., HvPIP2;1, HvTIP1;2, HvNIP1;1). This may be due to differences in the sensitivity of the techniques, innate inter-cultivar differences, and/or experimental factors (Golden Promise and Maythorpe salt-stressed for 5 days; Hindmarsh for 12 hours). However, HvTIP4;1 exhibited the largest change (Maythorpe shoot FC -2.50, Hindmarsh FC -15.89), reinforcing its importance.
Table 4. Analysis of reported microarray data for expression of barley aquaporins under salinity stress.
| Barley AQP | HarvEST Unigene No.^ | Probeset ID | Response to salinity stress | |||
|---|---|---|---|---|---|---|
| Golden Promise | Maythorpe | |||||
| Shoot | Root | Shoot | Root | |||
| PIPs | ||||||
| HvPIP1;1 | 13831 | Contig1225_s_at; HW09I11u_s_at | -1.43 | -1.51 | -1.79 | -1.50 |
| HvPIP1;2 | 13853 | Contig1228_s_at | -1.10 | -1.16 | +1.28 | -1.28 |
| HvPIP1;3 | 13837 | Contig1239_s_at | -1.14 | -1.00 | +1.16 | +1.02 |
| HvPIP1;4 | 13845 | Contig1219_s_at** | -1.08 | -1.00 | +1.19 | -1.19 |
| HvPIP1;5 | 13757 | Contig1230_at** | -1.10 | -1.26 | -1.22 | -1.52 |
| HvPIP2;1 | 13861 | Hv08C12u_x_at** | +1.03 | -1.05 | +1.01 | +1.02 |
| HvPIP2;2 | 13833 | Contig1216_s_at | -1.17 | -1.31 | -1.25 | -1.53 |
| HvPIP2;3 | 13857 | Contig1223_at | +1.40 | +1.03 | +1.95 | -1.12 |
| HvPIP2;4 | 13852 | EBem09_SQ003_F16_s_at | +1.06 | -1.00 | +1.40 | -1.08 |
| HvPIP2;5 | 13825 | Contig1222_s_at | -1.50 | -1.40 | -1.42 | -1.74 |
| HvPIP2;6 | 13694 | - | - | - | - | - |
| HvPIP2;7 | 8763 | Contig19393_at | +1.99 | -1.00 | +1.10 | -1.00 |
| HvPIP2;8 | 13870 | - | - | - | - | - |
| HvPIP2;9 | 43384 | HV_CEb0007N06r2_at | -1.22 | +1.01 | -1.08 | -1.03 |
| TIPs | ||||||
| HvTIP1;1 | 14110 | HS07J06u_s_at | -1.08 | -1.09 | -1.13 | -1.14 |
| HvTIP1;2 | 14114 | HVSMEf0019H18r2_at | +1.04 | -1.15 | +1.33 | -1.18 |
| HvTIP2;1 | 14113 | Contig1310_at** | +1.24 | -1.55 | +1.05 | -1.53 |
| HvTIP2;2 | 14105 | Contig1308_at | +1.13 | -1.76 | +1.10 | -1.87 |
| HvTIP2;3 | 14109 | Contig1315_s_at | -1.28 | -1.65 | -1.15 | -1.81 |
| HvTIP3;1 | 1488 | Contig3772_at; HT03K14r_s_at | +1.15 | +1.07 | +1.01 | -1.00 |
| HvTIP3;2 | - | EBem10_SQ003_I02_at | -1.17 | -1.04 | +1.08 | +1.08 |
| HvTIP4;1 | 16370 | Contig7377_s_at | +1.70 | +1.54 | -2.50 | -1.08 |
| HvTIP4;3 | 38155 | HF03B07r_at | +1.07 | +1.08 | +1.11 | -1.01 |
| HvTIP5;1 | 28056 | AF254799_CDS-2_at | +1.04 | -1.09 | -1.02 | +1.07 |
| NIPs | ||||||
| HvNIP1;1 | 5627 | Contig14229_at | +1.04 | +1.03 | +1.02 | +1.11 |
| HvNIP1;2 | 23039 | Contig19214_at | +1.17 | +1.50 | +1.09 | +1.50 |
| HvNIP2;1 | 31860 | - | - | - | - | - |
| HvNIP2;2 | 16339 | Contig5632_at Contig5632_s_at | -1.07 | -1.28 | +1.22 | -1.50 |
| HvNIP2;3 | 16340 | Contig5634_at | +1.17 | +1.07 | +1.23 | +1.16 |
| HvNIP3;1 | 7157 | Contig16901_at | -1.12 | +1.42 | +1.05 | +1.33 |
| HvNIP4;1 | - | Contig19489_at | +1.20 | +1.09 | +1.07 | +1.04 |
| SIPs | ||||||
| HvSIP1;1 | 2171 | Contig6340_at | -1.15 | +1.29 | +1.10 | +1.24 |
| HvSIP2;1 | - | Contig19630_at | -1.21 | +1.05 | -1.20 | -1.07 |
Microarray data obtained from Genevestigator (https://www.genevestigator.com/gv/plant.jsp).
^Unigene number obtained from HarvEST v.1.83;
**Probeset IDs obtained from [21], other probeset IDs acquired from HarvEST: barley. ‘-’ indicates no information available.
FCs indicative of notable up or down regulation (FC ≥ +1.5) are shown in bold, FCs indicative of no differential expression are shown in normal text. AQPs lacking microarray data are not shown.
Discussion
In recent years, the function of plant AQPs has been extended from transport of water alone to the transport of other substrates and responding to diverse physiological processes and environmental stresses (reviewed in [1]). However, while significant information exists on the AQP families in rice and soybean, a complete picture of this family is missing in barley and wheat, two of the most important cereal crops. In this work, we address this gap by providing comprehensive analysis of what we believe is the full gene set of the barley AQPs superfamily, including new annotations of sixteen transcripts, removal of some redundancies and possible inaccuracies of four annotations (HvNIP2;1, HvNIP2;2, HvTIP1;2 and HvTIP2;3) (Table 1). The annotation of HvNIP2;1 may be regarded as somewhat equivocal, as the sequences AB447482 (registration 14-JUL-2008; Ma, direct submission) and GQ496520 (18-AUG-2009; Sutton et al., direct submission) were annotated as HvNIP2;1, followed by registration of AB540229 (13-JAN-2010; Ligaba et al., direct submission;) also as HvNIP2;1. However, AB540229 is 100% identical to AB447484 and not to the former two. Our annotations of HvNIP2;1 and HvNIP2;2 are based on the dates of registration as well as % identities to the other annotated barley sequences (listed in Table 1) and to the orthologs in rice [37], maize [7] and/or sorghum (SbNIP2;1 - EF373651; SbNIP2;2 - EF408053). Hence we suggest that AB540229 represents HvNIP2;2, and likewise, AB710142 (annotated as HvNIP2;2; 02-APR-2012, Shibasaka et al, direct submission; also see [26]) represents HvNIP2;1. These suggestions are open to further reports and/or Genbank updates. In all, forty genes (nineteen PIPs, eleven TIPs, eight NIPs, two SIPs) were identified, making a significant advancement on the twenty-three (ten PIPs, eight TIPs, three NIPs, two SIPs) estimated by Katsuhara and Hanba [11] and twenty-five (eleven PIPs, eight TIPs, four NIPs, two SIPs) estimated by Besse et al. [21]. This family size is consistent with maize (36; [7]), rice (38; [8]) and soybean (66; [6]). Subject to confirmation, nine more sequences identified in the low confidence dataset may also belong. The AQP diversity in plants is attributed to the higher degree of compartmentalization of plant cells and the ability to fine-tune water control in situ, under different environmental conditions [34]. The gene identifications and analyses are also relevant to wheat, a genetically close relative; e.g., a previous study could not amplify TIP5 from wheat, with primers based on rice TIP5 [10]; the data on HvTIP5 will address this limitation. A wheat NIP is reported to be involved in salinity tolerance [38]; the present work will enable analysis of its orthologue in barley.
The regulation of AQPs under salinity stress has been well-reported (reviewed in [1, 8]). In barley, response to salinity has been studied mainly in PIPs [11–13]. The present study is unique in targeting all leaf AQPs together. Eighteen of the forty genes showed modulations considered meaningful (FC >1.5; [35]) (S1 Table). The changes in HvTIP4;1 and HvPIP1;2 were statistically significant (p-value < 0.05; S1 Table), indicative of their key roles in stress response, particularly noteworthy HvTIP4;1 which exhibited the largest change. The expression of HvPIP1;2 in the leaf was unclear in other studies (EST data, S4 Table; [21]). However, this isoform was found significantly up-regulated (S3 Fig; S1 Table), and predicted to be a CO2 transporter (Table 3). Pérez-López et al. [39] demonstrated that elevated CO2 moderates the effects of oxidative stress caused by salinity in barley. Therefore a surge in HvPIP1;2 may be protective, and other predicted (HvPIP1;1–1;5; HvPIP2;6, HvPIP2;7, HvPIP2;10; current study) and confirmed (HvPIP2;1–2;3, HvPIP2;5; [40]) CO2 transporters may also have similar roles. Their genes and expression patterns thus need to be compared in tolerant versus sensitive lines.
The isoform-specific response of barley AQPs noted here support the observations in maize [41]. Tissue-specific responses were also observed, e.g., HvPIP1;2 was down-regulated in the root [11, 13], but up-regulated in leaf mRNA-seq (S3 Fig; S1 Table). Differences were also noticed in results from different techniques; e.g., HvPIP2;1 was found largely unchanged in the microarray (Table 4), but down-regulated in roots [12, 13] and up-regulated in shoots by real-time PCR [12] and leaf mRNA-seq [19] and in IBGSC shoot + tillers RNA-Seq data (S5 Fig). In addition, the response of aquaporins to stimuli is often species-dependent. For example, PIP1;1 from rice was shown to be down-regulated in response to 150 mM NaCl [42], while its ortholog in barley (HvPIP1;1) was up-regulated (our current study). The expression of AQPs during water stress is reported to be dependent on cultivars [43] as well as time course and intensity of stress [41]. Thus investigations of wider germ-plasm for allelic differences are essential for assessing the genetic versus environmental factors (including experimental techniques) to such studies.
An N-terminal diacidic motif Asp-Ile-Glu in ZmPIP2;4 and ZmPIP2;5 [28] or Asp-Val-Glu in AtPIP2;1 [29], found important for exit of PIPs from the ER to plasma membrane, was noted in HvPIP2;1, HvPIP2;3 and HvPIP2;4, suggesting they may use this mechanism. It is unclear how the other HvPIPs lacking this motif but also predicted to be plasma membrane-located would be exported. For PIP1s, hetero-tetramer formation with PIP2s has been suggested as means of ER export [28]. The plant cell vacuole is associated with turgor regulation, osmotic adjustment, storage, pH regulation, cell signalling and protein degradation. HvTIP4;1 was predicted to be vacuolar (Table 3), similar to AtTIP4;1 [44]. OsTIP4;1 has been confirmed to be water-permeable and its transcripts fluctuate, possibly in response to turgor status [45], and the orthology suggests that HvTIP4;1 may play a similar role. In addition, other TIPs predicted to be localised to the vacuole could also play this role.
AQP activity can be regulated transcriptionally, and/or post-translationally by pH, phosphorylation and vesicle trafficking [1]. Protonation of a His in loop D of PIPs is shown to be important for pH-dependent gating during flooding (e.g., His 193 in SoPIP2;1; [30]). All HvPIPs except HvPIP2;7 and HvPIP2;10 possessed a His corresponding to His193 (S2 Fig), but the TIPs, NIPs and SIPs did not, suggesting HvPIPs may be regulated by pH, but not other isoforms. Tornroth-Horsefield et al. [30] also demonstrated the gating of SoPIP2;1 by phosphorylation of Ser115 and Ser274, with Leu197 as the key blocking residue. The equivalents of Ser115, Ser274, and Leu197 in most HvPIPs (S2 Fig) suggest a similar regulation. In contrast, most TIPs lacked these Sers, and Leu197 occurred only in HvTIP4;3 and HvTIP5;1, suggesting the gating may not apply. The Ser at Ser262 in GmNOD26 [46] was noted in all NIPs (except HvNIP3;2) suggesting their phosphorylation-mediated regulation also. The regulation of AQPs is critical in maintaining the plant water status during normal and stress conditions, and this ability could be employed in the development of transgenic plants. AtPIP2;1 is mono- and di-phosphorylated at two C-terminal sites, S280 (i.e. S274 of SoPIP2;1) and S283 under salt stress [31]. Alignments with AtPIP2;1 showed a conserved Ser at S280 and S283 in most HvPIP2s (S2 Fig), suggesting that these, although not exhibiting transcriptional modulations under salt stress, may be regulated post-translationally.
In recent years, plant AQPs, predominantly NIPs, have been shown to be permeable to twelve different substrates, many of which are of physiological significance, e.g., ammonia and CO2 [1], and the signalling molecule hydrogen peroxide [47], but some are toxic, e.g., arsenite. Hence, the involvement of the barley AQPs in transporting any non-aqua substrates was predicted using the specificity determining positions identified earlier [32]. Based on these predictions, a number of PIP isoforms have the potential to transport boron, CO2 and urea which are important for plant development (boron also being toxic in high concentrations), the TIPs share the urea and ammonia transport potential but not CO2, and transport of the signalling molecule H2O2 seems restricted to TIPs (Table 3). Some of these are experimentally proven substrates, e.g., boron for HvPIP1;3 and HvPIP1;4 [48] and HvNIP2;1 [49]; CO2 for HvPIP2;1–2;3 and HvPIP2;5 [50, 40]; arsenite for HvNIP1;2 [26], and silicon for HvNIP2;1 [51]. Many NIPs appear to have the potential to transport diverse substrates including the micronutrient silicon, but also the toxic arsenite. Further, in our previous work [32] the transport of non-aqua substrates was predicted to generally compromise the water permeability of such AQPs; however, this did not seem to apply to AtTIP1;1, AtTIP2;1, AtNIP1;1, HvPIP2;1, OsPIP2;4, OsPIP2;6, OsPIP2;7, OsTIP1;2, PtNIP1;1 and TaTIP2;2. Thus, their orthologs in barley may also be water transporters.
HvPIP1;3 was found up-regulated in response to boron, while HvPIP1;4 was unchanged [48]. In the leaf mRNA-seq data, both of these were up-regulated under salinity stress (S3 Fig; S1 Table). Down-regulation of HvNIP2;1 is suggested to induce boron tolerance [48]; this isoform was found down-regulated by salinity in our study [19]. Orthologs often retain their function during evolution; thus HvNIP2;1 may transport the substrates shown for OsNIP2;1, i.e., boron, silicon and urea [52] and arsenite and antimony [53], and may not necessarily show significant differential expression under salinity stress (alone) (see below). Boron toxicity and salinity both significantly reduce crop yield [54], and may occur together, e.g., in parts of Western Australia. Thus isoforms such as HvPIP1;3 and HvNIP2;1 would be invaluable for identification/development of lines tolerant to both stresses. HvPIP1;1, HvPIP1;2 and HvPIP2;1 are also of interest due to being salt-responsive and possible boron transporters, and HvPIP1;5, HvPIP2;3 HvPIP2;5, HvNIP2;2 and the newly identified HvNIP3;1 are also likely boron transporters. Urea is abundant in nature and application of urea fertilisers is a common agricultural practice; hence there is a need for plants to ‘load and unload’ urea [44], and some of the barley isoforms may be urea transporters. Silicon is found to alleviate abiotic and biotic stresses and improve light interception and canopy photosynthesis, and HvNIP2;1 and HvNIP2;2 being silicon transporters ([55] and within) was supported by our predictions. Isoforms that transport antimony or arsenite could be valuable in their detoxification. The rice orthologs could assist in testing the functions of barley genes, and the sequences and physical locations of barley genes are important for marker development and studying wheat orthologs.
Conclusions
Forty AQPs is the largest number reported for barley so far. The data acquired on their coding sequences, innate tissue and/or cultivar specific variations in expression, modulations under salinity, and the potential roles of various genes in transport of other substrates and in salinity and other stress responses, have collectively yielded a wealth of molecular information. This will be vital for assessing and/or transgenically developing germplasm of barley and possibly of wheat with improved abiotic stress tolerance and yield.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are most grateful to Dr Nils Stein (Leibniz Institute of Plant Genetics & Crop Plant Research (IPK), Gatersleben, Germany) for advice on analysis of the genomic sequence of some of the barley aquaporins. Thank you also to Mr Atul Kamboj (Swinburne University of Technology) for assistance with mRNA-seq data analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1. Maurel C, Verdoucq L, Luu DT, Santoni V (2008) Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol 59: 595–624. 10.1146/annurev.arplant.59.032607.092734 [DOI] [PubMed] [Google Scholar]
- 2. Gustavsson S, Lebrun A-S, Norden K, Chaumont F, Johanson U (2005) A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiol 139: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Danielson JA, Johanson U (2008) Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens . BMC Plant Biol 8: 45–60. 10.1186/1471-2229-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Danielson JA, Johanson U (2010) Phylogeny of major intrinsic proteins In: Jahn TP, Bienert GP (editors) MIPs and their role in the exchange of metalloids. Advances in Experimental Medicine and Biology, volume 679, pp 19–31. [DOI] [PubMed] [Google Scholar]
- 5. Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F (2011) Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J 66: 306–317. 10.1111/j.1365-313X.2011.04496.x [DOI] [PubMed] [Google Scholar]
- 6. Zhang DY, Ali Z, Wang CB, Xu L, Yi JX, Xu ZL, et al. (2013) Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 8: e56312 10.1371/journal.pone.0056312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol 125: 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forrest KL, Bhave M (2007) Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Funct Integr Genom 7: 263–289. [DOI] [PubMed] [Google Scholar]
- 9. Guo SW, Kaldenhoff R, Uehlein N, Sattelmacher B, Brueck H (2007) Relationship between water and nitrogen uptake in nitrate- and ammonium-supplied Phaseolus vulgaris L. plants. J Plant Nutr Soil Sc 170: 73–80. [Google Scholar]
- 10. Forrest KL, Bhave M (2008) The PIP and TIP aquaporins in wheat form a large and diverse family with unique gene structures and functionally important features. Funct Integr Genom 8: 115–133. [DOI] [PubMed] [Google Scholar]
- 11. Katsuhara M, Hanba YT (2008) Barley plasma membrane intrinsic proteins (PIP aquaporins) as water and CO2 transporters. Pflugers Arch Eur J Physiol 456: 687–691. 10.1007/s00424-007-0434-9 [DOI] [PubMed] [Google Scholar]
- 12. Katsuhara M, Akiyama Y, Koshio K, Shibasaka M, Kasamo K (2002) Functional analysis of water channels in barley roots. Plant Cell Physiol 43: 885–893. [DOI] [PubMed] [Google Scholar]
- 13. Horie T, Kaneko T, Sugimoto G, Sasano S, Panda SK, Shibasaka M, et al. (2011) Mechanism of water transport mediated PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell Physiol 52: 663–675. 10.1093/pcp/pcr027 [DOI] [PubMed] [Google Scholar]
- 14. Knipfer T, Besse M, Verdeil JL, Fricke W (2011) Aquaporin-facilitated water uptake in barley (Hordeum vulgare L.) roots. J Exp Bot 62: 4115–4126. 10.1093/jxb/err075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katsuhara M, Tsuji N, Shibasaka M, Panda SK (2014a) Osmotic stress decreases PIP aquaporin transcripts in barley roots but H2O2 is not involved in this process. J Plant Res 127: 787–792. 10.1007/s10265-014-0662-y [DOI] [PubMed] [Google Scholar]
- 16. Zohary D, Hopf M (2000) Domestication of plants in the old world: the origin and spread of cultivated plants in West Asia, Europe, and the Nile Valley, 3rd edn New York, Oxford University Press. [Google Scholar]
- 17. Ullrich S (2011) In: Ullrich SE (ed) Barley: Production, improvement and uses. John Wiley and sons, West Sussex, UK, pp 3–13. [Google Scholar]
- 18. Fricke W, Bijanadeh E, Emam Y, Knipfer T (2014) Root hydraulics in salt-stressed wheat. Funct Plant Biol 41: 366–378. [DOI] [PubMed] [Google Scholar]
- 19. Ziemann M, Kamboj A, Hove RM, Loveridge S, El-Osta A, Bhave M (2013) Analysis of the barley leaf transcriptome under salinity stress using mRNA-Seq. Acta Physiol Plant 35: 1915–1924. [Google Scholar]
- 20. The International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- 21. Besse M, Knipfer T, Miller T, Verdeil J-L, Jahn T, Fricke W (2011) Developmental pattern of aquaporin expression in barley (Hordeum vulgare L.) leaves. J Exp Bot 62: 4127–4142. 10.1093/jxb/err175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shibasaka M, Sasano S, Utsugi S, Katsuhara M (2012) Functional characterization of a novel plasma membrane intrinsic protein2 in barley. Plant Signal Behav 7: 1648–1652. 10.4161/psb.22294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Froger A, Tallur B, Thomas D, Delamarche C (1998) Prediction of functional residues in water channels and related proteins. Protein Sci 7: 1458–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suga S, Maeshima M (2004) Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol 45: 823–830. [DOI] [PubMed] [Google Scholar]
- 25. Zhang M, Lu S, Li G, Mao Z, Yu X, Sun W, et al. (2010) Identification of a residue in helix 2 of rice plasma membrane intrinsic proteins that influences water permeability. J Biol Chem 285: 41982–41992. 10.1074/jbc.M110.101790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katsuhara M, Sasano S, Horie T, Matsumoto T, Rhee J, Shibasaka M (2014b) Functional and molecular characteristics of rice and barley NIP aquaporins transporting water, hydrogen peroxide and arsenite. Plant Biotech 31: 213–219. 10.2108/zs130215 [DOI] [PubMed] [Google Scholar]
- 27. Bienert GP, Cavez D, Besserer A, Berny MC, Gilis D, Rooman M, et al. (2012) A conserved cysteine residue is involved in disulfide bond formation between plant plasma membrane aquaporin monomers. Biochem J 445: 101–111. 10.1042/BJ20111704 [DOI] [PubMed] [Google Scholar]
- 28. Zelazny E, Miecielica U, Borst JW, Hemminga MA, Chaumont F (2009) An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. Plant J 57: 346–355. 10.1111/j.1365-313X.2008.03691.x [DOI] [PubMed] [Google Scholar]
- 29. Sorieul M, Santoni V, Maurel C, Luu DT (2011) Mechanisms and effects of retention of over-expressed aquaporin AtPIP2;1 in the endoplasmic reticulum. Traffic 12: 473–482. 10.1111/j.1600-0854.2010.01154.x [DOI] [PubMed] [Google Scholar]
- 30. Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, et al. (2006) Structural mechanism of plant aquaporin gating. Nature 439: 688–694. [DOI] [PubMed] [Google Scholar]
- 31. Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, et al. (2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7: 1019–1030. 10.1074/mcp.M700566-MCP200 [DOI] [PubMed] [Google Scholar]
- 32. Hove RM, Bhave M (2011) Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol Biol 75: 413–430. 10.1007/s11103-011-9737-5 [DOI] [PubMed] [Google Scholar]
- 33. Kvam VM, Liu P, Si Y (2012) A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am J Bot 99: 248–256. 10.3732/ajb.1100340 [DOI] [PubMed] [Google Scholar]
- 34. Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, et al. (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59: 469–484. [DOI] [PubMed] [Google Scholar]
- 35. Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa . J Exp Bot 61: 4157–468. 10.1093/jxb/erq237 [DOI] [PubMed] [Google Scholar]
- 36. Walia H, Wilson C, Condamine P, Ismail AM, Xu J, Cui X, et al. (2007) Array-based genotyping and expression analysis of barley cv Maythorpe and Golden Promise. BMC Genomics 8: 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol 46: 1568–1577. [DOI] [PubMed] [Google Scholar]
- 38. Gao Z, He X, Zhao B, Zhou C, Liang Y, Ge R, et al. (2010) Overexpressing a putative aquaporin gene from wheat TaNIP, enhances salt tolerance in transgenic Arabidopsis. Plant Cell Physiol 51: 767–775. 10.1093/pcp/pcq036 [DOI] [PubMed] [Google Scholar]
- 39. Pérez-López U, Robredo A, Lacuesta M, Sgherri C, Munoz-Rueda A, Navari-Izzo F, et al. (2009) The oxidative stress caused by salinity in two barley cultivars is mitigated by elevated CO2 . Physiol Plant 135: 29–42. 10.1111/j.1399-3054.2008.01174.x [DOI] [PubMed] [Google Scholar]
- 40. Mori IC, Rhee J, Shibasaka M, Sasano S, Kaneko T, Horie T, et al. (2014) CO2 transport by PIP2 aquaporins of barley. Plant Cell Physiol 55: 251–257. 10.1093/pcp/pcu003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu C, Schraut D, Hartung W, Schaffner AR (2005) Differential responses of maize MIP genes to salt stress and ABA. J Exp Bot 56: 2971–2981. [DOI] [PubMed] [Google Scholar]
- 42. Li L, Li S, Tao Y, Kitagawa Y (2000) Molecular cloning of a novel water channel from rice: its products expression in Xenopus oocytes and involvement in chilling tolerance. Plant Sci 154: 43–51. [DOI] [PubMed] [Google Scholar]
- 43. Lian H-L, Yu X, Ye Q, Ding X, Kitagawa Y, Kwak S-S, et al. (2004) The role of aquaporin RWC3 in drought avoidance in rice. Plant Cell Physiol 45: 481–489. [DOI] [PubMed] [Google Scholar]
- 44. Liu L-H, Ludewig U, Gassert B, Frommer WB, von Wiren N (2003) Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol 133: 1220–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li G-W, Peng Y-H, Yu X, Zhang M-H, Cai W-M, Sun W-N, et al. (2008) Transport functions and expression analysis of vacuolar membrane aquaporins in response to various stresses in rice. J Plant Physiol 165: 1879–1888. 10.1016/j.jplph.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 46. Guenther JF, Chanmanivone N, Galetovic MP, Wallace IS, Cobb JA, Roberts DM (2003) Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell 15: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bienert GP, Chaumont F (2014) Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochem Biophys Acta General Subjects 1840: 1596–1604. 10.1016/j.bbagen.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 48. Fitzpatrick KL, Reid R (2009) The involvement of aquaglyceroporins in transport of boron in barley root. Plant Cell Envi 32: 1357–1365. 10.1111/j.1365-3040.2009.02003.x [DOI] [PubMed] [Google Scholar]
- 49. Schnurbusch T, Hayes J, Hrmova M, Baumann U, Ramesh S, Tyerman S, et al. (2010) Boron toxicity tolerance through reduced expression of the multifunctional aquaporin HvNIP2;1. Plant Physiol 153: 1706–1715. 10.1104/pp.110.158832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanba YT, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K, Terashima I, et al. (2004) Overexpression of the barley aquaporin HvPIP2;1 increases internal CO2 assimilation in the leaves of transgenic rice plants. Plant Cell Physiol 45: 521–529. [DOI] [PubMed] [Google Scholar]
- 51. Chiba Y, Mitani N, Yamaji N, Ma JF (2009) HvLsi1 is a silicon influx transporter in barley. Plant J 57: 810–818. 10.1111/j.1365-313X.2008.03728.x [DOI] [PubMed] [Google Scholar]
- 52. Mitani N, Yamaji N, Ma JF (2008) Characterization of substrate specificity of a rice silicon transporter, Lsi1 . Pflugers Arch Eur J Physiol 456: 679–786. 10.1007/s00424-007-0408-y [DOI] [PubMed] [Google Scholar]
- 53. Bienert GP, Thorsen M, Schüssler MD, Nilsson HR, Wagner A, Tamás MJ, et al. (2008) A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol 6: 26–41. 10.1186/1741-7007-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tanaka M, Fujiwara T (2008) Physiological roles and transport mechanism of boron: perspective from plants. Eur J Physiol 456: 671–677. [DOI] [PubMed] [Google Scholar]
- 55. Yamaji N, Chiba Y, Mitani-Ueno N, Ma JF (2012) Functional characterization of a silicon transporter gene implicated in Si distribution in barley. Plant Physiol 160: 1491–1497. 10.1104/pp.112.204578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.