Abstract
The Tat protein of human immunodeficiency virus type 1 (HIV-1) is essential for viral replication and activates RNA polymerase II transcriptional elongation through the association with a cellular protein kinase composed of Cdk9 and cyclin T1. Tat binds to this kinase complex through a direct protein-protein interaction with cyclin T1. Monocytes/macrophages are important targets of HIV-1 infection, and previous work has shown that cyclin T1 but not Cdk9 protein expression is low in monocytes isolated from blood. While Cdk9 expression is expressed at a high level during monocyte differentiation to macrophages in vitro, cyclin T1 expression is induced during the first few days of differentiation and is shut off after 1 to 2 weeks. We show here that the shutoff of cyclin T1 expression in late-differentiated macrophages involves proteasome-mediated proteolysis. We also show that cyclin T1 can be reinduced by a number of pathogen-associated molecular patterns that activate macrophages, indicating that up-regulation of cyclin T1 is part of an innate immune response. Furthermore, we found that HIV-1 infection early in macrophage differentiation results in sustained cyclin T1 expression, while infection at late times in differentiation results in the reinduction of cyclin T1. Expression of the viral Nef protein from an adenovirus vector suggests that Nef contributes to the HIV-1 induction of cyclin T1. These findings suggest that HIV-1 infection hijacks a component of the innate immune response in macrophages that results in enhancement rather than inhibition of viral replication.
Efficient RNA polymerase II (RNAP II) transcriptional elongation of the human immunodeficiency virus type 1 (HIV-1) provirus requires the viral Tat protein. Tat activates transcription through the recruitment of a cellular protein kinase named TAK/P-TEFb to the TAR RNA structure located at the 5′ end of nascent viral transcripts (reviewed in references 2, 9, 19, and 26). TAK/P-TEFb is composed of the cyclin T1 regulatory subunit and the Cdk9 catalytic subunit. Cdk9 is also found in complexes that contain cyclin subunits other than cyclin T1; these additional cyclins are cyclins T2a, T2b, and K (17). All Cdk9/cyclin complexes are collectively termed P-TEFb and activate transcriptional elongation through phosphorylation of the carboxyl-terminal domain of RNAP II and the Spt5 subunit of the DSIF negative factor (8, 11, 17). Tat makes direct protein-protein contact with cyclin T1 and therefore can only associate with the cyclin T1-containing P-TEFb complex (27). Recently, an snRNA known as 7SK snRNA has been found to associate with P-TEFb and negatively regulate kinase activity in vitro (15, 28). Interestingly, although 7SK snRNA has a negative function for P-TEFb in vitro, there is a large increase in the association of 7SK snRNA with P-TEFb upon activation of resting peripheral blood lymphocytes (PBLs), a system in which transcriptional elongation is increased globally (4).
The major target cells for HIV-1 infection are CD4+ T lymphocytes and macrophages, but the ability of HIV-1 to replicate in these two cell types is dependent upon the activation or maturation state of the cell. T-cell activation is required for HIV-1 replication in CD4+ T lymphocytes, while monocyte-derived macrophages (MDMs) are much more susceptible to infection than freshly isolated monocytes (20, 29). T-cell activation and macrophage differentiation are likely to enhance viral replication through induction of a number of cellular cofactors utilized by HIV-1 during its infectious cycle.
In a previous study of the effects of macrophage differentiation on TAK/P-TEFb function, we observed that Cdk9 levels are high in freshly isolated monocytes and remain high during differentiation (14). In contrast, cyclin T1 protein expression is very low in monocytes, increases strongly early during differentiation, and decreases to a very low level after 1 to 2 weeks in culture. This transient induction of cyclin T1 expression during differentiation involves posttranscriptional regulation, as reverse transcription (RT)-PCR assays revealed that cyclin T1 mRNA levels are high in monocytes and do not decrease when cyclin T1 protein expression is shut off late in differentiation. Additionally, we found that lipopolysaccharide (LPS), a component of the cell wall of gram-negative bacteria, can reinduce cyclin T1 expression in late-differentiated macrophages. These findings raised the possibility that HIV-1 replication rates in macrophages may be affected by changes in cyclin T1 protein expression.
The transient induction of cyclin T1 during macrophage differentiation was unexpected because HIV-1 generally replicates for extended periods in macrophages in culture (7, 16). In the present study, we investigated HIV-1 replication in macrophages under culture conditions in which cyclin T1 expression is regulated by differentiation. We found that HIV-1 infection early during differentiation results in sustained expression of cyclin T1. Additionally, we observed that infection can induce cyclin T1 after it has declined in late-differentiated macrophages, and the analysis of an adenovirus expression vector suggests that the viral Nef protein contributes to this induction. We found that cyclin T1 expression can be reinduced by multiple pathogen-associated molecular patterns (PAMPs) in addition to LPS. The use of proteasome inhibitors demonstrated that the down-regulation of cyclin T1 late in macrophages is mediated by proteasome-mediated proteolysis.
MATERIALS AND METHODS
Cells.
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy HIV-seronegative blood donors (obtained from the Gulf Coast Regional Blood Center) by density gradient centrifugation using Isolymph (Gallard/Schlesinger). Monocytes were isolated from PBMCs by an adherence method as described previously (14). Briefly, 2.5 × 106 PBMCs/ml in RPMI 1640 supplemented with 1% human serum (Sigma), penicillin, and streptomycin were seeded onto 10-cm-diameter plastic cell culture dishes (Sarstedt) and incubated at 37°C for 1 h. After removing nonadherent cells by washing with prewarmed phosphate-buffered saline (PBS), adherent cells were then incubated with RPMI 1640 complete medium for another 2 h. After additional washes with prewarmed PBS, cells were cultured in RPMI 1640 (Invitrogen Life Technologies) supplemented with 10% fetal bovine serum (FBS) and antibiotics (Invitrogen Life Technologies) plus granulocyte macrophage colony-stimulating factor (10 U/ml; R&D system). PBLs were isolated from PBMCs by depleting monocytes by two rounds of plastic adherence. To activate PBLs, cells were treated with phytohemagglutinin (PHA; 1 μg/ml; Sigma) and phorbol 12-myristate 13-acetate (PMA; 1 ng/ml; Sigma) for 48 h. Jurkat cells were cultured with RPMI 1640 supplemented with 10% FBS and antibiotics. 293T cells were cultured within Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics.
MDM treatment.
MDMs were treated with PAMPs at the following final concentrations: LPS, 0.1 μg/ml (Sigma); lipoteichoic acid (LTA), 10 μg/ml (Sigma); peptidoglycan (PGN), 10 μg/ml (Sigma); double-strand RNA poly(I:C), 50 μg/ml (Sigma); and CpG DNA, 2 μM (5′-phosphorothioate-TCG TCG TTT TGT CGT TTT GTC GTT; Invitrogen Life Technologies). The culture supernatants were collected and tumor necrosis factor alpha (TNF-α) was measured by enzyme-linked immunosorbent assay (ELISA) following the manufacturer's protocol (Biosource). The recombinant adenovirus expressing the HIV-1 Nef protein (25) used to infect MDMs was a generous gift from M. Stevenson. Proteasome inhibitors, MG101 (LLnL) and MG132 were purchased from Sigma and were used at a final concentration of 50 μM for 1 h.
HIV-1 infection.
Two HIV-1 proviral plasmids were used in this study: pBRU3Δenv (referred to as HIV-1Δenv; a generous gift from M. Emerman, Fred Hutchinson Cancer Research Center) and HIV-IRES-eGFP (expresses enhanced green fluorescent protein [GFP]). HIV-IRES-eGFP hereafter is referred to as HIV-GFP. HIV-GFP is Δnef Δvpr Δvpu Δvif Δenv (a generous gift from R. E. Sutton, Baylor College of Medicine). To generate HIV-1 stocks, proviral plasmids were cotransfected with a plasmid expressing vesicular stomatitis virus glycoprotein (VSV-G) and a plasmid expressing HIV-1 Tat into 293T cells by calcium phosphate coprecipitation. Culture supernatants were collected 3 days posttransfection and were concentrated by centrifugation at 23,000 rpm for 2.5 h in an SW28 rotor (23). The viral pellet was resuspended with PBS. Typically, the virus titer for HIV-GFP was from 107 to 108 infectious units/ml after concentration as determined by infections in 293T cells.
Infection of MDMs was carried out with 0.3 ml of concentrated virus in the presence of 2.5 ml of fresh medium and 6-μg/ml Polybrene for 8 h. In experiments with AZT (3′-azido-3′-deoxythymidine; Sigma), medium containing 20 μM AZT was used throughout experiments. The amount of virus applied to MDMs ranged from 10 to 100 infectious units as inferred from titers of HIV-GFP stock measured in infections of 293T cells. Flow cytometry analyses of MDM cultures infected at day 2 of differentiation with several concentrated stocks of HIV-1 Δenv indicated that from 10 to 20% of cells were positive for intracellular gag expression after 5 days of infection. To measure HIV-1 replication, portions of cell supernatants were collected at the indicated times and p24 assays were performed following the manufacturer's protocol (Beckman Coulter). Additionally, in some experiments, MDMs were lysed with lysis buffer (Beckman Coulter) and intracellular p24 levels were measured.
Immunoblots.
Cell extracts were prepared by incubating cells in lysis buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40) containing protease inhibitors (2-μg/ml aprotinin, 1-μg/ml leupeptin, 2.5 mM phenylmethylsulfonyl fluoride) as described previously (6). Protein concentrations were determined by a Bio-Rad protein assay, and 20 μg of total protein was loaded onto sodium dodecyl sulfate-9% polyacrylamide gels. The procedure for immunoblots using enhanced chemiluminescence for detection was described previously (5). Antibody to β-actin was purchased from Sigma, and other antibodies were purchased from Santa Cruz Biotechnology.
RESULTS
Multiple PAMPs reinduce cyclin T1 expression in late-differentiated macrophages.
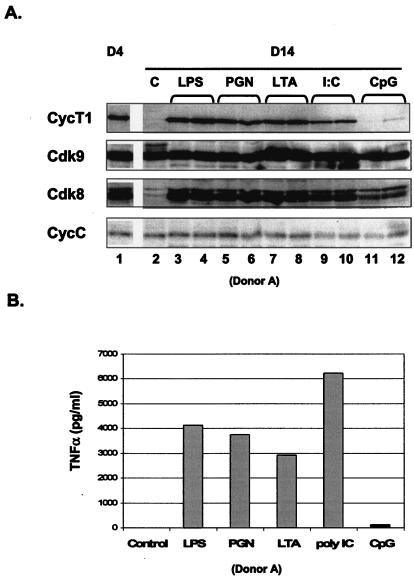
Previously, we showed that LPS can reinduce cyclin T1 expression after it has been shut off late in differentiation of MDMs (14). LPS is an example of a PAMP that signals through Toll-like receptors (TLRs) and activates innate immune functions of macrophages. To date, 10 TLRs have been identified, most of which can be detected on the cell surface of macrophages, with the exception of TLR9 (reviewed in references 12 and 13). To examine whether PAMPs in addition to LPS can reinduce cyclin T1 expression in late-differentiated macrophages, cultures were treated in duplicate with LPS, PGN, LTA, or poly(I:C), which signal through TLR4, -2/-6, -2, and -3, respectively. Bacterial CpG DNA, which is recognized by TLR9, was used as a negative control, as macrophages do not generally express TLR9. After 2 days of incubation with PAMPs, cell extracts were prepared and cyclin T1 expression was analyzed in immunoblots (Fig. 1A). TNF-α in the culture supernatant was analyzed by ELISA to verify that the PAMPs activated macrophages (Fig. 1B). As expected, TNF-α was induced by LPS, PGN, LTA, and poly(I:C), but not CpG DNA. The level of cyclin T1 protein in day 14 MDM was extremely low relative to its level in day 4 MDMs (compare lanes 1 and 2). However, all PAMPs except CpG DNA were able to reinduce cyclin T1 expression. Because cyclin T1 is induced by all PAMPs that were able to activate macrophages, the up-regulation of cyclin T1 appears to be a component of an innate immune response in macrophages.
FIG. 1.
Reinduction of cyclin T1 protein expression by multiple PAMPs. (A) Duplicate MDM cultures (donor A) at day 12 of differentiation were treated with LPS, PGN, LTA, poly(I:C) double-stranded RNA, or CpG DNA. A control culture (lane 2) was not treated with PAMPs. After 2 days of treatment (day 14 of differentiation [D14]), cell extracts were prepared and levels of the indicated proteins were determined by immunoblotting. A control extract was prepared from an MDM culture at day 4 of differentiation (D4). (B) Cultured supernatants from panel A were collected after 2 days of PAMP treatments and assayed for TNF-α by ELISA.
We observed previously that similar to cyclin T1, Cdk8 expression is reduced late in macrophage differentiation (14). Like P-TEFb complexes, the Cdk8/cyclin C complex is thought to regulate transcription by phosphorylation of the C-terminal domain (CTD) of RNAP II (21). All PAMPs that induce cyclin T1 also induced Cdk8 (Fig. 1A). PAMPs had no effects on the levels of Cdk9, the Cdk partner of cyclin T1, and cyclin C, the regulatory subunit of Cdk8. We observed that the induction of cyclin T1 and Cdk8 by PAMPs required 1 to 2 days of treatment (data not shown), in contrast to a rapid 1-h induction by proteasome inhibitors as discussed below.
Cyclin T1 protein is shut off by proteasome-mediated proteolysis.
We wished to examine mechanisms involved in the shutoff of cyclin T1 expression late in MDM differentiation. Cyclin T1 contains a PEST consensus sequence at its carboxyl terminus, which may be a signal for ubiquitination and proteasomal proteolysis (18, 27). To examine whether cyclin T1 shutoff may involve the proteasome pathway, we incubated MDMs at various time points of differentiation with proteasome inhibitor MG101 or MG132 (Fig. 2A). The proteasome inhibitors had minor effects on cyclin T1 protein levels in cells treated at day 4 of differentiation, when cyclin T1 level was high in the untreated culture (lanes 1 to 3). Cyclin T1 levels were increased by treatment with both proteasome inhibitors on day 6, at which time cyclin T1 protein was beginning to decrease in the untreated culture (lanes 4 to 6). At day 9, when cyclin T1 expression was largely shut off in the untreated culture dish, the proteasome inhibitors resulted in a dramatic increase in cyclin T1 expression. We observed similar results with proteasome inhibitors with MDM preparations from four additional donors. These results suggest that cyclin T1 shutoff late in differentiation is mediated by proteasomal degradation.
FIG. 2.
Cyclin T1 is degraded by proteasome-mediated proteolysis. (A) MDMs (donor E) were treated for 1 h with proteasome inhibitors (A, MG101; B, MG132) at the indicated days of differentiation, cell extracts were prepared, and the indicated proteins were detected by immunblotting. (B) Resting PBLs (control) and PBLs activated by PMA plus PHA were treated with MG132 for 1 h prior to preparation of cell extracts. Jurkat and 293T cells were incubated with MG132 for the indicated times prior to preparation of cell extracts. The levels of the indicated proteins were determined by immunoblotting.
MDM cultures treated with proteasome inhibitors showed a strong induction of Cdk8 at day 9 of differentiation (Fig. 2A, lanes 7 to 9). Of note, proteasome inhibitors had no effect on Cdk9, the Cdk partner of cyclin T1, and cyclin C, the regulatory subunit of Cdk8. Thus, the Cdk9/cyclin T1 and Cck8/cyclin C kinase activities are regulated similarly in late differentiated MDMs, although the mechanisms involved target the regulatory subunit of the Cdk9/cyclin T1 complex and the catalytic subunit of Cdk8/cyclin C complex.
We also investigated whether proteasome inhibitors would have an effect on cyclin T1 levels in other cell types. PBL, Jurkat (CD4+ T-cell line), and 293T (human embryonic kidney cell line) cultures were incubated with proteasome inhibitors and effects on cyclin T1 expression levels were examined (Fig. 2B). No significant effect of either proteasome inhibitor was observed in these cells, suggesting that the proteasome-mediated degradation of cyclin T1 is restricted to macrophages and perhaps a limited number of other cell types.
Cyclin T1 is induced by HIV-1 infection.
Since cyclin T1 is the direct target of the Tat protein, we wished to examine HIV-1 replication in macrophages under culture conditions in which cyclin T1 is shut off late in differentiation. MDM culture dishes were prepared from a healthy blood donor, and a dish was infected at day 2 of differentiation with an HIV-1 mutant with a deletion in the envelope (HIV-1Δenv, pseudotyped with VSV-G glycoprotein), and the culture supernatant was analyzed for the accumulation of secreted p24 (Fig. 3A). Because the HIV-1 used in this study has an envelope deletion, infection cannot spread through the culture and p24 is expressed only from the input integrated virus. Control extracts were prepared from uninfected MDM cultures from this donor at days 4 and 10 of differentiation, and immunoblots demonstrated the shutoff of cyclin T1 but not Cdk9 at day 10 (data not shown). Although cyclin T1 declined in the uninfected culture by day 10, p24 levels in the supernatant of the infected culture continued to accumulate in a linear fashion through day 20 of differentiation. We observed similar results to those shown in Fig. 3A with MDMs isolated from a number of other donors, assaying both intracellular and secreted p24. These results suggest that HIV-1 infection may overcome the shutoff of cyclin T1 expression in uninfected MDMs, perhaps through the induction of cyclin T1 expression.
FIG. 3.
HIV-1 infection induces cyclin T1 expression in MDMs. (A) An MDM culture (donor F) was infected on day 2 with an HIV-1Δenv, and accumulation of p24 in the culture supernatant was determined by ELISA. (B) MDM cultures (donor G) were infected with HIV-1Δenv or HIV-GFP or were mock infected at day 2 of differentiation. At day 18 of differentiation, cell extracts were prepared and the levels of cyclin T1 and Cdk9 were determined by immunoblotting. (C) MDM cultures (donor D) were allowed to differentiate until day 15, and cells were either mock infected (lane 1) or infected with HIV-1Δenv (lane 2). Cell extracts were prepared 3 days later at day 18 of differentiation, and cyclin T1 and Cdk9 levels were determined by immunoblotting. In an independent experiment, MDM cultures (donor H) were allowed to differentiate until day 30; an uninfected MDM cell extract was prepared as a control at day 30 (lanes 3). Culture dishes were infected with HIV-1Δenv at day 30 in the presence or absence of AZT (lanes 4 and 5), and 3 days later at day 33 of differentiation, cell extracts were prepared and analyzed by immunoblotting.
To examine if HIV-1 infection early in differentiation might result in sustained cyclin T1 expression, we infected an MDM culture with the HIV-1Δenv at day 2 of differentiation and examined cyclin T1 expression at day 18 of differentiation (Fig. 3B). Additionally, we infected a culture dish with an HIV-1 that expresses GFP and is defective for expression of Env, Vif, Vpr, Vpu, and Nef. In the uninfected culture, cyclin T1 but not Cdk9 was largely shut off at day 18. In the culture infected with the HIV-1Δenv, cyclin T1 was present at a relatively high level at day 18, indicating that HIV-1 infection results in sustained cyclin T1 expression over the course of MDM differentiation. As described in Materials and Methods, flow cytometry analyses of cultures infected with concentrated stocks of the HIV-1Δenv virus showed that from 10 to 20% of MDMs were positive for intracellular p24 expression under these conditions (data not shown). We were unable to detect an activity in the medium of infected MDMs that could induce cyclin T1 in uninfected MDMs. Because of the low background expression of cyclin T1 in mock-infected cultures, it is possible that the induction of cyclin T1 seen in Fig. 3 occurs in only the infected cells in the culture.
The culture infected with the HIV-GFP virus showed only a minor induction of cyclin T1, suggesting that one or more of the viral gene products mutated in this virus may contribute to the high level of cyclin T1 seen in the HIV-1Δenv-infected culture. However, the percentage of cells infected with the HIV-GFP virus may be lower than infections with the HIV-1 Δenv, and this may also contribute to the low level of cyclin T1 induction. Interestingly, the relative induction of Cdk8 by these two viral infections was similar to that of cyclin T1; infection with the HIV-GFP virus weakly induced Cdk8, while infection with the HIV-1Δenv virus resulted in a significantly higher induction of Cdk8.
To examine whether HIV-1 infection could reinduce cyclin T1 expression after it had been shut off, we infected MDM cultures at late times of differentiation (Fig. 3C). For donor D, culture dishes were either mock infected or infected with the HIV-1Δenv at day 15 of differentiation and extracts were prepared at day 18 of differentiation for immunoblot analysis. Cyclin T1 expression was shut off in the mock-infected culture at day 18, while it was expressed at a high level in the culture that has been infected 3 days previously (lanes 1 and 2). In an independent experiment (donor H), MDMs were allowed to differentiate for 30 days and then were infected with the HIV-1Δenv in the presence or absence of AZT. Prior to infection at day 30 in this MDM preparation, cyclin T1 was shut off (lane 3). After 3 days of infection (day 33), cyclin T1 had been reinduced by HIV-1 infection (lane 4). The infection in the presence of AZT showed only a low level of cyclin T1 (lane 5). The small increase in cyclin T1 in the presence of AZT may be due to a low residual level of RT in the presence of inhibitor or to factors present in the virion that can weakly induce cyclin T1. Control experiments using a concentrated supernatant from nontransfected 293T cells (used to generate viral stocks) showed no effect on cyclin T1 levels in uninfected MDMs (data not shown). The data shown in Fig. 3C indicate that HIV-1 infection can induce cyclin T1 after it has been shut off, and because AZT largely reduces this, the expression of one or more viral gene products is likely to be involved in the induction.
HIV-1 Nef protein expressed from an adenovirus vector induces cyclin T1 in MDMs.
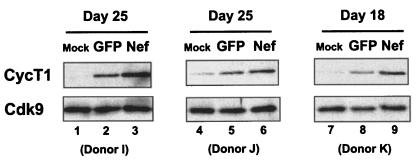
The HIV-1 Nef protein has been shown to activate signaling pathways in macrophages that enhance HIV-1 replication (24, 25). The HIV-GFP virus that is defective for Nef expression (and other accessory proteins) only weakly induced cyclin T1 in MDMs (Fig. 3B), and infections with the HIV-1 Δenv in the presence of AZT resulted in little cyclin T1 induction (Fig. 3C). We therefore used an adenovirus expressing Nef to examine whether Nef might play a role in the induction of cyclin T1. We infected three MDM preparations with a control adenovirus expressing GFP (Ad5-GFP) or the adenovirus expressing Nef (Ad5-Nef) at day 16 or 23 of differentiation and prepared cell extracts 2 days later for immunoblots. We observed in these experiments that MDM cultures infected with the Ad5-GFP were >90% GFP positive (data not shown). Expression of the Cdk9 protein was not affected by infection with either adenovirus. Cyclin T1 expression was slightly increased in the Ad5-GFP-infected MDM cultures (Fig. 4, lanes 2, 5, and 8), indicating that adenovirus infection itself can induce cyclin T1 expression to some extent, perhaps not unexpectedly, as a number of PAMPs induce cyclin T1 (Fig. 1). However, Ad5-Nef-infected cultures expressed significantly higher levels of cyclin T1 protein than the Ad5-GFP-infected culture (lanes 3, 6, and 9), suggesting that Nef may contribute to the induction of cyclin T1 expression during HIV-1 infection.
FIG. 4.
HIV-1 Nef protein induces cyclin T1 in MDMs. MDMs from donors I and J were infected at day 23 of differentiation with Ad5-GFP or Ad5-Nef at multiplicities of infection of 1,000 (donor I) or 100 (donor K), and cell extracts were prepared at day 25. Donor K was infected at day 16 of differentiation at a multiplicity of infection of 50, and cell extracts were prepared at day 18. Cell extracts were analyzed by immunoblotting.
DISCUSSION
Because cyclin T1 is an essential host factor utilized by the HIV-1 Tat protein, our previous observation that cyclin T1 is expressed only transiently during macrophage differentiation was unexpected, as HIV-1 generally replicates well in macrophages for extended periods of time in vitro. The data presented here demonstrate that HIV-1 infection induces cyclin T1 expression in macrophages, thereby providing an explanation for the ability of these cells to support HIV-1 long terminal repeat (LTR)-directed transcription and high levels of viral replication. Flow cytometry analyses of p24 expression in MDMs infected with the HIV-1Δenv virus indicated that from 10 to 20% of cells were infected under our experimental conditions (data not shown). We have been unable to detect an activity in the culture supernatants of infected MDMs that can induce cyclin T1 in uninfected MDMs. We therefore favor the notion that cyclin T1 is induced in only infected cells in the MDM cultures, although a definitive resolution of this issue will require additional work. Infections in the presence of AZT greatly reduced the induction of cyclin T1, indicating that expression of a viral gene product or products contributes to the up-regulation. The use of an adenovirus vector expressing Nef suggested that Nef is involved in the induction of cyclin T1, perhaps not unexpectedly, as Nef expression in macrophages has previously been shown to induce a number of factors that enhance HIV-1 replication in CD4+ T lymphocytes (24, 25). Because we did not analyze other viral gene products in this study, we cannot exclude a role for other HIV-1 proteins in the up-regulation of cyclin T1. A detailed analysis of mutant viruses unable to express individual gene products and combinations of gene products will be required to further investigate this issue.
We observed some induction of cyclin T1 with a control adenovirus expressing GFP, suggesting that adenovirus infection itself leads to induction of cyclin T1. We also observed a low level of induction of cyclin T1 in HIV-1 infections in the presence of AZT, raising the possibility that entry of the HIV-1 particle into macrophages can induce some cyclin T1. These observations, along with the finding that a number of PAMPs from bacteria or fungi induce cyclin T1, suggest that pathogens generate diverse signals that are recognized by macrophages and lead to up-regulation of cyclin T1. Such an induction may lead to elevated P-TEFb function and the subsequent transcriptional up-regulation of a number of cellular genes that may play important roles in the innate immune response. In the case of HIV-1, the virus replication strategy apparently hijacks an arm of this immune response to enhance viral replication, analogous to the HIV-1 replication strategy in CD4+ T lymphocytes in which an adaptive immune response leads to activation of CD4+ T lymphocytes and consequently a cellular environment highly permissive for viral replication.
It is unclear why cyclin T1 is normally shut off late in macrophage differentiation. Because inhibition of P-TEFb function in promonocytic cell lines results in increased apoptosis (1), it is possible that the shutoff of cyclin T1 may sensitize macrophages to apoptosis. The ability to readily undergo apoptosis is critical to macrophage homeostasis, as emigration of monocytes from blood into tissues requires that the preexisting macrophage population turnover at a relatively high rate; tissue macrophages have a half-life estimated to be no more than a few months (22). Because macrophages are resistant to the cytopathic effects of HIV-1 infection (7, 16), an intriguing possibility is that the induction of cyclin T1 by HIV-1 contributes to a resistance to apoptosis in infected tissue macrophages, thereby increasing the lifetime of these cells and the duration of viral production.
The shutoff of cyclin T1 expression late in differentiation involves proteasome-mediated proteolysis. Cyclin T1 contains a PEST sequence at its carboxyl terminus, a sequence that in some proteins can confer ubiquitination and proteolytic degradation via the proteasome (18, 27). Although there is as yet no direct evidence that cyclin T1 is ubiquitinated, it seems likely that an ubiquitination system becomes active in late-differentiated macrophages that leads to the ubiquitination and degradation of cyclin T1. Because proteasome inhibitors had no effect on cyclin T1 expression in other cell types that we examined (PBLs, Jurkat T cells, and 293T cells), the ubiquitination of cyclin T1 may be specific to primary macrophages and perhaps a limited number of other cell types. We note that a study in transformed cell lines has presented evidence that SCFSKP2 can be recruited to the Cdk9/cyclin T1 complex through recognition of the cyclin T1 PEST sequence, leading to ubiquitination and degradation of Cdk9 but not cyclin T1 (10). However, this report has been challenged by a recent publication that presented evidence that Cdk9 expression is not regulated in transformed cell lines by SCFSKP2 (3).
Here we found that the induction of cyclin T1 by PAMPs required 1 to 2 days of treatment, in contrast to the 1-h induction of cyclin T1 by proteasome inhibitors. It is therefore possible that the induction by PAMPs involves distinct mechanisms from those regulated by proteasome inhibitors. We have seen no significant induction of cyclin T1 by proteasome inhibitors in freshly isolated monocytes (unpublished result), suggesting that the low level of cyclin T1 expression in monocytes is not the result of active proteasome-mediated proteolysis, as is the case in late-differentiated macrophages. Thus, regulation of cyclin T1 expression in monocytes and macrophages may involve multiple mechanisms that are active at different stages of differentiation. It is presently unclear what mechanisms are involved in the induction of cyclin T1 during HIV-1 infection of late-differentiated macrophages. This induction may result from a viral block to proteasome-mediated proteolysis of cyclin T1 or some alternative mechanism that controls cyclin T1 protein levels. Future studies of mechanisms that regulate cyclin T1 expression in uninfected and infected macrophages may suggest strategies for manipulating cyclin T1 levels for therapeutic benefit in HIV-infected individuals.
Acknowledgments
We thank Michael Emerman (Fred Hutchinson Cancer Research Center) and Richard Sutton (Baylor College of Medicine) for plasmids and Mario Stevenson (University of Massachusetts) for the Nef-expressing adenovirus. We thank Richard Sutton for advice on preparing concentrated HIV stocks and Wade Harper for useful discussions of proteasome-mediated proteolysis.
This work was supported by NIH grants AI45374 (A.P.R.) and AI42558 (C.H.H.).
REFERENCES
- 1.Foskett, S. M., R. Ghose, D. N. Tang, D. E. Lewis, and A. P. Rice. 2001. Antiapoptotic function of Cdk9 (TAK/P-TEFb) in U937 promonocytic cells. J. Virol. 75:1220-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 3.Garriga, J., S. Bhattacharya, J. Calbó, R. M. Marshall, M. Truongcao, D. S. Haines, and X. Graña. 2003. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell. Biol. 23:5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haaland, R. E., C. H. Herrmann, and A. P. Rice. 2003. Increased association of 7SK snRNA with Tat co-factor P-TEFb following activation of peripheral blood lymphocytes. AIDS 17:2429-2436. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann, C. H., M. O. Gold, and A. P. Rice. 1996. Viral transactivators specifically target distinct cellular protein kinases that phosphorylate the RNA polymerase II C-terminal domain. Nucleic Acids Res. 24:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho, D. D., T. R. Rota, and M. S. Hirsch. 1986. Infection of monocyte/macrophages by human T lymphotropic virus type III. J. Clin. Investig. 77:1712-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 10.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbé, T. Lorca, K.-I. Nakayama, and M. Benkirane. 2001. Interaction between cyclin T1 and SCFSKP2 targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 21:7956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 12.Krieg, A. M. 2003. CpG motifs: the active ingredient in bacterial extracts? Nat. Med. 9:831-835. [DOI] [PubMed] [Google Scholar]
- 13.Lien, E., and R. R. Ingalls. 2002. Toll-like receptors. Crit. Care Med. 30:S1-S11. [PubMed] [Google Scholar]
- 14.Liou, L.-Y., C. H. Herrmann, and A. P. Rice. 2002. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J. Virol. 76:10579-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson, J. K., G. D. Cross, C. S. Callaway, and J. S. McDougal. 1986. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV). J. Immunol. 137:323-329. [PubMed] [Google Scholar]
- 17.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 19.Rice, A. P., and C. H. Herrmann. 2003. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr. HIV Res. 1:395-404. [DOI] [PubMed] [Google Scholar]
- 20.Rich, E. A., I. S. Chen, J. A. Zack, M. L. Leonard, and W. A. O'Brien. 1992. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J. Clin. Investig. 89:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riedl, T., and J. M. Egly. 2000. Phosphorylation in transcription: the CTD and more. Gene Expr. 9:3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross, J. A., and M. J. Auger. 2001. The biology of the macrophage, p. 1-72. In B. Burke and C. E. Lewis (ed.), The macrophage, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 23.Sutton, R. E., H. T. M. Wu, R. Rigg, E. Bohnlein, and P. O. Brown. 1998. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J. Virol. 72:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swingler, S., B. Brichacek, J. M. Jacque, C. Ulich, J. Zhou, and M. Stevenson. 2003. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature 424:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swingler, S., A. Mann, J. Jacque, B. Brichacek, V. G. Sasseville, K. Williams, A. A. Lackner, E. N. Janoff, R. Wang, D. Fisher, and M. Stevenson. 1999. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat. Med. 5:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 27.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 14:317-322. [DOI] [PubMed] [Google Scholar]
- 29.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]