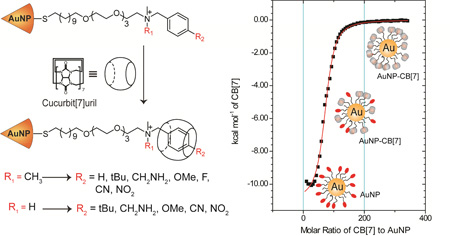
Abstract
Host-guest interactions between a synthetic receptor, cucurbit[7]uril (CB[7]), and gold nanoparticles (AuNPs) have been quantified using isothermal titration calorimetry. AuNPs were functionalized with ligands containing tertiary or quaternary benzylamine derivatives, with electron donating or withdrawing groups at the para position of the benzene ring. Analysis of binding interactions reveals that functional groups at the para position have no significant effect on binding constant. However, headgroups bearing a permanent positive charge increased the binding of AuNPs to CB[7] ten-fold compared to monomethyl counterparts.
Keywords: Gold nanoparticle, cucurbit[7]uril, isothermal titration calorimetry, host-guest interaction, supramolecular chemistry
Graphical Abstract
The goal of mimicking Nature’s use of non-covalent interactions has led the development of supramolecular chemistry.1–5 Molecular recognition,6, 7 self-assembly,8 lock-key modality9 and reversibility10 are among the most important features that enable supramolecular chemistry to be employed in many applications including catalysis,11, 12 delivery,13, 14 sensing,15–17 and imaging.18 These applications use building blocks assembled together by intermolecular interactions including electrostatic interactions,19 hydrogen bonding,20 π–π interactions,21 ion-dipole interactions,22, 23 hydrophobic or solvophobic effects.24
The choice of building blocks in host-guest chemistry is critical, as they determine the selectivity, reversibility and tunability of the supramolecular architectures. Gold nanoparticles (AuNPs) are useful platforms to play either the host or guest role due to their unique features such as tunable core size,25 monodispersity,26 large surface to volume ratio,27 and ready surface functionalization.28, 29 Rotello et al. have used AuNPs as host scaffolds to encapsulate hydrophobic guest molecules into the engineered monolayer.30 In addition to encapsulation, proper surface functionalization of AuNPs provides recognition by macrocyclic compound such as crown ethers,31, 32 calixarenes,33, 34 cyclodextrins,35, 36 pillararenes37 and cucurbiturils.38–40
Cucurbit[7]uril (CB[7])41, 42 is a member of cucurbituril family with a heptameric macrocyclic structure self-assembled from an acid catalyzed condensation reaction of glycoluril and formaldehyde. CB[7] is a water soluble molecule with well-established host properties.43, 44 It has a pumpkin-shaped structure45 consisting of a hydrophobic cavity of 7.3 Å diameter and two identical carbonyl-laced portals. While the hydrophobic interior provides an encapsulation site for hydrophobic guest molecules,46 the polar carbonyl groups at the portals allow CB[7] to bind ions47 and charged molecules48, 49 by forming charge-dipole and/or hydrogen bonding interactions.
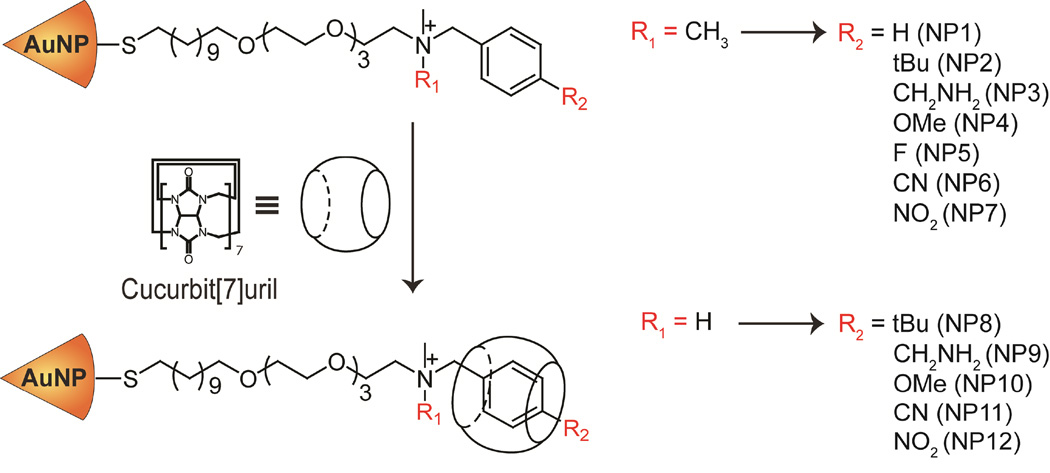
Here we quantify host-guest interactions between CB[7] molecule and AuNPs bearing ligands with a monomethyl-benzylammonium (MMBA) or dimethyl-benzylammonium (DMBA) head groups. We used AuNPs with core diameters of ~2.5 nm as the guest scaffold, while CB[7] served as a host molecule. We kept the monolayer design same with the exception of the headgroup functionality, allowing parametric comparison between systems. We have investigated the changes in the binding event considering two features: 1) the effect of different functionalities at the para position of the benzene ring and 2) the effect of carrying a permanent positive charge on the head group.
Result and Discussion
We have designed a series of nanoparticles with benzylammonium terminal group possessing different functionalities at the para position of the benzene ring, which is the least sterically hindered position for binding process based on the reported crystal structures of cucurbiturils.50, 51 Seven quaternary amine (DMBA) and five tertiary amine (MMBA) derivatives (Figure 1) were synthesized to represent a wide range of functionality that bring at least one of the following attributes to the system: hydrophobicity/hydrophilicity, electron withdrawing/donating, and steric bulk.
Figure 1.
Structures of DMBA and MMBA derivatives used in the study. The schematic represents the binding event between AuNP and CB[7].
Isothermal titration calorimetry (ITC) is an extremely powerful and sensitive technique that measures the heat taken up or evolved depending on the nature of the reaction when one solution is titrated against the other solution.52, 53 ITC has been widely used to investigate the binding of ligands, peptides and proteins to AuNP.54–56 ITC experiments were performed at 30 °C in 5 mM phosphate buffer at pH 7.4 using a MicroCal ITC200 system. During the experiment, the reference cell was filled with only 5 mM phosphate buffer and the sample cell was containing the AuNP solution (1 µM). Then, CB[7] solution (2 mM) was titrated into the sample cell. All AuNPs showed multiple host/guest bindings while free ligand (DMBA) showed a binding stoichiometry of 1:1.
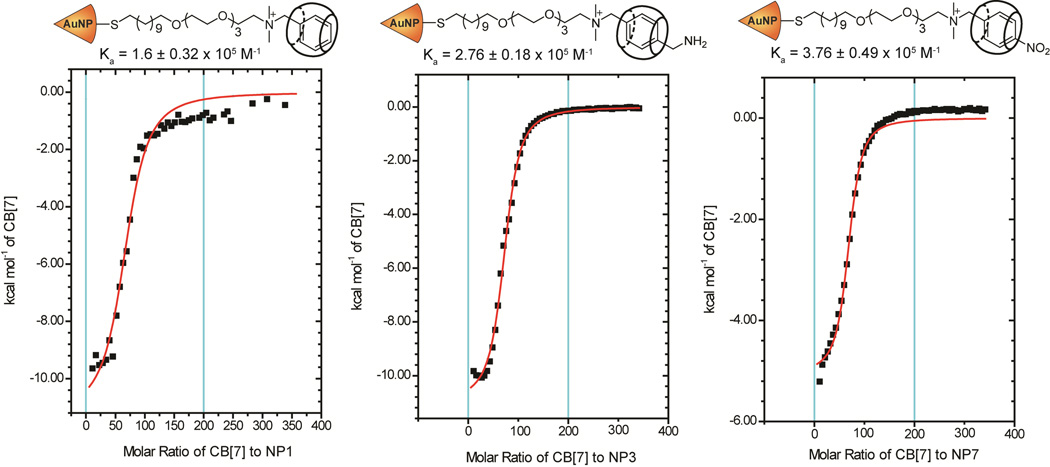
Binding affinities of DMBA head group bearing AuNPs (NP1-NP7) were in the order of 104 to 106 M−1 (Table 1). NP1 having H atom at the para position showed a binding constant of 1.6±0.32×105 M−1 (Figure 2).
Table 1.
Thermodynamic data of NP1-NP7 for binding to CB[7]
| Guest | Ka(M−1) | ΔG (kcal/mol) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
N |
|---|---|---|---|---|---|
| NP1:H | 1.6(±0.32)×105 | −7.23 | −11.29(±0.49) | 4.06 | 69.4(±2.2) |
| NP2:tBu | 7.23(±0.65)×104 | −6.74 | −9.06(±0.22) | 2.32 | 96.1(±1.68) |
| NP3:CH2NH2 | 2.76(±0.18)×105 | −6.61 | −10.1(±0.11) | 3.49 | 73.1(±0.54) |
| NP4:OMe | 2.99(±0.31)×105 | −7.59 | −6.6(±0.1) | −0.99 | 84.2 (±0.89) |
| NP5:F | 2(±0.19)×105 | −7.35 | −6.59(±0.14) | −0.76 | 54.2(±0.84) |
| NP6:CN | 1.32(±0.69)×106 | −8.47 | −4.17(±0.2) | −4.30 | 54.5(±1.74) |
| NP7:NO2 | 3.76(±0.49)×105 | −7.73 | −5.06(±0.09) | −2.67 | 68.5(±0.91) |
Figure 2.
ITC measurements of NP1, NP3, and NP7 bearing DMBA head group with H, CH2NH2 and NO2 functionalities at the para position of the benzene ring showing similar binding constants.
For the NPs with electron donating groups including CH2NH2 and OMe, binding affinities were similar to NP1 except for NP2, with a binding constant of 6.16±0.15x 104 M−1. The NP2 has tBu group located at the para position and although tBu functionality has weakly electron donating property, it also brings steric hindrance into the system and thus resulting in a lower binding affinity. On the other hand, the NP3 with CH2NH2 functionality demonstrated slightly higher association constant (Ka) compared to NP1 (Figure 2 and Table 1). This could be originated from the hydrogen bonding interactions between the carbonyl portals of CB[7] and amine moiety of CH2NH2 functionality. In accordance with our results, Urbach et al. have performed ITC experiments showing that amino acid and peptide derivatives bearing a CH2NH2 moiety resulted in a higher binding affinity to CB[7] compared to non-functionalized counterparts.57 Their data along with the calculated models supported that the benzene ring was fully buried inside the hydrophobic cavity of CB[7] in the presence of CH2NH2 functionality. However, in the presence of the bulky tBu group, benzene ring was only partially buried possibly due to steric hindrance. Hydrophobic interactions within the inner cavity of cucurbituril play an important role in the binding event. Electron donating groups make the benzene ring less electron deficient and therefore weaken the interaction of benzene ring with the inner cavity of CB[7]. However, this effect was not observed for NP4 carrying a stronger electron donating ‘methoxy group’.
Thermodynamic values for NP1-NP7 are listed in Table 1. Titration of CB[7] into NP solution is an exothermic process with negative enthalpy change (DH) as reported previously by Kim and co-workers.58 This negative enthalpy of complexation results from the robust van der Waals forces inside the hydrophobic cavity of CB[7] as well as additional forces including hydrogen bonding and ion-dipole interactions between the carbonyl portals of CB[7] and guest molecules. As the sites available on the surface of the AuNPs become progressively occupied by CB[7] during the titration, the exothermicity of the peaks decreases and eventually saturates (Figure 2). ΔH values for NP1-NP4 were similar to each other and varied between −6.6 and −11.29 kcal/mol. From the entropic standpoint, complexation of CB[7] with NP1, NP2, and NP3 is less favourable as -TΔS values were positive. However, overall entropy of complexation for NP4 is slightly favorable as -TΔS values were negative but very close to zero.
Electron withdrawing groups make the guest molecules electron deficient and therefore strengthen the binding of them to the inner cavity of host molecules due to increased π-stacking interactions with the host.59, 60 In agreement with the reported literature, AuNPs with electron withdrawing groups at the para position rendered the benzene ring more electron deficient, resulting in more stable inclusion complexes. Fluorine, nitrile, and nitro functionalities (NP5, NP6, and NP7) were used to bring electron withdrawing property into the system. All the three NPs showed somewhat higher binding towards CB[7] molecule compared to NP1 whereas the binding for NP5 and NP7 was not greatly so. However, NP6 bearing CN group exhibited an association constant of 1.32±0.69×106 M−1 which is about 10 times higher than that of NP1. The negative ΔH values for these NPs indicated an exothermic binding process. Numerically they were all quite similar to each other but smaller than that of NPs carrying electron donating groups. Positive entropy arising from desolvation indicated favorable complexation between CB[7] and NPs bearing electron withdrawing groups.58 The complexation process for NP1-NP7 was found spontaneous which was confirmed by negative Gibbs free energy change (ΔG).
Binding experiments of MMBA derivatives revealed a drop in the binding affinities; Ka values were in the order of 104 to 105 M−1. In accordance with the literature, this result indicated that a permanent positive charge played an essential role in the binding event as ion-dipole interactions between CB[7] and AuNP were dominating over the hydrogen bonding interactions.61 NP8 showed no complexation to CB[7] while both the absence of positive charge and bulkiness of the tBu group prevented the recognition event (Table 2). NP9 bearing CH2NH2 functionality showed a Ka of 1.33±0.32×105 M−1 which was a lower value compared to NP3 but a higher value compared to other MMBA derivatives (NP8 and NP10-NP12). This increase in the binding could be due to the additional interactions coming from the hydrogen bonding between the carbonyls of CB[7] and amine moiety of CH2NH2 functionality. On the other hand, NP10 with the methoxy group showed an unexpected Ka of 1.20±0.15×105 M−1. ΔH values for NP8-NP12 were very similar to each other, and negative showing exothermic binding events. Complexation between CB[7] and NP8-NP12 was found spontaneous as indicted by negative ΔG.
Table 2.
Thermodynamic data of NP8-NP12 for binding to CB[7]
| Guest | Ka (M−1) | ΔG (kcal/mol) |
ΔH (kcal/mol) | −TΔS (kcal/mol) |
N |
|---|---|---|---|---|---|
| NP8:tBu | N/A | N/A | N/A | N/A | N/A |
| NP9:CH2NH2 | 1.33(±0.32)x105 | −7.1 | −8.38(±0.52) | 1.28 | 64.6(±2.85) |
| NP10:OMe | 1.20(±0.15)x105 | −7.05 | −8.06(± 0.29) | 1.01 | 64 (±1.66) |
| NP11:CN | 4.32(±0.15)x104 | −6.43 | −8.92(±0.13) | 2.49 | 84.4(±0.92) |
| NP12:NO2 | 5.11(±0.21)x104 | −6.52 | −5.48(±0.09) | −1.04 | 72.5(±0.95) |
We have synthesized a ligand containing a DMBA head group same as in NP1 to investigate the behavior of free ligand. ITC measurement revealed a binding constant of 3.69±0.25×105 M−1 when 100 µM of ligand solution was titrated with 2 mM CB[7] in 5 mM phosphate buffer (Figure 3). This value was in the same order with the binding constant of NP1 (Ka = 1.6±0.32×105 M−1) but still 2.3 times higher in magnitude. This result revealed that attaching the ligand on AuNP showed no dramatic decrease in the binding process between CB[7] and head group of the ligand. However, this behavior could be affected by other parameters such as linker length. While longer ligands will achieve better flexibility and accessibility for the host molecules, shorter ligands may lead to steric crowding that may decrease the affinity of host molecules towards nanoparticle bound ligands.62, 63
Figure 3.
a) Schematic representation of binding between DMBA ligand and CB[7]. b) ITC measurement indicated a 1:1 binding stoichiometry between free ligand and CB[7].
Conclusion
We have used isothermal titration calorimetry to monitor the binding interactions of CB[7] with twelve different AuNPs. Binding studies revealed that NP1-NP7 possessing DMBA head groups showed a higher binding affinity towards CB[7] compared to NP8-NP12 having MMBA head groups. Insertion of tBu group at the para position of the benzene ring led to a lower binding due to the steric effect. Although not very strong difference in Ka constants was observed between electron donating and withdrawing groups, a slight decrease in the binding was monitored for electron donating groups compared to withdrawing derivatives. Methoxy functionality behaved differently than expected, computational studies may help to understand the process better.
Supplementary Material
Acknowledgements
This work was supported by the NIH (NIH R01 EB014277). T.M. is grateful to the Japan Society for the Promotion of Sciences (JSPS) for a Postdoctoral Fellowship for Research Abroad and for the Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Dedicated to the memory of Harry Wassserman, a true Renaissance man
Supplementary data
Supplementary data (synthesis and characterization) associated with this article can be found, in the online version, at ….
References
- 1.Lehn JM. Science. 1985;227:849–856. doi: 10.1126/science.227.4689.849. [DOI] [PubMed] [Google Scholar]
- 2.Lehn JM. Proc. Natl. Acad. Sci. U S A. 2002;99:4763–4768. doi: 10.1073/pnas.072065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fyfe MCT, Stoddart JF. Accounts Chem. Res. 1997;30:393–401. [Google Scholar]
- 4.Bradley J, Holliday BJ, Mirkin CA. Angew. Chem. Int. Ed. 2001;40:2022–2043. [PubMed] [Google Scholar]; Angew. Chem. 2001;113:2076–2097. [Google Scholar]
- 5.Mink D, Mecozzi S, Rebek J. Tetrahedron Lett. 1998;39:5709–5712. [Google Scholar]
- 6.Dong SY, Zheng B, Wang F, Huang FH. Accounts Chem. Res. 2014;47:1982–1994. doi: 10.1021/ar5000456. [DOI] [PubMed] [Google Scholar]
- 7.Chawla HM, Sahu SN, Shrivastava R, Kumar S. Tetrahedron Lett. 2012;53:2244–2247. [Google Scholar]
- 8.Rubio J, Alfonso I, Bru M, Burguete MI, Luis SV. Tetrahedron Lett. 2010;51:5861–5867. [Google Scholar]
- 9.Jordan BJ, Pollier MA, Ofir Y, Joubanian S, Mehtala JG, Sinkel C, Caldwell ST, Kennedy A, Rabani G, Cooke G, Rotello VM. Chem .Commun. 2008;14:1653–1655. doi: 10.1039/b718015b. [DOI] [PubMed] [Google Scholar]
- 10.Liu K, Kang YT, Wang ZQ, Zhang X. Adv. Mater. 2013;25:5530–5548. doi: 10.1002/adma201302015. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y, Wang Y, Yu T-Y, Liang Y-M, Xu P-F. Angew. Chem. Int. Ed. 2014;53:14128–14131. doi: 10.1002/anie.201406786. [DOI] [PubMed] [Google Scholar]
- 12.Gramage-Doria R, Hessels J, Leenders SHAM, Troppner O, Durr M, Ivanovic-Burmazovic I, Reek JNH. Angew. Chem. Int. Ed. 2014;53:13380–13384. doi: 10.1002/anie.201406415. [DOI] [PubMed] [Google Scholar]
- 13.Yao XM, Chen L, Chen XF, He CL, Zhang JP, Chen XS. Macromol. Rapid. Commun. 2014;35:1697–1705. doi: 10.1002/marc.201400291. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh PS, Kim CK, Han G, Forbes NS, Rotello VM. ACS Nano. 2008;2:2213–2218. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbrizzi L, Poggi A. Chem. Soc. Rev. 1995;24:197–202. [Google Scholar]
- 16.Elci SG, Moyano DF, Rana S, Tonga GY, Phillips RL, Bunz UHF, Rotello VM. Chem. Sci. 2013;4:2076–2080. [Google Scholar]
- 17.Biedermann F, Rauwald U, Cziferszky M, Williams KA, Gann LD, Guo BY, Urbach AR, Bielawski CW, Scherman OA. Chem. Eur. J. 2010;46:13716–13722. doi: 10.1002/chem.201002274. [DOI] [PubMed] [Google Scholar]
- 18.Carroll CN, Naleway JJ, Haley MM, Johnson DW. Chem. Soc. Rev. 2010;39:3875–3888. doi: 10.1039/b926231h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Rothberg L. Proc. Natl. Acad. Sci. U S A. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramani C, Yesilbag G, Jordan BJ, Li X, Khorasani A, Cooke G, Sanyal A, Rotello VM. Chem. Commun. 2010;46:2067–2069. doi: 10.1039/b926746h. [DOI] [PubMed] [Google Scholar]
- 21.Hunter CA, Meah MN, Sanders JKM. J. Am. Chem. Soc. 1990;112:5773–5780. [Google Scholar]
- 22.Kim K. Chem. Soc. Rev. 2002;31:96–107. doi: 10.1039/a900939f. [DOI] [PubMed] [Google Scholar]
- 23.Marquez C, Hudgins RR, Nau WM. J. Am. Chem. Soc. 2004;126:5806–5816. doi: 10.1021/ja0319846. [DOI] [PubMed] [Google Scholar]
- 24.Zhao D, Moore JS. Chem. Commun. 2003:807–818. doi: 10.1039/b207442g. [DOI] [PubMed] [Google Scholar]
- 25.Pengo P, Polizzi S, Battagliarin M, Pasquato L, Scrimin P. J. Mater. Chem. 2003;13:2471–2478. [Google Scholar]
- 26.Hussain I, Graham S, Wang ZX, Tan B, Sherrington DC, Rannard SP, Cooper AI, Brust M. J. Am. Chem. Soc. 2005;127:16398–16399. doi: 10.1021/ja055321v. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv. Drug Deliv. Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Kim ST, Saha K, Kim C, Rotello VM. Acc. Chem. Res. 2013;46:681–691. doi: 10.1021/ar3000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Rivera FG, Angurell I, Rossell O, Seco M, Llorca J. J. Organomet. Chem. 2012;715:13–18. [Google Scholar]
- 30.Kim CK, Ghosh P, Pagliuca C, Zhu Z-J, Menichetti S, Rotello VM. J. Am. Chem. Soc. 2009;131:1360–1361. doi: 10.1021/ja808137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuang H, Chen W, Yan WJ, Xu LG, Zhu YY, Liu LQ, Chu HQ, Peng CF, Wang LB, Kotov NA, Xu CL. Biosens. Bioelectron. 2011;26:2032–2037. doi: 10.1016/j.bios.2010.08.081. [DOI] [PubMed] [Google Scholar]
- 32.Chernikova E, Berdnikova D, Fedorov Y, Fedorova O, Peregudova A, Isaacs L. Chem. Commun. 2012;48:7256–7258. doi: 10.1039/c2cc33243d. [DOI] [PubMed] [Google Scholar]
- 33.Tshikhudo TR, Demuru D, Wang ZX, Brust M, Secchi A, Arduini A, Pochini A. Angew. Chem. Int. Ed. 2005;44:2913–2916. doi: 10.1002/anie.200462909. [DOI] [PubMed] [Google Scholar]
- 34.Rebek J. Chem. Commun. 2000:637–643. [Google Scholar]
- 35.Liu J, Mendoza S, Roman E, Lynn MJ, Xu RL, Kaifer AE. J. Am. Chem. Soc. 121:4304–4305. 199. [Google Scholar]
- 36.Liu J, Alvarez J, Ong W, Roman E, Kaifer AE. Langmuir. 2001;17:6762–6764. [Google Scholar]
- 37.Yao Y, Xue M, Zhang ZB, Zhang MM, Wang Y, Huang FH. Chem. Sci. 2013;4:3667–3672. [Google Scholar]
- 38.Yan B, Tonga GY, Hou S, Fedick PW, Yeh Y-C, Alfonso FS, Mizuhara T, Vachet RW, Rotello VM. Anal. Chem. 2014;86:6710–6714. doi: 10.1021/ac501682y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim C, Agasti SS, Zhu Z, Isaacs L, Rotello VM. Nat. Chem. 2010;2:962–966. doi: 10.1038/nchem.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones ST, Zayed JM, Scherman OA. Nanoscale. 2013;5:5299–5302. doi: 10.1039/c3nr01454a. [DOI] [PubMed] [Google Scholar]
- 41.Assaf KI, Nau WM. Chem. Soc. Rev. 2015;44:394–418. doi: 10.1039/c4cs00273c. [DOI] [PubMed] [Google Scholar]
- 42.Zhao N, Lloyd GO, Scherman OA. Chem. Commun. 2012;48:3070. doi: 10.1039/c2cc17433b. [DOI] [PubMed] [Google Scholar]
- 43.Kim K, Selvapalam N, Ko YH, Park KM, Kim D, Kim J. Chem. Soc. Rev. 2007;36:267–279. doi: 10.1039/b603088m. [DOI] [PubMed] [Google Scholar]
- 44.Jang Y, Natarajan R, Ko YH, Kim K. Angew. Chem. Int. Ed. 2013;53:1–6. doi: 10.1002/anie.201308879. [DOI] [PubMed] [Google Scholar]
- 45.Freeman WA, Mock WL, Shih N-Y. J. Am Chem. Soc. 1981;103:7367–7368. [Google Scholar]
- 46.Nau WM, Florea M, Assaf KI. Isr. J Chem. 2011;51:559–577. [Google Scholar]
- 47.Tang H, Fuentealba D, Ko YH, Selvapalam N, Kim K, Bohne O. J. Am. Chem. Soc. 2011;133:20623–20633. doi: 10.1021/ja209266x. [DOI] [PubMed] [Google Scholar]
- 48.Cao L, Isaacs L. Supramol. Chem. 2014;26:251–258. [Google Scholar]
- 49.Zhao J, Zhang YM, Sun HL, Chang XY, Liu Y. Chem. Eur. J. 2014;20:15108–15115. doi: 10.1002/chem.201404216. [DOI] [PubMed] [Google Scholar]
- 50.Heitmann LM, Taylor AB, Hart PJ, Urbach AR. J. Am. Chem. Soc. 2006;128:12574–12581. doi: 10.1021/ja064323s. [DOI] [PubMed] [Google Scholar]
- 51.Chinai JM, Taylor AB, Ryno LM, Hargreaves ND, Morris CA, Hart PJ, Urbach AR. J. Am. Chem. Soc. 2011;133:8810–8813. doi: 10.1021/ja201581x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joshi H, Shirude PS, Bansal V, Ganesh KN, Sastry M. J. Phys. Chem. B. 2004;108:11535–11540. [Google Scholar]
- 53.Roselin LS, Lin M-S, Lin P-H, Chang Y, Chen W-Y. Biotechnol. J. 2010;5:85–98. doi: 10.1002/biot.200900092. [DOI] [PubMed] [Google Scholar]
- 54.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc. Natl. Acad. Sci. U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fleischer CC, Payne CK. J. Phys. Chem. B. 2014;118:14017–14026. doi: 10.1021/jp502624n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang RX, Carney RP, Stellacci F, Lau BLT. Nanoscale. 2013;5:6928–6935. doi: 10.1039/c3nr02117c. [DOI] [PubMed] [Google Scholar]
- 57.Logsdon LA, Schardon CL, Ramalingam V, Kwee SK, Urbach AR. J. Am. Chem. Soc. 2011;133:17087–17092. doi: 10.1021/ja207825y. [DOI] [PubMed] [Google Scholar]
- 58.Rekharsky MV, Yamamura H, Inoue C, Kawai M, Osaka I, Arakawa R, Shiba K, Sato A, Ko YH, Selvapalam N, Kim K, Inoue Y. J. Am. Chem. Soc. 2006;128:14871–14880. doi: 10.1021/ja063323p. [DOI] [PubMed] [Google Scholar]
- 59.Li C, Xu Q, Li J, Yaoa F, Jia X. Org. Biomol. Chem. 2010;8:1568–1576. doi: 10.1039/b920146g. [DOI] [PubMed] [Google Scholar]
- 60.Biedermann F, Scherman OA. J. Phys. Chem. B. 2012;116:2842–2849. doi: 10.1021/jp2110067. [DOI] [PubMed] [Google Scholar]
- 61.Kim K, Selvapalam N, Oh DH. J. Incl. Phenom. Macro. 2004;50:31–36. [Google Scholar]
- 62.Chen W-Y, Huang H-M, Lin C-C, Lin F-Y, Chan Y-C. Langmuir. 2003;19:9395–9403. [Google Scholar]
- 63.M, Moreira JA, Garcia-Rio L. Chem. Eur. J. 2012;18:7931–7940. doi: 10.1002/chem.201103049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.