Abstract
The sub-3 Mbp genomes from microsporidian species of the Encephalitozoon genus are the smallest known among eukaryotes and paragons of genomic reduction and compaction in parasites. However, their diminutive stature is not characteristic of all Microsporidia, whose genome sizes vary by an order of magnitude. This large variability suggests that different evolutionary forces are applied on the group as a whole. In this study, we have compared the codon usage bias (CUB) between eight taxonomically distinct microsporidian genomes: Encephalitozoon intestinalis, Encephalitozoon cuniculi, Spraguea lophii, Trachipleistophora hominis, Enterocytozoon bieneusi, Nematocida parisii, Nosema bombycis and Nosema ceranae. While the CUB was found to be weak in all eight Microsporidia, nearly all (98%) of the optimal codons in S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis and N. ceranae are fond of A/U in third position whereas most (64.6%) optimal codons in the Encephalitozoon species E. intestinalis and E. cuniculi are biased towards G/C. Although nucleotide composition biases are likely the main factor driving the CUB in Microsporidia according to correlation analyses, directed mutational pressure also likely affects the CUB as suggested by ENc-plots, correspondence and neutrality analyses. Overall, the Encephalitozoon genomes were found to be markedly different from the other microsporidians and, despite being the first sequenced representatives of this lineage, are uncharacteristic of the group as a whole. The disparities observed cannot be attributed solely to differences in host specificity and we hypothesize that other forces are at play in the lineage leading to Encephalitozoon species.
Introduction
Microsporidia are spore-forming, single-celled fungal pathogens best known for their unique infection apparatus called the polar tube and for harboring species with the smallest reported nuclear genomes. Microsporidia as a group are highly diverse, with more than 1,500 distinct species infecting vertebrate and invertebrate hosts widely spread across the Tree of Life, and cause growing concerns due to their medical, environmental and economic relevance [1]. Their diversity is reflected at the genetic level, as the extreme levels of reduction encountered in the Encephalitozoon lineage [2–5] are not characteristic of the group. The microsporidian genetic paraphernalia vary by at least an order of magnitude, from as little as 2.3 Mbp [2] to more than 25 Mbp [6], with the underlying content and structure changing accordingly. These changes may reflect, at least in part, the different evolutionary pressures applied by the various host ranges with which each microsporidian species co-evolve, and a better understanding of these changes may lead to better predictions models about their zoonotic and lethal potentials.
The genetic code plays a critical role in living cells, but not all species use its built-in redundancy in the same way. Codon usage biases (CUB) are widespread across the Tree of Life and are affected by nucleotide composition [7], translation processes [8], tRNA abundance [9], gene function [10] and length [11], protein structure [12] and hydrophobicity [13], environment temperature [14] and other factors. In particular, the balance between gene mutation and natural selection determines the CUB [15]. The genetic code itself is not universal and deviations from the standard code can have profound impacts on the translational apparatus of the corresponding organisms. Conversely, irreversible modifications to a species’ translational apparatus can force it to adapt its CUB accordingly. With genome reduction often comes simplification and forced specialization at the expense of versatility.
Microsporidia have simpler ribosomes than their fungal relatives with sediment coefficients that are similar to that of prokaryotes [16]. This simplification could potentially limit the breadth of possible codon usage biases that they can adopt. While CUB in Microsporidia have been investigated to various degrees [4, 17–19], only one study addressed their CUB in a systematic, albeit succinct fashion [18]. Here, we expanded on previous studies by using a wider selection of eight taxonomically dispersed microsporidian representatives with available genomic sequences. By examining several statistics that characterize CUB, we identify several trends and uncover some implications for selection pressures affecting this idiomatic phylum.
Materials and Methods
Genomes and coding sequences
The annotated genomes of eight microsporidian species and their coding sequences (CDS) were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank). The choice of the genomes investigated was based on the overall quality of their respective annotations, the diversity of hosts infected by the selected species, and the wide distribution of the selected species across the microsporidian phylogenetic tree (see Vossbrink and Debrunner-Vossbrink 2005 [20]). Encephalitozoon intestinalis ATCC 50506 [2], Encephalitozoon cuniculi GB-M1 [3], Spraguea lophii 42_110 [21], Trachipleistophora hominis [18], Enterocytozoon bieneusi H348 [22], Nematocida parisii ERTm1 [23], Nosema bombycis CQ1 [19] and Nosema ceranae BRL01 [24] featured a total of 1939, 1996, 2499, 3212, 3632, 2661, 4468 and 2060 annotated CDS, respectively. To minimize outliers caused by small sizes, only CDS of at least 300 bp were kept for downstream analyses. Thus, a total of 1770, 1960, 2461, 2476, 2932, 2464, 3740 and 2022 CDS for E. intestinalis, E. cuniculi, S. lophii and T. hominis, E. bieneusi, N. parisii, N. bombycis and N. ceranae, respectively, were analyzed.
Nucleotide composition analyses
The GC content of the entire CDS (GCcds) as well as the first (P1), second (P2), and third (P3) codon position GC content were calculated using a custom PERL script (available on https://github.com/hxiang1019/calc_GC_content.git). To account for the inequality of α and γ at the third codon position [25], the three stop codons (UAA, UAG, and UGA) and the three codons for isoleucine (AUU, AUC, and AUA) were excluded in calculation of P3, and the two single codons for methionine (AUG) and tryptophan (UGG) were excluded from P1, P2, and P3. Neutrality plots were drawn using the average value of P1 and P2 (P12) as the vertical axis and the P3 as the horizontal axis. The nucleotide compositions of the third codon position (A3, U3, C3, and G3) were also obtained and used to calculate the AU-bias [A3/ (A3+U3)] and GC-bias [G3/ (G3+C3)]. The Parity rule 2 (PR2) plots were drawn based on AU-bias and GC-bias.
Codon usage indices and ENc-plot
The Codon Adaptation Index (CAI), the Effective Number of Codons (ENc), and the third synonymous codon position GC content (GC3s) were calculated using CodonW (John Peden, http://www.molbiol.ox.ac.uk/cu, version 1.4.2) using Saccharomyces cerevisiae as reference [16]. The ENc vs GC3s plots were generated from this data.
RSCU and correspondence analyses
The relative synonymous codon usage (RSCU) was calculated using CodonW. The high- and low-expression gene datasets were defined as genes in the upper and lower 5% of CAI values for each microsporidian species. RSCU values of these two datasets were compared through a chi-squared test, and the codons whose usage frequency in the high-expression genes was significantly higher (P-value < 0.05) than in the low-expression genes were identified as the optimal codons [26]. Codons with RSCU values less than 0.1 were classified as rare codons. A heat map was drawn with CIMMiner (http://discover.nci.nih.gov/cimminer) [27] and clustered the microsporidian RSCU values using a Euclidean distance method and an Average Linkage cluster algorithm. The correspondence analysis (COA) [28] was performed with CodonW utilizing the RSCU values to compare the intra-genomic variation of 59 informative codons, partitioned along 59 orthogonal axes with 41 degrees of freedom. Correlation analyses, ANOVA and significance tests were performed with Microsoft Excel and SPSS 18.0 (http://www.spss.com/).
Results
Codon usage biases
Codon usage patterns for microsporidian genomes were investigated by calculating RSCU values (Table 1). The RSCU is the observed frequency of a codon divided by the expected one. If the RSCU is close to 1, synonymous codons are used without apparent biases. When the RSCU value is greater or less than 1, the codons investigated are used more or less frequently than expected, respectively.
Table 1. The RSCU analysis of the preferred codons (codons with RSCU > 1), the optimal codons and the rare codons for microsporidian genomes.
| Amino acid | Codon | RCSU | |||||||
|---|---|---|---|---|---|---|---|---|---|
| E. intestinalis | E. cuniculi | S. lophii | T. hominis | Ent. bieneusi | Nem. parisii | N. bombycis | N. ceranae | ||
| Phe | UUU | 1.31* | 1.26* | 1.67* | 1.53* | 1.90* | 1.53* | 1.74* | 1.86* |
| UUC | 0.69 | 0.74 | 0.33 | 0.47 | 0.10 | 0.47 | 0.26 | 0.14 | |
| Leu | UUA | 0.22 | 0.20 | 4.02* | 2.04* | 4.33* | 3.21* | 3.70* | 3.33* |
| UUG | 0.92 | 0.77* | 0.40 | 1.35* | 0.49 | 0.47 | 0.65 | 0.71 | |
| CUU | 1.96* | 1.67 | 0.71 | 1.07 | 0.71* | 0.80* | 1.13* | 1.13 | |
| CUC | 0.66 | 0.91 | 0.15 | 0.34 | 0.04- | 0.12 | 0.20 | 0.08- | |
| CUA | 0.53 | 0.32 | 0.63* | 0.73* | 0.40* | 1.02* | 0.30 | 0.62 | |
| CUG | 1.71* | 2.14* | 0.09- | 0.47 | 0.03- | 0.38 | 0.03- | 0.13 | |
| Ile | AUU | 1.27* | 1.28* | 0.84 | 1.26* | 1.55* | 1.10* | 1.70* | 1.55 |
| AUC | 0.86 | 0.73 | 0.16 | 0.45 | 0.09- | 0.16 | 0.24 | 0.15 | |
| AUA | 0.88 | 0.99* | 2.01* | 1.29* | 1.36* | 1.74* | 1.06* | 1.31* | |
| Met | AUG | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Val | GUU | 1.26 | 1.28* | 1.07 | 1.49* | 1.85* | 1.03* | 1.56* | 1.53 |
| GUC | 0.45 | 0.56 | 0.04- | 0.39 | 0.11 | 0.18 | 0.24 | 0.20 | |
| GUA | 0.50 | 0.37 | 2.54* | 1.28* | 1.90* | 2.09* | 1.88* | 1.96* | |
| GUG | 1.78* | 1.79* | 0.35 | 0.83 | 0.15 | 0.70 | 0.32 | 0.31 | |
| Tyr | UAU | 1.01 | 0.98* | 1.82* | 1.48* | 1.87* | 1.58* | 1.55* | 1.54* |
| UAC | 0.99* | 1.02 | 0.18 | 0.52 | 0.13 | 0.42 | 0.45 | 0.46 | |
| Stop | UAA | 1.16 | 0.00- | 2.32 | 1.57 | 2.42 | 2.16 | 1.75 | 2.02 |
| UAG | 0.41 | 0.00- | 0.32 | 0.54 | 0.32 | 0.57 | 0.43 | 0.50 | |
| UGA | 1.44 | 0.00- | 0.37 | 0.90 | 0.25 | 0.27 | 0.81 | 0.48 | |
| His | CAU | 1.13 | 0.96* | 1.80* | 1.53* | 1.84* | 1.53* | 1.68* | 1.61* |
| CAC | 0.87* | 1.04 | 0.20 | 0.47 | 0.16 | 0.47 | 0.32 | 0.39 | |
| Gln | CAA | 0.59 | 0.36 | 1.50 | 1.47* | 1.87* | 1.23* | 1.86* | 1.53* |
| CAG | 1.41* | 1.64* | 0.50 | 0.53 | 0.13 | 0.77 | 0.14 | 0.47 | |
| Asn | AAU | 0.83 | 0.95* | 1.67* | 1.43* | 1.85* | 1.68* | 1.79* | 1.73* |
| AAC | 1.17* | 1.05 | 0.33 | 0.57 | 0.15 | 0.32 | 0.21 | 0.27 | |
| Lys | AAA | 0.37 | 0.29 | 1.55* | 1.49* | 1.85* | 1.44* | 1.62* | 1.67* |
| AAG | 1.63* | 1.71* | 0.45 | 0.51 | 0.15 | 0.56 | 0.38 | 0.33 | |
| Asp | GAU | 1.00 | 0.98* | 1.91* | 1.59* | 1.85* | 1.60* | 1.88* | 1.71@ |
| GAC | 1.00* | 1.02 | 0.09- | 0.41 | 0.15 | 0.40 | 0.12 | 0.29 | |
| Glu | GAA | 0.64 | 0.45 | 1.67 | 1.53* | 1.87* | 1.53* | 1.67* | 1.70* |
| GAG | 1.36* | 1.55* | 0.33 | 0.47 | 0.13 | 0.47 | 0.33 | 0.30 | |
| Ser | UCU | 1.27 | 1.30 | 1.49 | 1.18* | 1.65* | 1.37* | 2.22* | 1.93 |
| UCC | 0.51 | 0.46 | 0.14 | 0.42 | 0.20 | 0.30 | 0.46 | 0.21 | |
| UCA | 0.46 | 0.28 | 0.82 | 1.60* | 2.23* | 1.39* | 1.23* | 1.38 | |
| UCG | 1.23* | 1.09* | 0.09- | 0.65 | 0.12 | 0.18 | 0.13 | 0.26 | |
| AGU | 1.04* | 1.27* | 3.15* | 1.41* | 1.58* | 2.31* | 1.90* | 1.82* | |
| AGC | 1.49* | 1.60* | 0.32 | 0.74 | 0.21 | 0.44 | 0.06- | 0.40 | |
| Pro | CCU | 1.24 | 1.28* | 2.15* | 1.22* | 1.56* | 1.20* | 2.12* | 1.93* |
| CCC | 0.67 | 0.74 | 0.31 | 0.37 | 0.13 | 0.38 | 0.39 | 0.28 | |
| CCA | 0.99 | 0.65 | 1.43 | 1.85* | 2.20* | 2.03* | 1.38* | 1.54 | |
| CCG | 1.11* | 1.33* | 0.10 | 0.55 | 0.10 | 0.38 | 0.11 | 0.25 | |
| Thr | ACU | 0.63 | 0.84 | 1.89* | 1.03* | 1.69* | 1.39* | 2.03* | 1.35 |
| ACC | 0.54 | 0.63 | 0.56 | 0.53 | 0.18 | 0.32 | 0.33 | 0.26 | |
| ACA | 1.75 | 0.91 | 1.44 | 1.81* | 2.00* | 2.00* | 1.39* | 2.19* | |
| ACG | 1.08* | 1.61* | 0.11 | 0.63 | 0.13 | 0.29 | 0.24 | 0.20 | |
| Ala | GCU | 0.58 | 0.75 | 1.81* | 1.27* | 1.74* | 1.07* | 2.24* | 1.31 |
| GCC | 0.69 | 0.66 | 0.18 | 0.48 | 0.15 | 0.41 | 0.43 | 0.27 | |
| GCA | 2.09* | 1.48 | 1.80 | 1.71* | 2.02* | 2.25* | 1.29* | 2.17* | |
| GCG | 0.64* | 1.11* | 0.22 | 0.54 | 0.09- | 0.27 | 0.04- | 0.26 | |
| Cys | UGU | 0.99 | 0.93* | 1.87* | 1.41* | 1.72* | 1.51* | 1.91* | 1.61 |
| UGC | 1.01 | 1.07 | 0.13 | 0.59 | 0.28 | 0.49 | 0.09- | 0.39 | |
| Trp | UGG | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Arg | CGU | 0.18 | 0.23 | 0.40 | 0.85 | 0.77 | 0.53* | 0.27 | 0.39 |
| CGC | 0.14 | 0.19 | 0.03- | 0.34 | 0.08- | 0.16 | 0.00- | 0.21 | |
| CGA | 0.67* | 0.22 | 0.11 | 1.09* | 1.42* | 0.17 | 0.25 | 0.46 | |
| CGG | 0.81* | 1.08* | 0.02- | 0.33 | 0.11 | 0.22 | 0.05- | 0.06- | |
| AGA | 1.95 | 1.53 | 4.97* | 2.51* | 3.17* | 3.72* | 3.45* | 4.22* | |
| AGG | 2.25* | 2.76* | 0.46 | 0.87 | 0.46 | 1.21 | 1.97* | 0.66 | |
| Gly | GGU | 0.17 | 0.34 | 2.66* | 1.60* | 1.19* | 1.37* | 1.13* | 1.27 |
| GGC | 0.23 | 0.65 | 0.11 | 0.52 | 0.12 | 0.59 | 0.06- | 0.34 | |
| GGA | 2.01 | 1.27 | 1.09 | 1.53* | 2.53* | 1.32 | 1.81* | 2.12* | |
| GGG | 1.59* | 1.74* | 0.14 | 0.36 | 0.16 | 0.73 | 1.00* | 0.27 | |
Both the sign * (P-value < 0.01) and @ (0.01 < P-value < 0.05) represent the optimal codons, while the sign - (RSCU < 0.10) denotes the rarely used codons. The preferred codons (RSCU > 1) are in bold.
The preferred codons (RSCU > 1, Table 1; in bold) in S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis, and N. ceranae are strongly biased towards A/U bases in third position, in contrast to E. intestinalis and E. cuniculi where more than half of the codons end with G or C (Table 2). Optimal codons (shown in * or @, Table 1) identified by chi-squared tests are similarly biased. Nearly all of the optimal codons in S. lophii (17 A/U-end in 17 optimal codons), T. hominis (28 A/U-end in 29 optimal codons), E. bieneusi (29 A/U-end in 29 optimal codons), N. parisii (28 A/U-end in 28 optimal codons), N. bombycis (27 A/U-end in 29 optimal codons), and N. ceranae (17 A/U-end in 17 optimal codons) are A/U-end whereas more than half of the optimal codons in E. intestinalis (17 G/C-end in 23 optimal codons) and E. cuniculi (14 G/C-end in 25 optimal codons) are G/C-end (Table 2). When clustering these biases according to a heat map (Fig 1), these values display a remarkable difference between the Encephalitozoon species and the other six microsporidians.
Table 2. The summary of the preferred codons, the optimal codons and the rare codons for microsporidian genomes.
| Codon type | Codon 3rd base | E. intestinalis | E. cuniculi | S. lophii | T. hominis | Ent. bieneusi | Nem. parisii | N. bombycis | N. ceranae |
|---|---|---|---|---|---|---|---|---|---|
| RSCU > 1 | A/U/G/C | 28 | 28 | 25 | 30 | 28 | 29 | 29 | 28 |
| A/U | 15 | 10 | 25 | 29 | 28 | 28 | 28 | 28 | |
| G/C | 13 | 18 | 0 | 1 | 0 | 1 | 1 | 0 | |
| A | 6 | 3 | 12 | 14 | 14 | 14 | 13 | 13 | |
| U | 9 | 7 | 13 | 15 | 14 | 14 | 15 | 15 | |
| G | 10 | 12 | 0 | 1 | 0 | 1 | 1 | 0 | |
| C | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Optimal | A/U/G/C | 23 | 25 | 17 | 29 | 29 | 28 | 29 | 17 |
| A/U | 6 | 11 | 17 | 28 | 29 | 28 | 27 | 17 | |
| G/C | 17 | 14 | 0 | 1 | 0 | 0 | 2 | 0 | |
| A | 2 | 1 | 6 | 14 | 14 | 12 | 12 | 10 | |
| U | 4 | 10 | 11 | 14 | 15 | 16 | 15 | 7 | |
| G | 12 | 13 | 0 | 1 | 0 | 0 | 2 | 0 | |
| C | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rare | A/U/G/C | 0 | 3 | 6 | 0 | 5 | 0 | 7 | 2 |
Fig 1. Heat map of RSCU values in microsporidian genomes.

The heat-map was drawn with CIMminer, using the quantile binning method. Bigger RSCU values, suggesting more frequent codon usage, are represented with darker shades of red. Six Microsporidia (S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis, N. ceranae) strongly prefer the A/U-end codons, while Encephalitozoon genus (E. intestinalis, E. cuniculi) displays a more varied distribution.
Correlation analyses
Nucleotide composition is an important factor influencing CUB, and the mean values of all of the microsporidian GCcds are similar to their reported overall genomic GC content (Table 3). The GCcds of S. lophii (25.62%), T. hominis (39.60%), E. bieneusi (32.07%), N. parisii (36.37%), N. bombycis (30.57%), and N. ceranae (27.23%) are low, while the GCcds of E. intestinalis (42.01%) and E. cuniculi (47.59%) are notably higher. The correlation analysis (Table 4) shows that the GCcds, P1, P2 and P3 are significantly related to each other for all eight Microsporidia.
Table 3. Genome features (Genome size, GCgenome, No. of predicted gene and CDS) obtained from the genome database of NCBI (http://www.ncbi.nlm.nih.gov/genome/), and GC contents calculated in this paper for microsporidia genomes.
| Organism | E. intestinalis | E. cuniculi | S. lophii | T. hominis | Ent. bieneusi | Nem. parisii | N. bombycis | N. ceranae |
|---|---|---|---|---|---|---|---|---|
| Size (Mbp) | 2.22 | 2.50 | 4.98 | 8.50 | 3.86 | 4.07 | 15.69 | 7.86 |
| GCgenome | 41.5% | 47.3% | 23.4% | 34.1% | 33.7% | 34.4% | 30.8% | 25.3% |
| Genes | 2,011 | 2,029 | 2,596 | 3,253 | 3,806 | 2,724 | 4,468 | 2,678 |
| CDS | 1,939 | 1,996 | 2,499 | 3,212 | 3,632 | 2,661 | 4,468 | 2,060 |
| CDSused | 1,770 | 1,960 | 2,461 | 2,476 | 2,932 | 2,464 | 3,740 | 2,022 |
| GCcds | 42.01% | 47.59% | 25.62% | 39.60% | 32.07% | 36.37% | 30.57% | 27.23% |
| GC3s | 44.23% | 55.25% | 15.50% | 41.63% | 30.69% | 32.72% | 23.69% | 18.11% |
| P1 | 47.42% | 51.03% | 34.55% | 45.48% | 42.28% | 40.36% | 39.03% | 36.28% |
| P2 | 34.37% | 37.73% | 25.74% | 32.14% | 30.53% | 34.13% | 28.79% | 27.08% |
| P12 | 40.90% | 44.38% | 30.15% | 38.81% | 36.41% | 37.25% | 33.91% | 31.68% |
| P3 | 45.82% | 56.90% | 16.49% | 42.93% | 31.64% | 34.83% | 24.51% | 19.26% |
| ENc | 50.41 | 50.49 | 37.04 | 51.91 | 44.96 | 47.37 | 42.29 | 40.86 |
Table 4. Correlation analysis among GCcds, P1, P2, P12, P3 and ENc for eight Microsporidia.
| GCcds | P1 | P2 | P12 | P3 | |
|---|---|---|---|---|---|
| P1 | 0.583*/0.615* 0.816*/0.725* 0.896*/0.666* 0.717*/0.768* |
||||
| P2 | 0.515*/0.534* 0.793*/0.595* 0.812*/0.699* 0.747*/0.805* |
0.102*/0.209* 0.476*/0.370* 0.674*/0.226* 0.377*/0.458* |
|||
| P12 | 0.747*/0.742* 0.946*/0.810* 0.940*/0.877* 0.889*/0.930* |
0.694*/0.726* 0.872*/0.819* 0.932*/0.812* 0.821*/0.856* |
0.745*/0.787* 0.827*/0.810* 0.888*/0.722* 0.813*/0.835* |
||
| P3 | 0.671*/0.580* 0.507*/0.792* 0.939*/0.792* 0.543*/0.539* |
0.160*/0.103* 0.190*/0.454* 0.767*/0.420* 0.130*/0.160* |
-0.023/-0.120* 0.219*/0.239* 0.646*/0.340* 0.166*/0.263* |
0.082*/-0.021 0.246*/0.420* 0.781*/0.481* 0.168*/0.254* |
|
| ENc | 0.016/-0.287* 0.446*/0.143* 0.611*/0.486* 0.234*/0.423* |
-0.017/-0.068* 0.252*/0.095* 0.538*/0.265* 0.061*/0.191* |
-0.102*/-0.033 0.230*/0.041* 0.352*/0.274* 0.010/0.209* |
-0.080*/-0.061* 0.291*/0.079* 0.503*/0.338* 0.048*/0.240* |
0.101*/-0.385* 0.627*/0.194* 0.643*/0.545* 0.471*/0.663* |
The eight Spearman’s rank correlation coefficients (ρ) are the results of E. intestinalis / E. cuniculi, S. lophii / T. hominis, Ent. bieneusi / Nem. parisii, N. bombycis / N. ceranae, respectively. P-values < 0.01 are indicated by asteriskes (*).
In addition, correlations between ENc and GCcds, P1, P2 and P3 were also investigated. ENc relates the overall synonymous codon usage, ranging from only 20 codons being used for each of the 20 amino acids, to all 61 codons being used randomly [29]. In Table 4, the ENc is significantly correlated to GCcds, P1, P2 and P3 for Microsporidia. Correlations between ENc and GCcds, P1, P2 and P3 were either negative or null for Encephalitozoon species but positive for the other six Microsporidia.
To judge whether the nucleotide composition is the only factor to influence CUB for Microsporidia, the ENc-plot (Fig 2) was drawn. If genes follow the standard curve ENc = 2+GC3s+29/[GC3s 2+(1-GC3s)2], the microsporidian CUB is determined primarily by the nucleotide composition [29]. In Fig 2, the distribution of most genes below the standard curve indicates that there are other factors acting on microsporidian CUB.
Fig 2. The ENc vs. GC3s plots of microsporidian genomes.
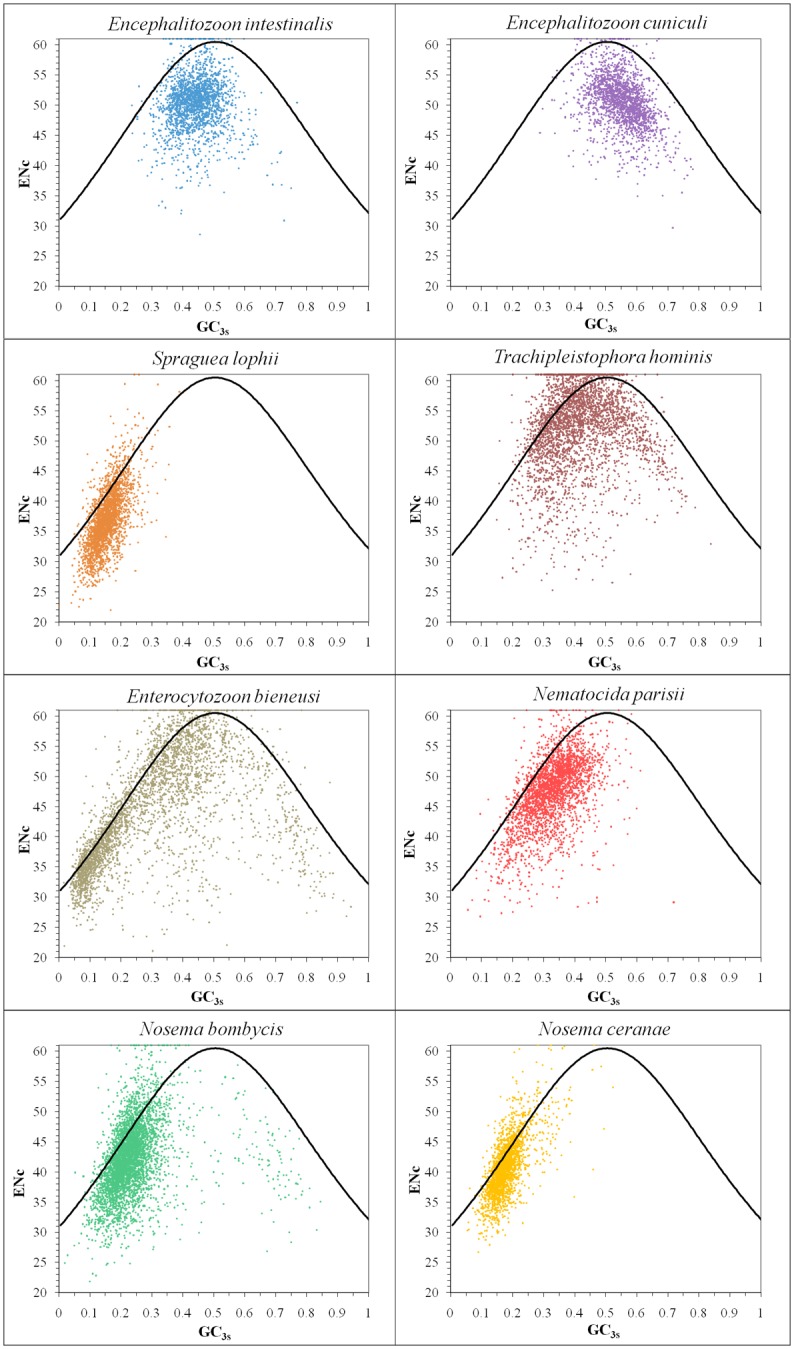
The standard curve ENc = 2+GC3s+29/[GC3s 2+(1-GC3s)2] represents the expected ENc to GC3s. Most microsporidian genes are far away from the curve, showing that their codon usage pattern might be affected by other factors besides nucleotide composition. Some genes with the ENc score of 61 display no bias and use all the 61 sense codons.
Parity Rule 2 plot analyses
Examining the codons with RSCU > 1 and the optimal codons, species from the Encephalitozoon genus prefer G-end over C-end, despite the other microsporidians preferring A and U ends to a roughly equivalent degree (Table 2). Because of this bias of guanine over cytosine, all codons were examined by PR2 plot analysis (Fig 3). In the PR2 plot, the mean GC-biases [G3/(G3+C3)] of E. intestinalis, E. cuniculi, S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis, and N. ceranae are 0.560, 0.558, 0.574, 0.569, 0.514, 0.538, 0.526, and 0.564; while their mean AU-biases [A3/(A3+U3)] are 0.517, 0.497, 0.538, 0.503, 0.514, 0.554, 0.485, and 0.502, respectively. In several of the species the PR2 plot highlights a slight preference for third position G over C. In a gene where CUB is only influenced by nucleotide composition, the third positions should have the identical distribution between G3 and C3 as well as A3 and U3 [30]. Thus it appears there are other factors besides nucleotide composition affecting microsporidian codon usage.
Fig 3. The PR2-bias plots of microsporidian genomes.
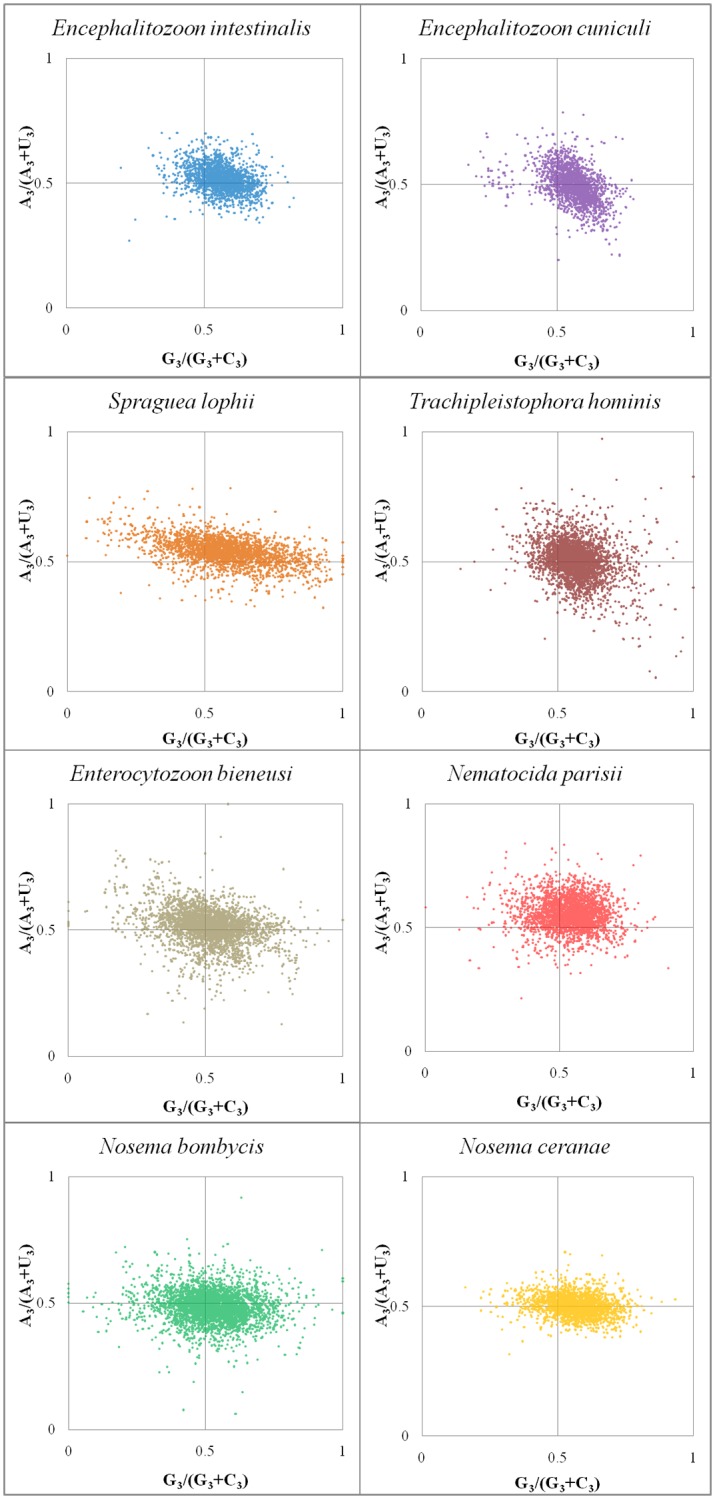
Genes are plotted based on their GC bias [G3/(G3+C3)] and AU bias[A3/(A3+U3)] in the third codon position. The mean GC-biases of E. intestinalis, E. cuniculi, S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis, and N. ceranae are 0.560, 0.558, 0.574, 0.569, 0.514, 0.538, 0.526, and 0.564, respectively; while their mean AU-biases are 0.517, 0.497, 0.538, 0.503, 0.514, 0.554, 0.485, and 0.502, respectively.
Neutrality plot analyses
The neutrality plot analysis (Fig 4) was carried out to characterize the correlation among the three codon positions, and then identify the presence of selective mutation on CUB [25]. In the neutrality plot, if a gene is located on the slope of unity there is a significant correlation between its P12 and P3, meaning the gene is under neutral mutation via random selection pressure. If the gene is under a directed mutational pressure it should fall below the slope of unity, closer to the X-axis. Thus a regression line with a slope less than 1 would indicate a whole genome trend of non-neutral mutational pressure [31]. The Encephalitozoon species have regression slopes of 0.0589 and -0.0048, and correlation coefficients of 0.082 and -0.021 respectively. Their extremely low relative neutralities (5.9% and 4.8%) might suggest a large amount of directed mutational pressure, although the low Spearman correlations combined with P-values > 0.05 make these unreliable (Fig 4 and Table 4). S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis and N. ceranae all have relative neutralities ranging from 20–38% and significant correlation coefficients with P-values < 0.01 (Fig 4 and Table 4), indicating that directed mutational pressure plays an important role in shaping CUB for these Microsporidia.
Fig 4. Neutrality plots of microsporidian genomes.
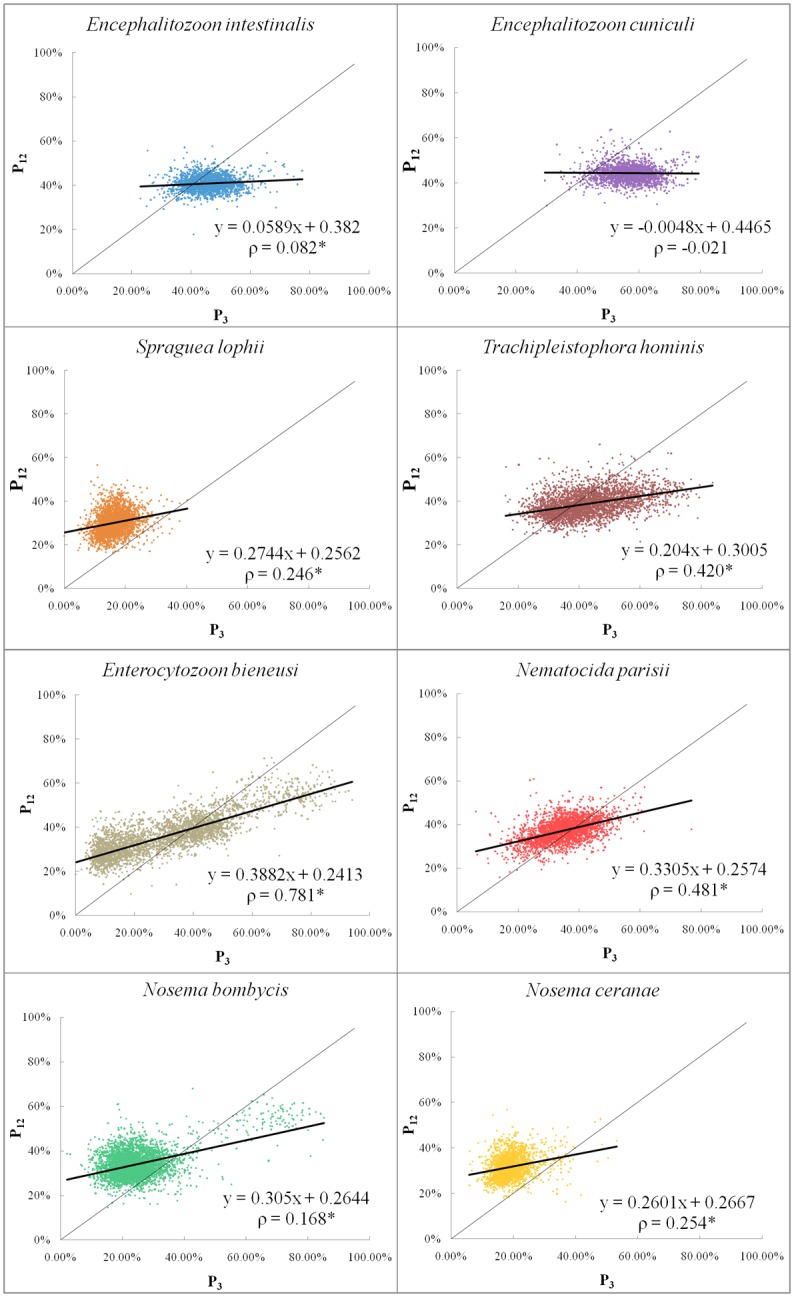
Individual genes are plotted based on the mean GC content in the first and second codon position (P12) versus the GC content of the third codon position (P3). Regression lines and Spearman’s rank correlation coefficients (ρ) are shown, with the asterisk (*) denoting P-values < 0.01.
Correspondence analyses
The correspondence analysis was used to check what other factors shape the microsporidian CUB. This multivariate statistical method surveys the variation of RSCU values within the genome [28]. The correspondence analysis shows the distribution of genes and reflects the distribution of their corresponding codons, unveiling potential influences on CUB [13]. In the correspondence analysis, a series of orthogonal axes were produced to represent the factors responsible for CUB (Fig 5). For E. intestinalis, E. cuniculi, S. lophii, T. hominis, E. bieneusi, N. parisii, N. bombycis, N. ceranae, Axis 1 accounted for 9.81%, 11.98%, 12.44%, 16.42%, 18.06%, 12.61%, 12.29%, 9.59% of their respective total variation; Axis 2 accounted for 8.81%, 8.69%, 7.33%, 6.33%, 8.32%, 10.45%, 8.98%, 6.67% of variation, respectively; and the first four axes combined (Axes 1 through 4) accounted for 26.51%, 31.01%, 31.08%, 33.29%, 35.24%, 34.92%, 31.31%, 24.54%, respectively. This suggests that the first axis is the primary factor (9–18% of the overall variation), which was also found to be significantly correlated with CAI, ENc, and GC3s. However, other factors are also responsible for the codon usage variation based on Axes 2 to 4.
Fig 5. The correspondence analysis (COA) of the genes in microsporidian genomes.
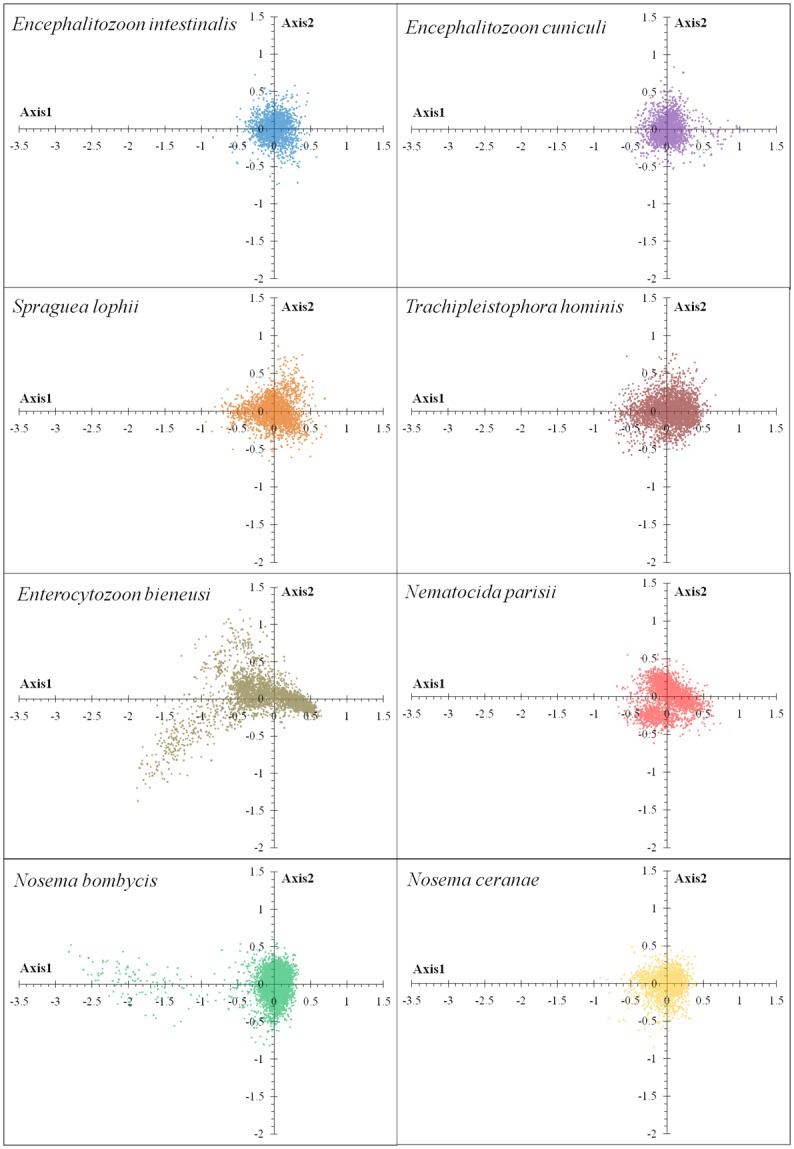
Each point represents a gene corresponding to the coordinates of the first and second axes of variation generated from the correspondence analysis. Some E. bieneusi and N. bombycis genes scattered to the left of Axis 1 might have distinct codon usage biases.
Codon usage indices
The notable differences observed in CUB between Encephalitozoon species and the other microsporidians (Fig 1) were confirmed by one-way ANOVA (F-value: 1734.906 in CAI, 1919.114 in ENc; and P-value < 0.01 in both) based on the CAI and ENc (Fig 6) and by a T-test (P-value < 0.01). CAI is a ratio of the synonymous codon bias in a gene to a highly expressed reference gene. With values that range between 0 and 1, a higher CAI value indicates a stronger bias of synonymous codon usage and a potentially higher gene expression [32]. For microsporidian genomes, their mean CAI values are less than 0.15, and their mean ENc values are larger than 37 (Fig 6). These indicate the presence of mild synonymous codon usage bias across microsporidian genomes as a whole, with Encephalitozoon species showing the highest degree of randomization (Fig 6).
Fig 6. The variance analysis of codon indices (CAI and ENc) among microsporidian genomes.

The one-way ANOVA shows high F-values (1734.906 in CAI, 1919.114 in ENc) and significant P-values (0.000 in both), strongly supporting the differences among microsporidian genomes.
Discussion
The codon usage bias, an important feature of species that can reflect the evolutionary patterns of their genome, has been reported in numerous organisms [33]. Here, the CUB in Microsporidia was studied based on eight genomes and 22,467 coding sequences, with optimal codons identified by RSCU values. The optimal codon usage pattern was found to be significantly different between species from the genus Encephalitozoon and those from other microsporidian lineages. Nearly all of the optimal codons in S. lophii, T. hominis, N. parisii, E. bieneusi, N. bombycis, N. ceranae feature a biased A/U third codon position, while the Encephalitozoon species (E. intestinalis, E. cuniculi) have a more balanced nucleotide distribution, yet slightly biased towards G/C. Although the microsporidian CUB are mild according to CAI and ENc values, these statistics are likely the result of a larger distribution of varying biases for individual genes averaging out to a less significant overall genome bias. This has previously been described [3, 5], where Encephalitozoon GC content smoothly arced across the chromosomes, but averaged an unremarkable GC%. Still, nucleotide composition is clearly a factor in microsporidian CUB, which was confirmed by correlation analysis.
Besides nucleotide composition, the microsporidian CUB is also influenced by other factors including directional mutation pressure, which appears to play a much larger role than selection in the CUB of Microsporidia according to neutrality analyses. While the neutrality plots do suggest an even larger directional mutation pressure for the Encephalitozoon species, the observed correlation coefficients undermine its overall significance. Being the first completely sequenced microsporidian genome, Encephalitozoon cuniculi has long been regarded as the model Microsporidia. Its reduction and compaction were for a time thought to be typical traits of microsporidian genomes. However, comparative analyses of recently released microsporidian genomes [6] rather indicates that Encephalitozoons are the exception rather than the norm, and that the evolutionary trends they have followed are not characteristic of the group. This is corroborated by our analyses. As the Encephalitozoon species are known as the smallest eukaryotic genomes (assembled genomic sizes of 2.22 and 2.50 Mbp; Table 3), they are likely under stronger reductionist pressure (directional mutation pressure) than the larger microsporidian genomes, of which many have expanded by genome duplication, horizontal gene transfer and transposable elements proliferation [1].
Intuitively, different hosts respond differently to parasitic infections, and while arthropods do possess an innate immunity, they lack the adaptive immune response found in mammals. However, the Encephalitozoon species represented here are not the only microsporidians infecting mammals, with both E. bieneusi and T. hominis reported as human pathogens [18, 22]. If host specificity was the sole factor involved, we would expect these four species to display similar trends, which is clearly not the case. Unfortunately, it is unknown how long these species have been coevolving with their host, and the observed disparities may be due to differences in duration rather than in the relative strengths of the underlying evolutionary pressure applied. Here, we hypothesize that the Encephalitozoon species may have been under mammalian host pressure for longer evolutionary periods, which would explain why they are so markedly different. A caveat of this is that the host specificity of these organisms is unclear. In fact, the Encephalitozoon species E. romaleae infects grasshoppers but also most likely can pester mammalian cells based on its uncanny genomic similarities with is sister species E. hellem [4], and we don’t know if its presumed absence in humans is real or rather due to limited sampling. Alternatively, the observed differences may be a direct consequence of the strongly reduced metabolic potential inherent to their Lilliputian gene repertoire, and it would be interesting to revisit the genome of the closest known relative of Encephalitozoon species, Ordospora colligata [6], which has been released during the latter stages of publication of this manuscript. O. colligata infects the water flea Daphnia (an arthropod [34]) and displays a similarly reduced genome, whose overall characteristics may help better delineate what has happened in the lineage leading to the Encephalitozoon species.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 31302036 and 30930067), the Ph.D. Programs Foundation of Ministry of Education of China (No. 20110182120008), the Fundamental Research Funds for the Central Universities (Nos. XDJK2011C027 and XDJK2015B024), the Ph.D. Programs Foundation of Southwest University (No. SWU110047), and the China Scholarship Council (No. 201206995026).
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Nos. 31302036 and 30930067), the Ph.D. Programs Foundation of Ministry of Education of China (No. 20110182120008), the Fundamental Research Funds for the Central Universities (Nos. XDJK2011C027 and XDJK2015B024), the Ph.D. Programs Foundation of Southwest University (No. SWU110047), and the China Scholarship Council (No. 201206995026). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Peyretaillade E, Boucher D, Parisot N, Gasc C, Butler R, Pombert J, et al. Exploiting the architecture and the features of the microsporidian genomes to investigate diversity and impact of these parasites on ecosystems. Heredity. 2014. 10.1038/hdy.2014.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis . Nature Commun. 2010;1:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi . Nature. 2001;414(6862):450–3. [DOI] [PubMed] [Google Scholar]
- 4. Pombert J-F, Selman M, Burki F, Bardell FT, Farinelli L, Solter LF, et al. Gain and loss of multiple functionally related, horizontally transferred genes in the reduced genomes of two microsporidian parasites. Proc Natl Acad Sci USA. 2012;109(31):12638–43. 10.1073/pnas.1205020109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pombert J-F, Xu J, Smith DR, Heiman D, Young S, Cuomo CA, et al. Complete genome sequences from three genetically distinct strains reveal high intraspecies genetic diversity in the microsporidian Encephalitozoon cuniculi . Eukaryotic cell. 2013;12(4):503–11. 10.1128/EC.00312-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pombert J-F, Haag KL, Beidas S, Ebert D, Keeling PJ. The Ordospora colligata Genome: Evolution of Extreme Reduction in Microsporidia and Host-To-Parasite Horizontal Gene Transfer. mBio. 2015;6(1):e02400–14. 10.1128/mBio.02400-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osawa S, Ohama T, Yamao F, Muto A, Jukes TH, Ozeki H, et al. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci USA. 1988;85(4):1124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharp PM, Li W-H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’codons. Nucleic Acids Res. 1986;14(19):7737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151(3):389–409. [DOI] [PubMed] [Google Scholar]
- 10. Chiapello H, Lisacek F, Caboche M, Hénaut A. Codon usage and gene function are related in sequences of Arabidopsis thaliana . Gene. 1998;209(1):1–38. [DOI] [PubMed] [Google Scholar]
- 11. Moriyama EN, Powell JR. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli . Nucleic Acids Res. 1998;26(13):3188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orešič M, Shalloway D. Specific correlations between relative synonymous codon usage and protein secondary structure. J Mol Biol. 1998;281(1):31–48. [DOI] [PubMed] [Google Scholar]
- 13. Romero H, Zavala A, Musto H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 2000;28(10):2084–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sau K, Deb A. Temperature influences synonymous codon and amino acid usage biases in the phages infecting extremely thermophilic prokaryotes. In silico biology. 2009;9(1):1–9. [PubMed] [Google Scholar]
- 15. Morton BR. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J Mol Evol. 2003;56(5):616–29. [DOI] [PubMed] [Google Scholar]
- 16. Sharp PM, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae . Yeast. 1991;7(7):657–78. [DOI] [PubMed] [Google Scholar]
- 17. Zhang R, Xiang H, Pan G, Li T, Zhou Z. Comparison of the codon bias between two microsporidian genomes. Newsl Sericultural Sci. 2011;1(31):1–8. [Google Scholar]
- 18. Heinz E, Williams TA, Nakjang S, Noël CJ, Swan DC, Goldberg AV, et al. The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 2012;8(10):e1002979 10.1371/journal.ppat.1002979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan G, Xu J, Li T, Xia Q, Liu S-L, Zhang G, et al. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC genomics. 2013;14(1):186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vossbrinck CR, Debrunner-Vossbrinck BA. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia parasitologica. 2005;52(1–2):131–42. [DOI] [PubMed] [Google Scholar]
- 21. Campbell SE, Williams TA, Yousuf A, Soanes DM, Paszkiewicz KH, Williams BA. The genome of Spraguea lophii and the basis of host-microsporidian interactions. PLoS genetics. 2013;9(8):e1003676 10.1371/journal.pgen.1003676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akiyoshi DE, Morrison HG, Lei S, Feng X, Zhang Q, Corradi N, et al. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi . PLoS Pathog. 2009;5(1):e1000261 10.1371/journal.ppat.1000261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012;22(12):2478–88. 10.1101/gr.142802.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5(6):e1000466 10.1371/journal.ppat.1000466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci USA. 1988;85(8):2653–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Q. Analysis of codon usage pattern in the radioresistant bacterium Deinococcus radiodurans . Biosystems. 2006;85(2):99–106. [DOI] [PubMed] [Google Scholar]
- 27. Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Kohn KW, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275(5298):343–9. [DOI] [PubMed] [Google Scholar]
- 28. Greenacre MJ. Theory and applications of correspondence analysis. London: Academic Press; 1984. [Google Scholar]
- 29. Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87(1):23–9. [DOI] [PubMed] [Google Scholar]
- 30. Sueoka N. Translation-coupled violation of Parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G + C content of third codon position. Gene. 1999;238(1):53–8. [DOI] [PubMed] [Google Scholar]
- 31. Sueoka N, Kawanishi Y. DNA G + C content of the third codon position and codon usage biases of human genes. Gene. 2000;261(1):53–62. [DOI] [PubMed] [Google Scholar]
- 32. Sharp PM, Li WH. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16(17):8207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebert D. Introduction to the Ecology, Epidemiology, and Evolution of Parasitism in Daphnia 2005. Available: http://www.ncbi.nlm.nih.gov/books/NBK2036/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


