Abstract
Flaviviruses have a spherical capsid that is composed of multiple copies of a single capsid protein and, in contrast to the viral envelope, apparently does not have an icosahedral structure. So far, attempts to isolate distinct particulate capsids and soluble forms of the capsid protein from purified virions as well as to assemble capsid-like particles in vitro have been largely unsuccessful. Here we describe the isolation of nucleocapsids from tick-borne encephalitis (TBE) virus and their disintegration into a capsid protein dimer by high-salt treatment. Purified capsid protein dimers could be assembled in vitro into capsid-like particles when combined with in vitro transcribed viral RNA. Particulate structures could also be obtained when single-stranded DNA oligonucleotides were used. These data suggest that the dimeric capsid protein functions as a basic building block in the assembly process of flaviviruses.
The genus Flavivirus in the family Flaviviridae comprises about 70 distinct viruses, most of which are transmitted by mosquitoes or ticks to their vertebrate hosts. Several of these viruses are important human pathogens, including yellow fever virus, dengue virus, Japanese encephalitis virus, West Nile virus, and tick-borne encephalitis (TBE) virus. Flaviviruses are spherical enveloped positive-stranded RNA viruses with a diameter of 50 nm that are composed of multiple copies of only three different proteins, designated C (capsid), E (envelope), and prM/M (precursor of membrane protein and membrane protein, respectively) (21). The genomic RNA is approximately 11,000 nucleotides long and functions as the sole viral mRNA in infected cells. Translation of the single long open reading frame gives rise to the structural and nonstructural (NS) viral proteins, with the structural proteins encoded in the 5′-terminal third of the genome in the order C-prM/M-E. The intracellular assembly of flaviviruses is not precisely understood but is believed to take place at the endoplasmic reticulum, because in infected cells, viral particles first become visible in this compartment by electron microscopy (EM) (5, 10, 15, 30, 35, 36). The primary assembly products are immature particles that contain the prM protein and are noninfectious (6, 9, 13, 32, 41). Infectivity is generated through the proteolytic cleavage of prM by a cellular protease (presumably furin [37]) in the trans-Golgi network shortly before the release of virions by exocytosis. The role of the 11-kDa capsid protein in virus assembly is ill defined and still a matter of speculation (26). It is first synthesized as a membrane-associated protein (anchored C), and the final cytoplasmic form is generated by cleavage of the membrane anchor by the viral NS2A/NS3 protease (2, 19, 22, 31, 43). The processed C protein is believed to remain membrane associated through an internal hydrophobic sequence (26). The high content of basic amino acids is consistent with its potential to interact with RNA in the process of viral core formation. Since fully formed capsids are almost never found in flavivirus-infected cells, it has been suggested that virus particle formation is a coordinated process that is driven by interactions between the membrane-associated C protein (probably coupled to the RNA replication complex) and the two membrane proteins (21). The mechanism of this process is believed to be regulated by proteolytic cleavage events in the membrane-associated polyprotein precursor (19, 34).
A wealth of new information concerning the structure of flaviviruses has become available recently. This includes the atomic structures of the E proteins of TBE virus and dengue virus (27, 33) as well as lower-resolution structures obtained by cryoelectron microscopy (cryo-EM) of mature and immature dengue virus (18, 44, 45), West Nile virus (29), and a recombinant subviral particle from TBE virus (7). The cryo-EM structures demonstrated an icosahedral organization of the viral envelope but did not reveal a well-defined, ordered organization of the nucleocapsid, and no details of C protein-RNA and C protein-envelope proteins interactions are known. Predictions that the flavivirus C protein has a large alpha-helical content (16) were recently confirmed by the structural analysis of the capsid proteins from yellow fever and dengue virus expressed in Escherichia coli (14). Attempts to generate capsid-like particles (CLPs) in vitro by using these recombinant C proteins have been unsuccessful (14).
So far, very little information is available on the characteristics of authentic flavivirus capsids and capsid proteins isolated from virions. We therefore conducted a study on the isolation and characterization of the capsid and the capsid protein from purified TBE virions. In this investigation we show that capsids can be isolated from virions under certain conditions, and although they have a very strong tendency to aggregate, the extent to which this occurs is dependent on the detergent used for membrane solubilization. Capsids-also in the form of insoluble precipitates-can be completely disintegrated into C protein dimers by increasing the salt concentration, suggesting that dimers are the basic building blocks for capsid formation. By using these dimers together with in vitro transcribed viral RNA or single-stranded DNA (ssDNA) oligonucleotides, CLPs could be assembled in vitro.
MATERIALS AND METHODS
Materials.
The following detergents were used for virus solubilization: Triton X-100 (TX-100; Merck); n-octyl-β-d-glucopyranoside (n-OG; Bachem); n-dodecyl-β-d-maltoside (DDM; Sigma); and 3-[(3-cholamidopropyl)dimethylammonio]-1-propane-sulfonate (CHAPS; Sigma).
The ΔNS5 cDNA clone is a derivative of the full-length infectious cDNA clone of TBE virus strain Neudoerfl with a deletion of 495 nucleotides. This deletion removed from the carboxy terminus approximately one fifth of the NS5 protein and essentially the entire variable region of the 3′ noncoding sequence (16). RNA transcribed from this clone is, therefore, not infectious.
For the in vitro assembly assay in the presence of ssDNA, we used two different, randomly chosen oligonucleotides: 36-mer (5′-ACGTGCTGAGCCTACTATCCGATGCTGCTCCCTTTT-3′) and 46-mer (5′-AGTCAGTCAGTCGCGGCCGCTACTATCCGATGCTGCTCCCTTTTTG-3′).
Virus growth and purification.
TBE virus strain Neudoerfl was grown in primary chicken embryo cells and purified by two cycles of sucrose density centrifugation as described previously (12).
Detergent treatment of virions.
Purified TBE virus at a final protein concentration of 100 μg/ml in TAN buffer (0.05 M triethanolamine [pH 8.0], 0.1 M NaCl) was mixed with detergent at the concentrations specified below and incubated for 1 h at 25°C. The following final concentrations were used for the different detergents: TX-100, 0.2%; n-OG, 1.4%; CHAPS, 0.8%; and DDM, 1%. After the 1-h incubation period, soluble and aggregated material was separated by centrifugation for 20 min at 14,000 rpm and 4°C (Eppendorf microcentrifuge). The aggregated material was resuspended in TAN buffer. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (20), the samples were precipitated by trichloroacetic acid as described previously (37) and redissolved in sample buffer by heating for 5 min at 95°C. Standard proteins (Pharmacia) were used as molecular mass markers, and staining was done with PhastGel Blue R (Pharmacia).
Sedimentation analysis. (i) Sedimentation analysis of detergent-solubilized virus.
Virus solubilized with TX-100 (0.2%) was subjected to rate zonal density gradient centrifugation by using 5 to 30% (wt/wt) continuous sucrose gradients made in TAN buffer containing 0.2% TX-100. As a control, untreated virus was applied to the same gradient in the absence of detergent.
(ii) Sedimentation analysis of in vitro-assembled CLPs.
The samples of the in vitro assembly reactions were applied to 5 to 30% (wt/wt) continuous sucrose gradients in TAN buffer.
The gradients were centrifuged for 90 min in an SW-40 rotor (Beckman) at 38,000 rpm and 4°C. Fractions were collected by upward displacement with an ISCO model 640 fraction collector. Aliquots of each fraction were precipitated by trichloroacetic acid (37) and analyzed by SDS-PAGE.
(iii) Sedimentation analysis of high-salt treated nucleocapsids.
The disintegrated nucleocapsids were applied to a 5 to 30% (wt/wt) continuous sucrose gradient made in 50 mM triethanolamine (pH 8.0) containing 1.5 M NaCl. The gradients were centrifuged for 38 h in an SW-40 rotor (Beckman) at 38,000 rpm and 4°C. The fractions were collected and treated as described above.
Disintegration of aggregated nucleocapsids.
Aggregated nucleocapsids obtained by centrifugation of detergent-treated virions were washed with 50 mM triethanolamine (pH 8.0) and resuspended at a protein concentration of 100 μg/ml in 50 μl of 50 mM triethanolamine (pH 8.0) containing 0, 0.5, 1.0, 1.5, or 2.0 M NaCl. The samples were incubated for 1 h at 25°C, and the soluble material was separated from aggregates by centrifugation for 20 min at 14,000 rpm and 4°C in an Eppendorf microcentrifuge. The material was precipitated with trichloroacetic acid (37) and analyzed by SDS-PAGE.
Size-exclusion chromatography.
Aggregated nucleocapsid particles (250 μg) obtained by detergent treatment of virions were treated with 500 μl of 0.05 M triethanolamine (pH 8.0) containing 1.5 M NaCl, 2% n-OG, and 5 μg of RNase A (Sigma) and incubated for 3 h at 4°C. Insoluble material was removed by centrifugation for 20 min (Eppendorf) at 14,000 rpm and 4°C. The clarified sample was loaded onto a Superdex 75 HR 10/30 column (Pharmacia) equilibrated with a buffer consisting of 50 mM triethanolamine (pH 8.0), 1.5 M NaCl, and 0.8% n-OG, which was also used as the running buffer. Fractions containing the C protein were pooled and stored at −80°C.
Chemical cross-linking.
The pooled C protein-containing fractions from the size exclusion chromatography were subjected to a chemical cross-linking reaction with dimethylsuberimidate (DMS; Pierce) as described in detail elsewhere (1). The cross-linked samples were separated by gel electrophoresis in 5% SDS-polyacrylamide gels by using a continuous phosphate-buffered system as described by Maizel (23).
In vitro assembly reaction. (i) Viral RNA.
A total of 7.5 μg of C protein dimers (final concentration, 10 μg/ml), purified by size exclusion chromatography, in 0.05 M triethanolamine (pH 8.0) containing 0.15 M NaCl and 0.08% n-OG was mixed with in vitro synthesized RNA from TBE virus cDNA clone ΔNS5 (see above) at a molar RNA-to-protein ratio of 1:300. In vitro RNA synthesis from the cDNA clone was performed with a commercially available system (Ambion) as described previously (25).
(ii) ssDNA oligonucleotides.
For the assembly reaction with ssDNA oligonucleotides, 22.5 μg of C protein dimers (final concentration, 10 μg/ml), purified by size exclusion chromatography, in 0.05 M triethanolamine (pH 8.0) containing 0.15 M NaCl and 0.08% n-OG was mixed with a 36-mer random oligonucleotide at a molar DNA-to-protein ratio of 1:1.1.
The samples were incubated at 37°C for 30 min, concentrated fivefold by membrane filtration (Vivascience) according to the manufacturer's instructions, and the formation of CLPs was analyzed by sucrose gradient sedimentation.
Ultrastructural analysis.
For negative staining, 200-mesh copper grids, coated with Pioloform film, stabilized by carbon evaporation, and freshly glow discharged, were floated for 2 min, face down, on the surface of approximately 10 μl of sample. After a short washing passage on 1 drop of water, the samples were stained on 1 drop of a 1% uranyl acetate solution in water for 2 min. After removal of the excess stain by blotting with filter paper, the grids were air dried (11). The negatively stained specimens were viewed in a Philips CM12 electron microscope (Eindhoven, The Netherlands), operating at 80 kV. The magnification was calibrated by using negatively stained catalase crystals.
RESULTS
Virus solubilization and isolation of capsids.
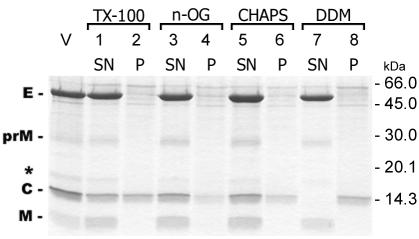
In order to isolate and characterize nucleocapsids from purified TBE virions, we investigated the suitability of four different detergents for solubilizing the viral membrane. These included TX-100, n-OG, CHAPS, and DDM (see Materials and Methods). The virus preparation (final protein concentration, 100 μg/ml) was mixed with the detergents at the indicated concentrations and allowed to stand for 1 h at 25°C. All of the samples were subjected to low-speed centrifugation for the removal of aggregated material, although a visible precipitate was formed only in the case of DDM. Aliquots of the supernatants as well as the pellets were analyzed by SDS-PAGE, and the results are shown in Fig. 1. With each of the detergents, at least a certain fraction of C protein devoid of envelope proteins was recovered from the pellet obtained by low-speed centrifugation, indicating that the protein was insoluble under these conditions. However, significant quantitative differences were observed between the different detergents. When DDM was used for solubilization, apparently all of the C protein was insoluble (lane 8), whereas an estimated 50, 80, and 70% remained soluble in the presence of TX-100, n-OG, and CHAPS, respectively.
FIG. 1.
SDS-PAGE of samples from TBE virus solubilized with TX-100 (lanes 1 and 2), n-OG (lanes 3 and 4), CHAPS (lanes 5 and 6), and DDM (lanes 7 and 8). Soluble and aggregated material was separated by low-speed centrifugation, and aliquots of the supernatant (SN) and the pellet (P) were analyzed by SDS-PAGE and stained with PhastGel Blue R. V, untreated virus control. The positions of the E, prM, C, and M proteins and molecular mass markers are labeled, and a band previously identified as an M dimer (37) is indicated by an asterisk.
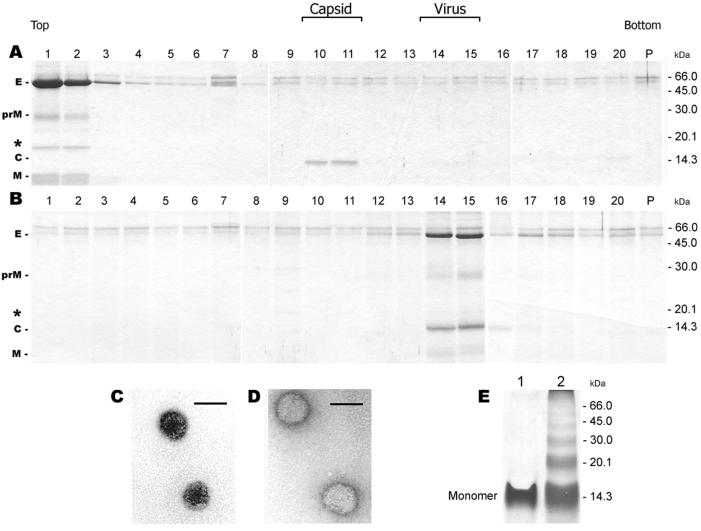
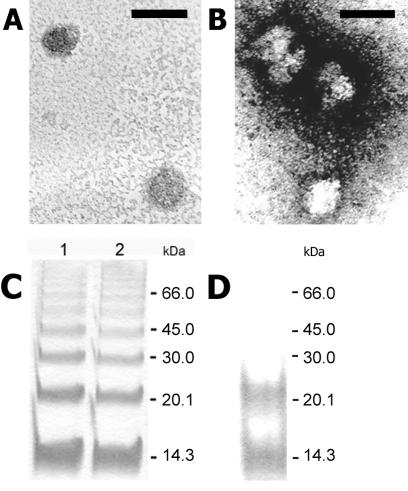
To define the physical state of the nonprecipitated forms of the C protein, the detergent-containing supernatants were subjected to sucrose density gradient ultracentrifugation in the presence of the corresponding detergents. In the case of n-OG and CHAPS (data not shown), the C protein was not detected in any of the gradient fractions. With TX-100, however, the C protein was found to be present in a particulate structure that sedimented to fractions 10 and 11, whereas the solubilized membrane proteins remained at the top of the gradient (Fig. 2A). Under the same centrifugation conditions but in the absence of detergent, the untreated control virus was recovered in fractions 14 and 15 (Fig. 2B). EM of the C protein-containing fractions from TX-100-treated samples (Fig. 2A, fractions 10 and 11) revealed spherical particles with a diameter of approximately 33 nm (Fig. 2C), whereas untreated whole virions had a diameter of 50 nm (Fig. 2D). The cross-linking of fractions 10 and 11 of the gradient shown in Fig. 2A with DMS followed by SDS-PAGE yielded a series of oligomeric C protein bands (Fig. 2E), consistent with the organization of the C protein in a particulate oligomeric complex. These experiments show that distinct nucleocapsids can be isolated from solubilized virions under appropriate conditions.
FIG. 2.
Sedimentation analysis of TX-100-solubilized (A) and untreated (B) TBE virus in sucrose gradients (5 to 30%, wt/wt). The sedimentation direction is from left to right. The proteins in each fraction were separated by SDS-PAGE and stained with PhastGel Blue R. In the case of solubilized TBE virus (panel A) the gradient also contained 0.2% TX-100 to prevent aggregation. The positions of the E, prM, C, and M proteins and molecular mass markers are labeled, and the M dimer band (37) is indicated by an asterisk. Electron micrographs of negatively stained, isolated nucleocapsids (C) and of TBE virus (D). Bar, 50 nm. (E) Cross-linking analysis of the pooled fractions 10 and 11 in Fig. 2A with the chemical cross-linker DMS. The samples were analyzed by SDS-PAGE and stained with PhastGel Blue R. Lane 1, no cross-linker; lane 2, cross-linker added. The position of the C monomer is indicated.
Isolation of a soluble form of the C protein from capsid aggregates.
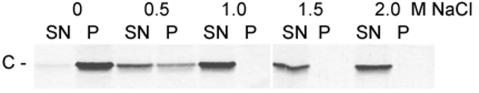
Although the use of DDM for virus solubilization did not allow the isolation of the capsid in a nonaggregated form, the insolubility and precipitation of the capsid under these conditions provided a fast and easy method for its quantitative and clean separation from the envelope proteins (Fig. 1, lanes 7 and 8). Using these aggregates as starting material, we attempted to generate a soluble form of the C protein by disintegrating the capsids by increasing the salt concentration. For this purpose, the precipitate was washed in TAN buffer and resuspended at a protein concentration of 100 μg/ml in 0.05 M triethanolamine (pH 8.0) containing different concentrations of NaCl. After an incubation of 1 h at 25°C, the samples were subjected to low-speed centrifugation for separating soluble from insoluble material, and aliquots of the supernatant and the pellets were analyzed by SDS-PAGE. As shown in Fig. 3, all of the C protein was recovered from the supernatant at NaCl concentrations of 1.0 M and higher.
FIG. 3.
SDS-PAGE of insoluble capsid aggregates treated with increasing concentrations of NaCl. Nucleocapsids aggregated with DDM were treated with 0, 0.5, 1.0, 1.5, and 2.0 M NaCl. Soluble and insoluble material was separated by low-speed centrifugation separated by SDS-PAGE, and stained with PhastGel Blue R. SN, supernatant; P, pellet.
The soluble form of the C protein is a dimer.
For determining the oligomeric state of the C protein obtained by NaCl-induced disintegration of the nucleocapsids, the material was subjected to size exclusion chromatography in the presence of high salt concentrations. Preliminary experiments had shown that in the absence of detergent, all of the C protein was nonspecifically retained on the column. This could be avoided by the use of n-OG, which was chosen because it is also a suitable detergent for crystallographic studies (8). This detergent was added to the running buffer in all subsequent experiments at a concentration of 0.8%. We also observed that at protein concentrations higher than 200 μg/ml, capsid disintegration was incomplete at 1.0 M NaCl. The NaCl concentration was therefore raised to 1.5 M, both in the buffer for capsid disintegration and in the running buffer for column chromatography.
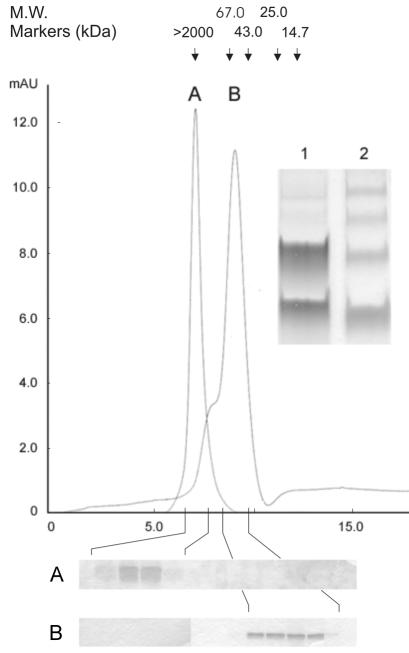
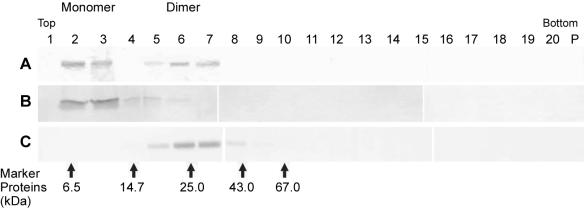
As revealed by the chromatogram shown in Fig. 4, about 75% of the C protein eluted in a major peak (peak B) at a position which, compared to the positions of standard proteins, would correspond to a molecular mass of 60 kDa. Cross-linking of the peak fractions with DMS and analysis by SDS PAGE, however, yielded almost exclusively C protein monomers and dimers (inset, lane 1), as expected for a dimeric molecule, rather than a ladder of higher oligomeric species (inset, lane 2), as was observed with a control sample of capsids isolated by TX-100 solubilization of the virions that eluted in the void volume (peak A). The dimeric nature of the C protein eluted from the column and the lack of higher oligomeric structures were also confirmed by sedimentation analysis in sucrose gradients. As shown in Fig. 5A, the column-purified material sedimented as two separate peaks. The position of the faster-sedimenting species corresponded to that expected for a C homodimer (∼25 kDa), based on a comparison to standard marker proteins. The smaller species sedimented identically to a monomer control made by heat treatment (Fig. 5B), suggesting that some of the C dimers had fallen apart during storage or centrifugation. Chemical cross-linking with DMS (1) of the eluted material before applying it to the gradient revealed an oligomeric structure that corresponded to the larger C protein oligomer in Fig. 5A. No higher-order oligomers were observed either with or without cross-linking (Fig. 5A and C). After weighing all of the data, we conclude that, under these conditions, the C protein is, in fact, a homodimer and that it exhibits an anomalous elution behavior on the size exclusion column, causing molecular mass estimates based on retention volume to be erroneous.
FIG. 4.
Size exclusion chromatography of DMS-cross-linked whole capsids isolated by TX-100 solubilization of virions (peak A) and of soluble C protein obtained by high-salt treatment of nucleocapsid aggregates (peak B). The running buffer contained 50 mM triethanolamine (pH 8.0), 1.5 M NaCl, and 0.8% n-OG. The absorbance of the eluate was monitored at 280 nm, and the arrows indicate the elution positions of molecular weight markers. The fractions in the peak region were analyzed by SDS-PAGE (panels below the chromatogram) and stained with PhastGel Blue R. (Inset) SDS-PAGE of aliquots of the pooled fractions of peak B (soluble protein) cross-linked with DMS (lane 1) and of peak A (lane 2, DMS-cross-linked whole capsids).
FIG. 5.
Sedimentation analysis of soluble C protein obtained by high-salt treatment of nucleocapsid aggregates in sucrose gradients (5 to 30%, wt/wt) containing 1.5 M NaCl. The sedimentation direction is from left to right. Proteins in each fraction were analyzed by SDS-PAGE and PhastGel Blue R staining. (A) Aliquot of the pooled peak fraction from the size exclusion column (Fig. 4). (B) Aliquot of the pooled peak fraction from the size exclusion column (Fig. 4) treated at 95°C for 5 min before loading onto the gradient. (C) Aliquot of the pooled peak fractions from the size exclusion column (Fig. 4) treated with DMS cross-linker before loading onto the gradient. The band corresponding to the cross-linked C dimer is shown. The arrows indicate the positions of marker proteins.
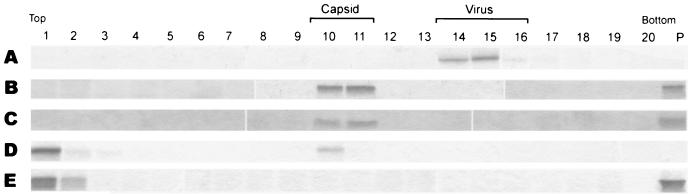
In vitro assembly of capsid-like structures. Using the purified C protein dimer (Fig. 4) and in vitro transcribed viral RNA as well as ssDNA oligonucleotides, we attempted to reconstitute CLPs in vitro under conditions comparable to those that had been applied for the in vitro assembly of alphavirus cores (38, 40). The conditions of the in vitro assembly reactions are described in detail in Materials and Methods. Particulate structures sedimenting to the same position as authentic cores isolated from virions by TX-100 solubilization (Fig. 6B) were obtained when ssDNA oligonucleotides were included in the assembly reaction (Fig. 6C), as well as when in vitro transcribed viral RNA was used (Fig. 6D). In the absence of any nucleic acid, however, no particles were observed (Fig. 6E). Particulate structures were also obtained with 46-mer ssDNA oligonucleotides (data not shown). In the case of in vitro transcribed viral RNA, a considerable amount of C protein (approximately 64%) was recovered from the top of the gradient, whereas in the case of ssDNA, all of the input material was found to sediment either to a position corresponding to monodisperse capsids (about 70%) or to the bottom of the gradient because of aggregation (about 30%). The apparent lower efficiency of particle formation in the presence of in vitro transcribed viral RNA compared to ssDNA could be due to RNA degradation by residual RNase A from the capsid purification procedure. EM of the structures assembled with in vitro transcribed viral RNA (Fig. 6D, fraction 10) revealed spherical particles with an average diameter of 32 nm (Fig. 7 A). Particles assembled in the presence of ssDNA (Fig. 6C) were heterogeneous in size and shape (Fig. 7B). Cross-linking of these materials with DMS followed by SDS-PAGE revealed the formation of a series of oligomeric C protein bands (Fig. 7C) similar to those formed after the cross-linking of capsids isolated from virions (Fig. 2E). The formation of higher oligomeric complexes did not occur in the absence of nucleic acid (Fig. 7D).
FIG. 6.
Sedimentation analysis of in vitro assembled capsid-like particles in 5 to 30% (wt/wt) sucrose gradients. Purified C protein dimers were assembled in vitro in the presence of in vitro transcribed viral RNA and ssDNA of an arbitrarily chosen sequence. The sedimentation direction is from left to right, and the fractions were analyzed by SDS-PAGE and PhastGel Blue R staining. (A) Untreated virus. (B) Nucleocapsids isolated with TX-100. (C) C protein dimers assembled with ssDNA. (D) C protein dimers assembled with in vitro transcribed viral RNA. (E) C protein dimers assembled without nucleic acid. In the case of solubilized TBE virus (panel B) the gradient also contained 0.2% TX-100.
FIG. 7.
Electron micrographs of negatively stained in vitro assembled CLPs. (A) Particles assembled in the presence of in vitro transcribed viral RNA. (B) Particles assembled in the presence of ssDNA 36-mer oligonucleotides. Bar, 50 nm. (C) DMS cross-linking and SDS-PAGE of CLPs. C protein dimers assembled in vitro in the presence of in vitro transcribed viral RNA (lane 1) and in the presence of ssDNA 36-mer oligonucleotides (lane 2) were analyzed by SDS-PAGE and stained with PhastGel Blue R. (D) DMS cross-linking and SDS-PAGE of C protein dimers treated under the same conditions as in panel C but in the absence of nucleic acid.
DISCUSSION
Recent structure determinations of mature and immature virions by cryo-EM and three-dimensional image reconstructions have revealed that flaviviruses possess an icosahedral envelope organization and contain a spherical nucleocapsid core with an outer radius of 15 nm (18, 29, 44, 45). The core is separated from the membrane by a gap of about 3 nm. With respect to their overall particle organization, flaviviruses thus exhibit several similarities with alphaviruses. However, in contrast to the alphaviruses, no evidence for icosahedral symmetry in the flavivirus capsid has been obtained so far.
Our attempts to isolate nucleocapsids from purified TBE virions in a monodisperse form after solubilization of the membrane with different detergents were unsuccessful with most of the detergents tested. Even in the presence of detergent, the viral capsids had a strong tendency to aggregate, and only when TX-100 was used, were about 50% of the particles shown to sediment in a monomeric nonaggregated form. The apparent hydrophobicity of the capsids is likely to be due to the surface exposure of specific sequence elements that have been implicated in hydrophobic interactions of the capsid protein with the membrane of the endoplasmic reticulum in the course of the intracellular assembly process (16, 17, 26). These structures could correspond to the density found in image reconstructions of immature virions (i.e., the primary product of virus assembly) that extends from the core and contacts the inner layer of the membrane (45). The observed hydrophobicity appears to be a significant feature that distinguishes the nucleocapsids of flaviviruses from those of alphaviruses, which can be easily isolated and purified in monodisperse form by membrane solubilization (3).
Although the tendency of TBE virus nucleocapsids to aggregate first appeared to be an experimental drawback, it turned out to form the basis for an easy method to separate the capsid from the envelope proteins. Disintegration of these aggregated capsids into a dimeric C protein subunit was achieved by increasing the concentration of NaCl, consistent with the assumption that flavivirus capsids are primarily stabilized by ionic interactions. Jones et al. (14) have recently characterized the C proteins of dengue and yellow fever viruses that were expressed as recombinant proteins in E. coli. Using cross-linking, analytical ultracentrifugation, and nuclear magnetic resonance studies, they provided evidence that at moderate concentrations these recombinant C proteins exist predominantly in a dimeric state. This finding is consistent with the assumption that the basic protein building block for the formation of flavivirus capsids is a C protein dimer. Both the cross-linking and sedimentation data in this study provide strong evidence that the C protein species isolated from native TBE virions is also a dimer in solution. However, size estimation by size-exclusion chromatography resulted in an apparent molecular mass corresponding to a hexameric or pentameric state. This inconsistent result is possibly explainable by interactions of the C protein with detergent micelles in the high-salt running buffer. The formation of a protein-micelle complex would, therefore, mimic a higher oligomeric state of the protein, resulting in an anomalous elution volume. The existence of unstable higher oligomers at that stage of the purification procedure is also conceivable, although unlikely.
It was previously hypothesized that interactions between alpha helices in the flavivirus C protein subunits could be responsible for oligomerization (16), as has been shown for the alpha-helical class of C proteins found in hepadnavirus and retroviruses (28, 42). This hypothesis was indeed confirmed recently by the elucidation by nuclear magnetic resonance study of the structure of the recombinant dengue virus C protein (22a). Also in the case of alphavirus cores, a C protein dimer containing one monomer from each of two adjacent capsomers is believed to be the initial intermediate in the nucleocapsid assembly mechanism (39). In contrast to flaviviruses, however, the alphavirus C protein is monomeric in solution, and dimerization depends on the interaction with nucleic acid (39).
The in vitro assembly of alphavirus core-like particles has been shown both with C proteins isolated from virions as well as with recombinant proteins in combination with different single-stranded nucleic acids and even with nonnucleic acid polyanionic substances (38, 40). Similar attempts at in vitro assembly of flavivirus CLPs by using C proteins from dengue and yellow fever virus expressed in E. coli have proven unsuccessful so far (14). However, using a dimeric C protein isolated from TBE virions and in vitro transcribed viral RNA as well as ssDNA oligonucleotides, we were able to assemble particulate structures in vitro that exhibited a sedimentation behavior in sucrose gradients similar to that of capsids isolated from virions. At present, it is not clear whether the different results obtained relate to intrinsic differences between the TBE, the dengue, and yellow fever C proteins, the source of the proteins (recombinant versus isolated from virions), or details of the experimental procedures used. In contrast to alphaviruses, in which the icosahedral arrangement of the viral envelope proteins is coordinated with that of the internal core structure (4, 24), the flavivirus core appears to be partially disordered and, unlike the viral envelope, does not exhibit an icosahedral symmetry (18, 29, 44, 45). The absence of a distinct capsid protein density together with the unusually low nucleocapsid density in cryo-EM reconstructions of immature virions have led to the speculation that the size of the assembled virus might be determined by the size of the genomic RNA (45). As we have shown, CLPs with similar sedimentation behavior could be generated not only with in vitro transcribed viral RNA but also with short ssDNA oligonucleotides. The latter, however, appeared to be less well defined and less homogeneous. Despite the demonstration of in vitro CLP formation with C protein dimers and nucleic acids only, it has to be kept in mind that flavivirus assembly in vivo does not proceed via preformed capsids and arrays of envelope proteins at the site of maturation, as is the case with alphaviruses, but apparently instead as a complex process involving membrane-associated precursor proteins and finely tuned proteolytic cleavage events (21).
Acknowledgments
We thank Angela Dohnal and Walter Holzer for technical assistance and Karin Stiasny and Steven Allison for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg, S. M., A. Nestorowicz, D. W. McCourt, and C. M. Rice. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 68:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boege, U., G. Wengler, G. Wengler, and B. Wittmann-Liebold. 1980. Partial amino acid sequences of Sindbis and Semliki Forest virus-specific core proteins. Virology 103:178-190. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H. K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deubel, V., J. P. Digoutte, X. Mattei, and D. Pandare. 1981. Morphogenesis of yellow fever virus in Aedes aegypti cultured cells. II. An ultrastructural study. Am. J. Trop. Med. Hyg. 30:1071-1077. [DOI] [PubMed] [Google Scholar]
- 6.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183-191. [DOI] [PubMed] [Google Scholar]
- 7.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 8.Garavito, R. M., D. Picot, and P. J. Loll. 1996. Strategies for crystallizing membrane proteins. J. Bioenerg. Biomembr. 28:13-27. [PubMed] [Google Scholar]
- 9.Guirakhoo, F., R. A. Bolin, and J. T. Roehrig. 1992. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191:921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hase, T., P. L. Summers, K. H. Eckels, and W. B. Baze. 1987. Maturation process of Japanese encephalitis virus in cultured mosquito cells in vitro and mouse brain cells in vivo. Arch. Virol. 96:135-151. [DOI] [PubMed] [Google Scholar]
- 11.Hayat, M. A., and S. E. Miller. 1990. Negative staining. McGraw-Hill, New York, N.Y.
- 12.Heinz, F. X., and C. Kunz. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57:263-274. [DOI] [PubMed] [Google Scholar]
- 13.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 14.Jones, C. T., L. Ma, J. W. Burgner, T. D. Groesch, C. B. Post, and R. J. Kuhn. 2003. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 77:7143-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko, K. K., A. Igarashi, and K. Fukai. 1979. Electron microscopic observations on Aedes albopictus cells infected with dengue viruses. Arch. Virol. 62:41-52. [DOI] [PubMed] [Google Scholar]
- 16.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kofler, R. M., A. Leitner, G. O'Riordain, F. X. Heinz, and C. W. Mandl. 2003. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 77:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus. Implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 21.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields Virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 22.Lobigs, M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc. Natl. Acad. Sci. USA 90:6218-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Ma, L., C. T. Jones, T. D. Groesch, R. J. Kuhn, and C. B. Post. 2004. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA 101:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maizel, J. V., Jr. 1971. Polyacrylamide gel electrophoresis of viral proteins. Methods Virol. 5:179-246. [Google Scholar]
- 24.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 25.Mandl, C. W., M. Ecker, H. Holzmann, C. Kunz, and F. X. Heinz. 1997. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 78:1049-1057. [DOI] [PubMed] [Google Scholar]
- 26.Markoff, L., B. Falgout, and A. Chang. 1997. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233:105-117. [DOI] [PubMed] [Google Scholar]
- 27.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 30.Ng, M. L. 1987. Ultrastructural studies of Kunjin virus-infected Aedes albopictus cells. J. Gen. Virol. 68:577-582. [DOI] [PubMed] [Google Scholar]
- 31.Nowak, T., P. M. Farber, G. Wengler, and G. Wengler. 1989. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology 169:365-376. [DOI] [PubMed] [Google Scholar]
- 32.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174:450-458. [DOI] [PubMed] [Google Scholar]
- 33.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 34.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 35.Sriurairatna, S., and N. Bhamarapravati. 1977. Replication of dengue-2 virus in Aedes albopictus mosquitoes. An electron microscopic study. Am. J. Trop. Med. Hyg. 26:1199-1205. [DOI] [PubMed] [Google Scholar]
- 36.Sriurairatna, S., N. Bhamarapravati, and O. Phalavadhtana. 1973. Dengue virus infection of mice: morphology and morphogenesis of dengue type-2 virus in suckling mouse neurones. Infect. Immun. 8:1017-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wengler, G., U. Boege, G. Wengler, H. Bischoff, and K. Wahn. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 118:401-410. [DOI] [PubMed] [Google Scholar]
- 41.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 43.Yamshchikov, V. F., and R. W. Compans. 1993. Regulation of the late events in flavivirus protein processing and maturation. Virology 192:38-51. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]