Contact-dependent growth inhibition (CDI) is an important mechanism of intercellular competition between Gram-negative bacteria. The X-ray crystal structure of an immunity protein from N. meningitidis MC58 is presented together with a structural model of the toxin–immunity protein complex.
Keywords: contact-dependent growth inhibition, CdiA-CT toxin domain, CdiI immunity protein, toxin–immunity protein complex, Neisseria meningitidis, docking studies
Abstract
Contact-dependent growth inhibition (CDI) is an important mechanism of intercellular competition between neighboring Gram-negative bacteria. CDI systems encode large surface-exposed CdiA effector proteins that carry a variety of C-terminal toxin domains (CdiA-CTs). All CDI+ bacteria also produce CdiI immunity proteins that specifically bind to the cognate CdiA-CT and neutralize its toxin activity to prevent auto-inhibition. Here, the X-ray crystal structure of a CdiI immunity protein from Neisseria meningitidis MC58 is presented at 1.45 Å resolution. The CdiI protein has structural homology to the Whirly family of RNA-binding proteins, but appears to lack the characteristic nucleic acid-binding motif of this family. Sequence homology suggests that the cognate CdiA-CT is related to the eukaryotic EndoU family of RNA-processing enzymes. A homology model is presented of the CdiA-CT based on the structure of the XendoU nuclease from Xenopus laevis. Molecular-docking simulations predict that the CdiA-CT toxin active site is occluded upon binding to the CdiI immunity protein. Together, these observations suggest that the immunity protein neutralizes toxin activity by preventing access to RNA substrates.
1. Introduction
Bacteria have developed several complex mechanisms to interact and communicate with neighboring microbes in the environment. One such mechanism is contact-dependent growth inhibition (CDI), a form of interbacterial competition found in several important human pathogens including uropathogenic Escherichia coli, Burkholderia pseudomallei and Neisseria meningitidis (Aoki et al., 2011 ▶). CDI is mediated by the CdiB/CdiA family of two-partner secretion proteins. CdiB is an outer membrane β-barrel protein that exports and displays the CdiA effector protein on the surface of CDI+ inhibitor cells (Aoki et al., 2005 ▶). CdiA proteins are very large, ranging from 180 kDa to over 600 kDa depending on the bacterial species, and are characterized by hemagglutinin-peptide repeats that suggest a filamentous structure (Kajava et al., 2001 ▶). CdiA proteins are predicted to extend several hundred angstroms from the surface of inhibitor cells to interact with specific receptors on the surface of susceptible target bacteria. Upon contact with its receptor, CdiA delivers a toxin domain derived from its extreme C-terminus (CdiA-CT) into the target bacterium (Aoki et al., 2010 ▶; Ruhe et al., 2014 ▶). CdiA-CT toxins vary considerably between bacteria and even between different strains of the same species (Aoki et al., 2010 ▶). This sequence diversity corresponds to a variety of toxin activities ranging from the formation of membrane pores to the degradation of ribosomal RNA (Ruhe et al., 2013 ▶). CDI+ bacteria protect themselves from auto-inhibition by expressing CdiI immunity proteins, which bind to the CdiA-CT domain and neutralize its toxin activity. CdiI immunity proteins are specific for their cognate CdiA-CT and do not protect cells from the toxins of other CDI+ bacteria. Thus, CDI systems encode a complex network of toxin–immunity protein pairs that are deployed for intercellular competition.
N. meningitidis is a parasitic, aerobic, Gram-negative bacterium responsible for pyogenic meningitis and meningococcal septicemia. It is a major cause of disease worldwide, resulting in hearing loss, brain damage and death in 4–10% of sufferers (Thigpen et al., 2011 ▶; Nikulin et al., 2006 ▶). Every year, approximately 3000–4000 cases of N. meningitidis-linked meningitis are reported in the United States (Thigpen et al., 2011 ▶). Because this pathogen poses a serious threat to global health, a greater understanding of its growth control could be leveraged to develop novel therapeutics targeted specifically to Neisseria. All N. meningitidis isolates carry at least one CDI system, and some strains have multiple complex loci that contain two cdiA genes and tandem arrays of ‘orphan’ cdiA-CT/cdiI gene pairs (Bentley et al., 2007 ▶; Poole et al., 2011 ▶). Orphan cdiA-CT gene fragments often share significant regions of homology with the upstream cdiA gene and therefore can undergo homologous recombination to fuse the orphan cdiA-CT/cdiI module onto cdiA. This process can abruptly change the toxin deployed by the cell (Koskiniemi et al., 2014 ▶). The large number of CDI-associated toxin/immunity genes carried by N. meningitidis suggests that these systems mediate interstrain competition. This hypothesis is supported by a recent study by Tommassen and coworkers (Arenas et al., 2013 ▶). Here, we report the crystal structure of CdiIo2 MC58-1, an orphan CDI immunity protein from N. meningitidis MC58. In addition, we have generated structural models for the cognate CdiA-CTo2 MC58-1 toxin and its corresponding toxin–immunity protein complex.
2. Materials and methods
2.1. Cloning of the N. meningitidis CdiA-CTo2 MC58-1/CdiIo2 MC58-1 genes
A fragment containing NMB0502 and NBM0503 (encoding CdiA-CTo2 MC58-1 and CdiIo2 MC58-1, respectively) was amplified from N. meningitidis MC58 genomic DNA using 5′ -GTC TCT CCC ATG GTG AAA AAT AAT CAG CTT AGC GAC AAA GAG as the forward primer and 5′ -TGG TGG TGC CCA GCG GTT TCA TGC AGG CTA CAG TTT GTT TGA as the reverse primer. The gel-purified PCR product was treated with phage T4 DNA polymerase and dTTP as described previously (Eschenfeldt et al., 2010 ▶) and ligated to plasmid pMCSG58, which appends a noncleavable His6 tag to the C-terminus of CdiIo2 MC58-1 (Eschenfeldt et al., 2013 ▶). The identity of the cloned insert was confirmed by DNA sequencing.
2.2. Expression and purification of N. meningitidis CdiIo2 MC58-1
The construct was introduced into E. coli BL21 (DE3) cells for overexpression and protein purification. The cells were grown at 37°C in LB medium supplemented with 100 µg ml−1 ampicillin. After the cells had grown to an optical density at 600 nm of ∼0.6, the culture was cooled to 18°C and protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight. Under these growth conditions, only the CdiIo2 MC58-1 immunity protein was overproduced. The cells were harvested by centrifugation, resuspended in 50 mM Tris pH 8.0, 500 mM NaCl, 10 mM β-mercaptoethanol (BME), 10% glycerol and lysed with Fast Break reagent (Promega) containing 10 µg ml−1 lysozyme and protease-inhibitor cocktail (Roche). The cell lysate was centrifuged at 10 000 rev min−1 for 1 h and the supernatant was passed through a 0.22 µm filter. The clarified lysate was then loaded onto an Ni2+-Sepharose HisTrap column (GE Healthcare) and proteins were eluted with a 20–250 mM linear gradient of imidazole in resuspension buffer. Fractions were pooled and loaded onto a HiLoad 26/60 Superdex 75 size-exclusion column equilibrated with 20 mM Tris pH 7.5, 150 mM NaCl, 2 mM dithiothreitol. Fractions containing purified CdiIo2 MC58-1 immunity protein were pooled and concentrated for crystallization using an Amicon Ultra centrifugal filter device with a 3000 Da cutoff (Millipore).
2.3. Size-exclusion chromatography
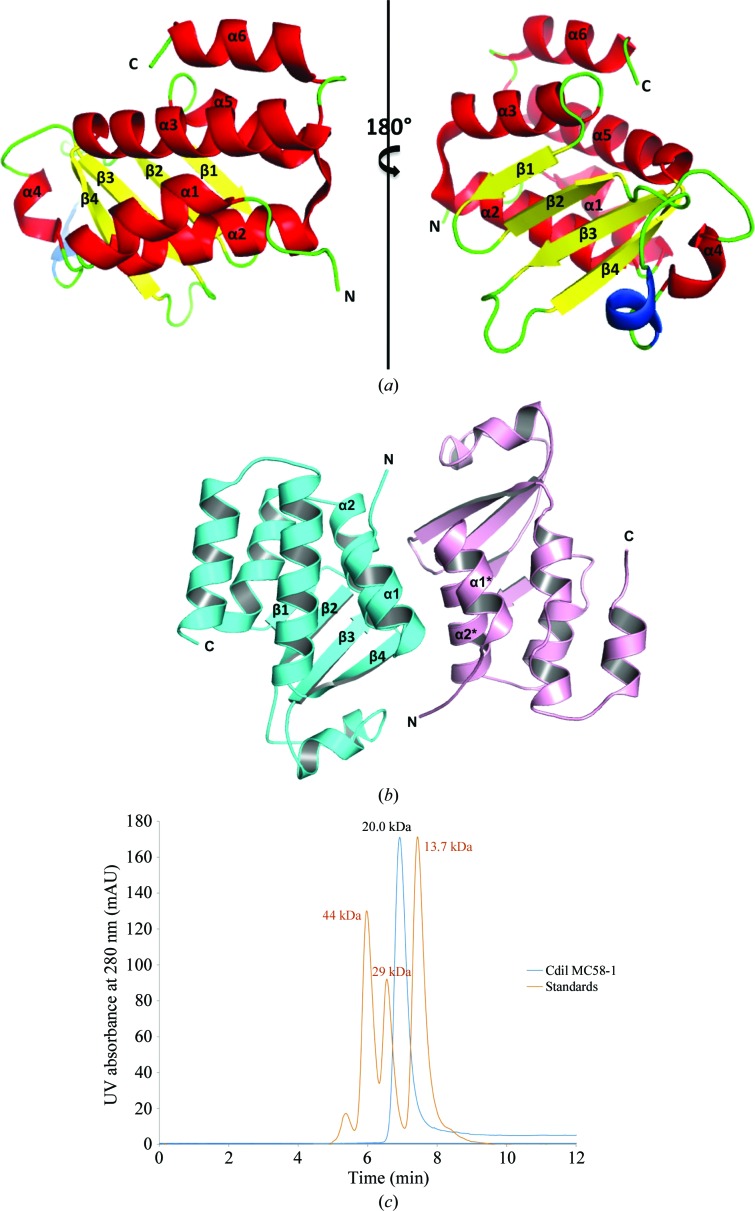
Analysis of the purified CdiIo2 MC58-1 was performed using a Dionex HPLC system with an analytical size-exclusion column from Sepax (SRT-SEC-150, Sepax Technologies). CdiIo2 MC58-1 was diluted to 5 mg ml−1 in standard running buffer (20 mM Tris pH 7.8, 150 mM NaCl, 2 mM dithiothreitol). The sample-injection volume was 20 µl and the flow rate of the analysis was 1.0 ml min−1. CdiIo2 MC58-1 was run in duplicate. Each run took approximately 15 min. The molecular-weight determination of CdiIo2 MC58-1 was calculated using linear regression data analysis with ovalbumin (44 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa) as migration standards.
2.4. Crystallization of the CdiIo2 MC58-1 immunity protein
Native CdiIo2 MC58-1 crystals were grown at 4°C using sitting drops that consisted of 10 mg ml−1 protein in 0.2 M MgCl2, 0.1 M bis-tris pH 5.5, 20% PEG 3350. Bromide derivatives were prepared by dipping crystals into a solution of 1.0 M KBr, 0.2 M MgCl2, 0.1 M bis-tris pH 5.5, 20% PEG 3350, 15% glycerol for approximately 10 s. Bromide-derivatized crystals were subsequently cryocooled in liquid nitrogen and used to collect X-ray diffraction data for phase determination (Dauter et al., 2000 ▶).
2.5. X-ray data collection, structure determination and refinement
A set of single-wavelength anomalous diffraction (SAD) data was collected near the bromine absorption peak (12.40 keV) at 100 K from one CdiIo2 MC58-1 crystal. Data were obtained on the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory using the SBCcollect program (Rosenbaum et al., 2006 ▶). Data-set intensities were integrated, scaled and merged using the HKL-3000 program suite (Minor et al., 2006 ▶; Table 1 ▶). From the Matthews correlation coefficient, two CdiIo2 MC58-1 molecules were predicted in one asymmetric unit. 17 Br sites were located using SHELXD (Sheldrick, 2008 ▶) and were used for phasing with MLPHARE from CCP4 (Winn et al., 2011 ▶). After density modification, a partial model of 138 residues (46% of a dimer) without side chains was built in three cycles of ARP/wARP model building (Cohen et al., 2004 ▶). All of the abovementioned programs are integrated within the HKL-3000 suite (Minor et al., 2006 ▶). The final CdiIo2 MC58-1 model was completed manually using Coot (Emsley & Cowtan, 2004 ▶) and was refined with phenix.refine (Afonine et al., 2012 ▶) (Table 1 ▶).
Table 1. Data-collection and crystallographic statistics for CdiIo2 MC58-1 .
Values in parentheses are for the last resolution bin.
| Data collection | |
| Space group | P21 |
| Unit-cell parameters (, ) | a = 45.38, b = 53.53, c = 59.70, = 98.04 |
| Molecular weight† (Da) | 16727 |
| No. of residues† | 143 |
| Molecules in asymmetric unit | 2 |
| Wavelength () | 0.9193 [Br peak] |
| Resolution () | 30.01.45 (1.481.45) |
| No. of unique reflections | 49973‡ |
| Multiplicity | 3.4 (2.2) |
| Completeness (%) | 99.5 (94.9) |
| R merge (%) | 10.7 (38.4) |
| I/(I) | 24.4 (2.3) |
| Solvent content (%) | 41.7 |
| Phasing | |
| R Cullis (anomalous) (%) | 89 |
| Figure of merit (%) | 19.4§ |
| Refinement | |
| Resolution () | 30.01.45 |
| No. of reflections (work/test) | 47390/2544 |
| R cryst/R free (%) | 14.9/18.9 |
| R.m.s. deviations from ideal geometry | |
| Bond lengths () | 0.005 |
| Bond angles () | 0.967 |
| No. of atoms | |
| Protein | 2229 |
| Heteroatoms | 300 |
| Mean B value (2) | |
| Main chain | 11.25 |
| Side chain | 14.24 |
| Ramachandran plot statistics, residues in (%) | |
| Most favored regions | 93.0 |
| Additional allowed region | 7.0 |
| Generously allowed regions | 0 |
| Disallowed region | 0 |
| PDB code | 4q7o |
Not including a three-residue N-terminal tag, SNA.
Including Bijvoet pairs.
Before density modification.
2.6. Expression and purification of the N. meningitidis CdiA-CTo2 MC58-1–CdiIo2 MC58-1 complex
The construct from §2.1 was introduced into E. coli BL21 (DE3) cells and grown at 37°C in LB medium supplemented with 100 µg ml−1 ampicillin. After the cells had grown to an optical density at 600 nm of ∼0.8, protein expression was induced with 1.5 mM IPTG for 2.5 h at 37°C. The cells were harvested by centrifugation and resuspended in 20 mM Tris pH 8.0, 150 mM NaCl, 10 mM BME, 1 mM phenylmethylsulfonyl fluoride, 10 µg ml−1 lysozyme. The cells were lysed using a microfluidizer and centrifuged at 10 000 rev min−1 for 1 h and the supernatant was passed through a 0.22 µm filter. The clarified lysate was then loaded onto Ni2+–NTA resin (GE Healthcare) and nonspecifically bound proteins were eluted with resuspension buffer with no BME and 20 mM imidazole under gravity. The imidazole concentration was increased to 250 mM to elute the toxin–immunity protein complex.
2.7. Docking of predicted toxin and immunity proteins
A model of the CdiA-CTo2 MC58-1–CdiIo2 MC58-1 binding interaction was generated through docking simulations. A computational model of the CdiA-CTo2 MC58-1 structure was generated with Sculptor (Birmanns et al., 2011 ▶) using the three-dimensional structure of the homologous XendoU nuclease (PDB entry 2c1w; Renzi et al., 2006) as a guide. The CdiA-CTo2 MC58-1 sequence was fitted into the XendoU structure while maintaining the overall fold and alignment of the predicted active-site histidine residues.
Hex 8.0 (Macindoe et al., 2010 ▶) was used to dock the CdiIo2 MC58-1 immunity protein onto the Sculptor-modeled CdiA-CTo2 MC58-1 structure. The proteins were oriented and the origins were set to allow free rotation of the two molecules during the search for low-energy binding interactions based on complementary shape and electrostatics. Energies for each model were calculated by adding all intermolecular interactions after a round of molecular-mechanics energy minimization. Typical Hex simulations produce binding energies of −600 to −1000 kcal mol−1 (Gupta et al., 2013 ▶). A control docking simulation using CdiA-CTEC536 toxin and CysK, which are known to interact (Diner et al., 2012 ▶), generated a low energy of interaction of −988 kcal mol−1. Simulations for proteins that do not interact (E. coli CysK and B. pseudomallei E479 CdiI; Nikolakakis et al., 2012 ▶) yielded a much higher energy of −368 kcal mol−1.
3. Results and discussion
3.1. Overall structure of CdiIo2 MC58-1
CdiIo2 MC58-1 crystallized in space group P21 with two molecules in the asymmetric unit. The structure was determined using a bromide derivative and SAD phasing, and the final model was refined to a resolution of 1.45 Å (Table 1 ▶).
The CdiIo2 MC58-1 structure represents an α/β-fold comprising a four-stranded antiparallel β-sheet, against which a five-helix bundle is packed (Fig. 1 ▶ a). The helical bundle includes three helices (α1–α3) from the N-terminus and two (α5 and α6) from the C-terminus. Additionally, there are two consecutive helices with one short 310-helix (colored blue in Fig. 1 ▶ a) followed by another short α-helix (α4) within a large loop that connects strands β2 and β3. Helix α4 is located on the edge of the β-sheet on the face opposite to the helical bundle and close to the C-terminus. With the exception of the surface interacting with the large loop, the remainder of one face of the β-sheet is completely exposed to solvent.
Figure 1.
(a) Ribbon cartoon of the CdiIo2 MC58-1 structure with α-helices, β-strands and the 310-helix colored red, yellow and blue, respectively. (b) The interface produced by helices α1 and α2 from each monomer in the asymmetric unit. (c) Analytical size-exclusion chromatography using an SRT-SEC-150 column suggests that CdiIo2 MC58-1 (blue trace; standards, orange trace) is monomeric in solution.
Within the asymmetric unit, CdiIo2 MC58-1 appears to form a nearly perfect noncrystallographic twofold-symmetric dimer, with helices α1 and α2 from each monomer packed against each other in an antiparallel mode (Fig. 1 ▶ b). This type of helical bundle is a common structural motif at protein–protein interfaces (Norel et al., 1995 ▶). The buried surface area owing to dimerization is about 1065 Å2 per monomer as determined by PDBePISA (Krissinel & Henrick, 2007 ▶). To test whether CdiIo2 MC58-1 is dimeric in solution, we analyzed the immunity protein by analytical size-exclusion chromatography and found it to be predominately monomeric (Fig. 1 ▶ c). Thus, the dimeric assembly observed in the crystal structure is perhaps an artifact of crystallization.
3.2. Structural comparison
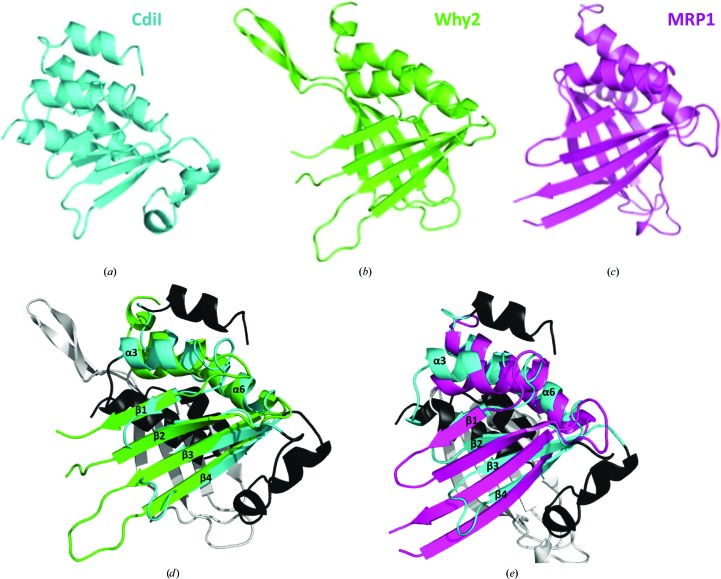
CdiIo2 MC58-1 has moderate structural similarity to two eukaryotic nucleic acid-binding proteins. The closest structural homolog, as determined using the DALI server (Holm et al., 2008 ▶), is a mitochondrial Whirly protein (Why2; PDB entry 4kop) from Arabidopsis thaliana (Cappadocia et al., 2013 ▶), and CdiIo2 MC58-1 superimposes upon Why2 with an r.m.s.d. of 2.5 Å over 80 of 145 Cα atoms (Figs. 2 ▶ a, 2 ▶ b and 2 ▶ d). Whirly family members are single-stranded DNA-binding proteins that modulate DNA repair in plant chloroplasts (Cappadocia et al., 2010 ▶; Desveaux et al., 2005 ▶). The next closest structural homolog is mitochondrial RNA-binding protein 1 (MRP1; PDB entry 2gia) from Trypanosoma brucei (Schumacher et al., 2006 ▶), and MRP1 superimposes onto CdiIo2 MC58-1 with an r.m.s.d. of 3.0 Å over 85 of 132 Cα atoms (Figs. 2 ▶ c and 2 ▶ e). MRP1 forms a heterotetramer with MRP2, and together the two proteins function in RNA editing by promoting the hybridization to guide RNAs to their target mRNAs (Aphasizhev et al., 2003 ▶). Although the top structural homologs are nucleic acid-binding proteins, CdiIo2 MC58-1 lacks the structural elements used by these proteins to bind DNA or RNA. These observations suggest that CdiIo2 MC58-1 is unlikely to bind nucleic acids.
Figure 2.
CdiIo2 MC58-1 structural homologs in ribbon representation. (a) CdiIo2 MC58-1 (PDB entry 4q7o), (b) Why2 (PDB entry 4kop) and (c) MRP1 (PDB entry 2gia). (d) Superimposition of CdiIo2 MC58-1 and Why2. Secondary-structure elements of CdiIo2 MC58-1 and Why2 that superimpose with good agreement are colored cyan and green, respectively. Structural elements that do not superimpose are colored black and white for CdiIo2 MC58-1 and Why2, respectively. (e) Superimposition of CdiIo2 MC58-1 and MRP1. Secondary-structure elements of CdiIo2 MC58-1 and MRP1 that superimpose with good agreement are colored cyan and pink, respectively. Structural elements that do not superimpose are colored black and white for CdiIo2 MC58-1 and MRP1, respectively.
3.3. Predicted function of the CdiA-CTo2 MC58-1 toxin
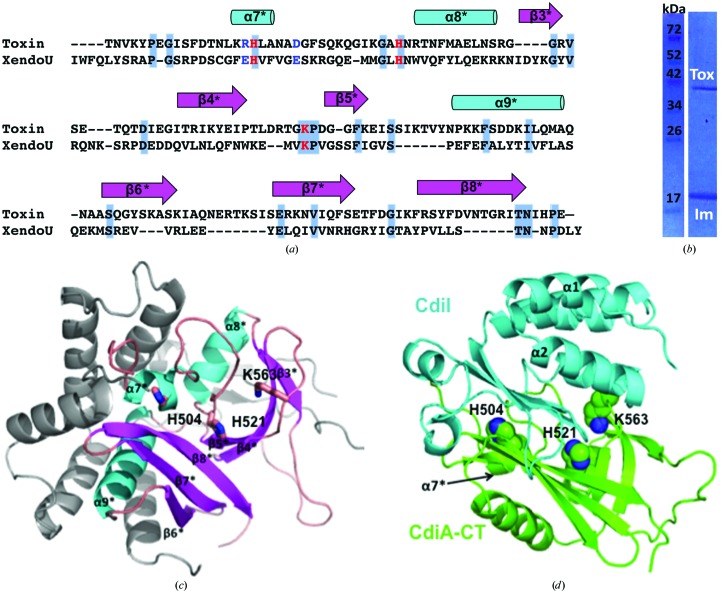
No experimental or structural information is available for CdiA-CTo2 MC58-1, which is the predicted toxin encoded by the adjacent NMB0502 gene. Aravind and coworkers have predicted that the C-terminal domain of CdiA-CTo2 MC58-1 is related to the EndoU nucleases (Zhang et al., 2012 ▶), which comprise a superfamily of Mn2+-dependent RNA-processing enzymes found mostly in eukaryotes, although family members are also found in the cyanobacterium Nostoc punctiforme (Renzi et al., 2006 ▶) and at the C-terminus of a MafB RNase toxin from N. meningitidis (Jamet et al., 2015 ▶). CdiA-CTo2 MC58-1 has diverged substantially from the eukaryotic enzymes and shares only 16% sequence identity with the C-terminal nuclease domain (residues Ile142–Tyr292) of XendoU, a poly(U)-specific endoribonuclease from Xenopus laevis (Fig. 3 ▶ a; Laneve et al., 2003 ▶). The crystal structure of XendoU has been solved (Renzi et al., 2006 ▶), and therefore we used it as a guide to generate a model of CdiA-CTo2 MC58-1 (Fig. 3 ▶ c). The resulting model shows that the XendoU active-site residues His162, His178 and Lys224 correspond to His504, His521 and Lys563 in CdiA-CTo2 MC58-1 (Renzi et al., 2006 ▶; Gioia et al., 2005 ▶). Together, these observations suggest that CdiA-CTo2 MC58-1 may possess a similar Mn2+-dependent RNA-processing/degrading activity to other members of the EndoU family.
Figure 3.
Modeled structures of CdiA-CTo2 MC58-1 and its complex with CdiIo2 MC58-1. (a) Sequence alignment by ClustalW of the C-terminal region of CdiA-CTo2 MC58-1 with the C-terminal domain of XendoU (PDB entry 2c1w). Residues on a gray/blue background are conserved, catalytic residues are colored red and RNA-binding residues are colored blue. (b) SDS–PAGE of the Ni2+-affinity purified complex of untagged CdiA-CTo2 MC58-1 (Tox) with His6-tagged CdiIo2 MC58-1 (Im). (c) Modeled structure of the C-terminal domain of CdiA-CTo2 MC58-1 with α-helices colored cyan and β-strands colored salmon. Elements in light gray are those of the N-terminal domain of the XendoU structure that have no sequence homology to CdiA-CTo2 MC58-1. The predicted active-site residues of CdiA-CTo2 MC58-1 are shown in stick representation with C, O and N atoms colored pink, red and blue, respectively. (d) The docked complex structure in ribbon representation of the toxin (colored green, secondary-structure elements indicated with asterisks) with the immunity protein (colored cyan). The predicted CdiA-CTo2 MC58-1 active-site residues, shown in sphere representation with C and N atoms colored green and blue, respectively, are occluded from solvent by the docked immunity protein CdiIo2 MC58-1.
3.4. Modeling of the CdiA-CTo2 MC58-1–CdiIo2 MC58-1 complex
To gain insight into the toxin–immunity protein binding interactions, we first tested whether CdiA-CTo2 MC58-1 forms a complex with CdiIo2 MC58-1. We co-expressed the toxin with His6-tagged CdiIo2 MC58-1 (as described in §2.6) and then purified the immunity protein by Ni2+-affinity chromatography. The untagged toxin co-eluted with His6-tagged CdiIo2 MC58-1 (Fig. 3 ▶ b), indicating that CdiA-CTo2 MC58-1 and CdiIo2 MC58-1 do indeed form a complex. We then conducted docking simulations of the monomeric CdiIo2 MC58-1 immunity protein structure onto the CdiA-CTo2 MC58-1 model. The lowest interaction energy obtained from these simulations was −776.7 kcal mol−1, which is considerably lower than the energy calculated for non-interacting proteins as described in §2. The Hex-generated model predicts that CdiIo2 MC58-1 binds directly over the active site of CdiA-CTo2 MC58-1 (Fig. 3 ▶ d), likely neutralizing the toxin by preventing access to RNA substrates.
4. Conclusions
We have elucidated the structure of CdiIo2 MC58-1, a predicted immunity protein encoded within the CDI-1 locus of N. meningitidis MC58. CdiIo2 MC58-1 has moderate structural homology to Whirly-like proteins found in plastids, but appears to lack the characteristic Whirly RNA-binding site. In addition, we modeled the structure of the associated CdiA-CTo2 MC58-1 toxin domain, which is proposed to have a similar active-site motif and RNA-processing activity as eukaryotic EndoU nucleases. Molecular-docking simulations predict that CdiIo2 MC58-1 occludes the active site of CdiA-CTo2 MC58-1 in the toxin–immunity protein complex. Experiments to test the biochemical activity of CdiA-CTo2 MC58-1 and its proposed active-site residues are under way.
Supplementary Material
PDB reference: CdiI immunity protein, 4q7o
Acknowledgments
This research was supported by National Institutes of Health grants GM102318 (to CWG, CSH and subcontract to Argonne) and GM094585 (to AJ). The use of SBC beamlines was supported by the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
References
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Aoki, S. K., Diner, E. J., t’Kint de Roodenbeke, C., Burgess, B. R., Poole, S. J., Braaten, B. A., Jones, A. M., Webb, J. S., Hayes, C. S., Cotter, P. A. & Low, D. A. (2010). Nature (London), 468, 439–442. [DOI] [PMC free article] [PubMed]
- Aoki, S. K., Pamma, R., Hernday, A. D., Bickham, J. E., Braaten, B. A. & Low, D. A. (2005). Science, 309, 1245–1248. [DOI] [PubMed]
- Aoki, S. K., Poole, S. J., Hayes, C. S. & Low, D. A. (2011). Virulence, 2, 356–359. [DOI] [PMC free article] [PubMed]
- Aphasizhev, R., Aphasizheva, I., Nelson, R. E. & Simpson, L. (2003). RNA, 9, 62–76. [DOI] [PMC free article] [PubMed]
- Arenas, J., Schipper, K., van Ulsen, P., van der Ende, A. & Tommassen, J. (2013). BMC Genomics, 14, 622. [DOI] [PMC free article] [PubMed]
- Bentley, S. D. et al. (2007). PLoS Genet. 3, e23. [DOI] [PMC free article] [PubMed]
- Birmanns, S., Rusu, M. & Wriggers, W. (2011). J. Struct. Biol. 173, 428–435. [DOI] [PMC free article] [PubMed]
- Cappadocia, L., Maréchal, A., Parent, J.-S., Lepage, E., Sygusch, J. & Brisson, N. (2010). Plant Cell, 22, 1849–1867. [DOI] [PMC free article] [PubMed]
- Cappadocia, L., Parent, J.-S., Sygusch, J. & Brisson, N. (2013). Acta Cryst. F69, 1207–1211. [DOI] [PMC free article] [PubMed]
- Cohen, S. X., Morris, R. J., Fernandez, F. J., Ben Jelloul, M., Kakaris, M., Parthasarathy, V., Lamzin, V. S., Kleywegt, G. J. & Perrakis, A. (2004). Acta Cryst. D60, 2222–2229. [DOI] [PubMed]
- Dauter, Z., Dauter, M. & Rajashankar, K. R. (2000). Acta Cryst. D56, 232–237. [DOI] [PubMed]
- Desveaux, D., Maréchal, A. & Brisson, N. (2005). Trends Plant Sci. 10, 95–102. [DOI] [PubMed]
- Diner, E. J., Beck, C. M., Webb, J. S., Low, D. A. & Hayes, C. S. (2012). Genes Dev. 26, 515–525. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Eschenfeldt, W. H., Makowska-Grzyska, M., Stols, L., Donnelly, M. I., Jedrzejczak, R. & Joachimiak, A. (2013). J. Struct. Funct. Genomics, 14, 135–144. [DOI] [PMC free article] [PubMed]
- Eschenfeldt, W. H., Maltseva, N., Stols, L., Donnelly, M. I., Gu, M., Nocek, B., Tan, K., Kim, Y. & Joachimiak, A. (2010). J. Struct. Funct. Genomics, 11, 31–39. [DOI] [PMC free article] [PubMed]
- Gioia, U., Laneve, P., Dlakic, M., Arceci, M., Bozzoni, I. & Caffarelli, E. (2005). J. Biol. Chem. 280, 18996–19002. [DOI] [PubMed]
- Gupta, U. K., Mahanta, S. & Paul, S. (2013). Med. Hypotheses, 81, 853–861. [DOI] [PubMed]
- Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Jamet, A., Jousset, A. B., Euphrasie, D., Mukorako, P., Boucharlat, A., Ducousso, A., Charbit, A. & Nassif, X. (2015). PLoS Pathog. 11, e1004592. [DOI] [PMC free article] [PubMed]
- Kajava, A. V., Cheng, N., Cleaver, R., Kessel, M., Simon, M. N., Willery, E., Jacob-Dubuisson, F., Locht, C. & Steven, A. C. (2001). Mol. Microbiol. 42, 279–292. [DOI] [PubMed]
- Koskiniemi, S., Garza-Sánchez, F., Sandegren, L., Webb, J. S., Braaten, B. A., Poole, S. J., Andersson, D. I., Hayes, C. S. & Low, D. A. (2014). PLoS Genet. 10, e1004255. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Laneve, P., Altieri, F., Fiori, M. E., Scaloni, A., Bozzoni, I. & Caffarelli, E. (2003). J. Biol. Chem. 278, 13026–13032. [DOI] [PubMed]
- Macindoe, G., Mavridis, L., Venkatraman, V., Devignes, M. D. & Ritchie, D. W. (2010). Nucleic Acids Res. 38, W445–W449. [DOI] [PMC free article] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Nikolakakis, K., Amber, S., Wilbur, J. S., Diner, E. J., Aoki, S. K., Poole, S. J., Tuanyok, A., Keim, P. S., Peacock, S., Hayes, C. S. & Low, D. A. (2012). Mol. Microbiol. 84, 516–529. [DOI] [PMC free article] [PubMed]
- Nikulin, J., Panzner, U., Frosch, M. & Schubert-Unkmeir, A. (2006). Int. J. Med. Microbiol. 296, 553–558. [DOI] [PubMed]
- Norel, R., Lin, S. L., Wolfson, H. J. & Nussinov, R. (1995). J. Mol. Biol. 252, 263–273. [DOI] [PubMed]
- Poole, S. J., Diner, E. J., Aoki, S. K., Braaten, B. A., t’Kint de Roodenbeke, C., Low, D. A. & Hayes, C. S. (2011). PLoS Genet. 7, e1002217. [DOI] [PMC free article] [PubMed]
- Renzi, F., Caffarelli, E., Laneve, P., Bozzoni, I., Brunori, M. & Vallone, B. (2006). Proc. Natl Acad. Sci. USA, 103, 12365–12370. [DOI] [PMC free article] [PubMed]
- Rosenbaum, G. et al. (2006). J. Synchrotron Rad. 13, 30–45.
- Ruhe, Z. C., Low, D. A. & Hayes, C. S. (2013). Trends Microbiol. 21, 230–237. [DOI] [PMC free article] [PubMed]
- Ruhe, Z. C., Nguyen, J. Y., Beck, C. M., Low, D. A. & Hayes, C. S. (2014). Mol. Microbiol. 94, 466–481. [DOI] [PMC free article] [PubMed]
- Schumacher, M. A., Karamooz, E., Zíková, A., Trantírek, L. & Lukeš, J. (2006). Cell, 126, 701–711. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Thigpen, M. C. et al. (2011). N. Engl. J. Med. 364, 2016–2025.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M. & Aravind, L. (2012). Biol. Direct, 7, 18. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: CdiI immunity protein, 4q7o