The X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state was determined at 2.0 Å resolution. This structure shows that a cyanide ion bridges the two metals in a nearly symmetric end-on fashion.
Keywords: membrane protein, cytochrome c oxidase
Abstract
The X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state was determined at 2.0 Å resolution. The structure reveals that the peroxide that bridges the two metals in the fully oxidized state is replaced by a cyanide ion bound in a nearly symmetric end-on fashion without significantly changing the protein conformation outside the two metal sites.
1. Introduction
Cytochrome c oxidase (CcO), the terminal oxidase of the respiratory chain, resides in the mitochondrial inner membrane or the bacterial cytoplasmic membrane (Ferguson-Miller & Babcock, 1996 ▶). CcO contains four redox-active metal sites: haem a, haem a 3, CuA and CuB. CuA is in the extramembrane space of the positive side, whereas haem a, haem a 3 and CuB are in the transmembrane region (Tsukihara et al., 1995 ▶). The Fea 3–CuB site catalyzes the reduction of molecular oxygen to two water molecules. Substrate O2 is transferred to the Fea 3–CuB site through a hydrophobic channel in the transmembrane region (Shinzawa-Itoh et al., 2007 ▶). Electrons are transmitted to the dioxygen-reduction site from cytochrome c on the positive-side surface of CcO through the CuA site and haem a, whereas protons are taken up from the negative side and transferred through specific hydrogen-bond networks in the transmembrane region (Tsukihara et al., 1996 ▶). In addition to the protons used for water production, additional protons are transferred through CcO across the membrane, generating the electrochemical proton gradient.
Previously, we determined the crystal structure of fully oxidized CcO at 1.8 Å resolution (Aoyama et al., 2009 ▶). The peak electron density in the vicinity of the hydroxide group of Tyr244 in the oxidized enzyme increases in proportion to the period of X-ray exposure, whereas the density of the peroxide group between Fea 3 and CuB decreases (Aoyama et al., 2009 ▶). This is because haem irons are reduced to the ferrous state to activate the oxygen-reduction centre and generate a water molecule from the peroxide anion. The water molecules generated in this manner accumulate at a site near the OH of Tyr244. The location and occupancy of the intrinsic (not artificially produced) water molecules inside the O2-reduction site, which are likely to play critical roles in the reaction mechanism of CcO, must be carefully examined in various oxidation and ligand-binding states.
Cyanide anion (CN−), which is a probe for the O2-reduction site, inhibits the catalytic activity of CcO. Because cyanide anion in the CN−-bound enzyme, which has an extremely high affinity for oxidized haem a 3 (Hirota et al., 1996 ▶), completely removes the bound peroxide at the oxygen-reduction centre, X-ray reduction of the haem a 3–CuB site does not effect the production of water molecules. In this study, we determined the structure of the CN−-bound fully oxidized form at 2.0 Å resolution. It should be noted that the X-ray structure of CN−-bound mammalian CcO in the fully reduced state, which was distinctively different from the fully oxidized mammalian CcO in the structure around the haem a 3–CuB, including Tyr244, has previously been reported (Muramoto et al., 2010 ▶), but to date the structure of the CN−-bound fully oxidized state has not been reported. Comparison of this structure with that of the fully oxidized enzyme previously determined revealed that X-ray irradiation, which induces the accumulation of water in the vicinity of Tyr244, does not induce any significant change in the protein structure.
2. Materials and methods
2.1. Preparation of crystals of CN−-bound bovine heart CcO in the fully oxidized state
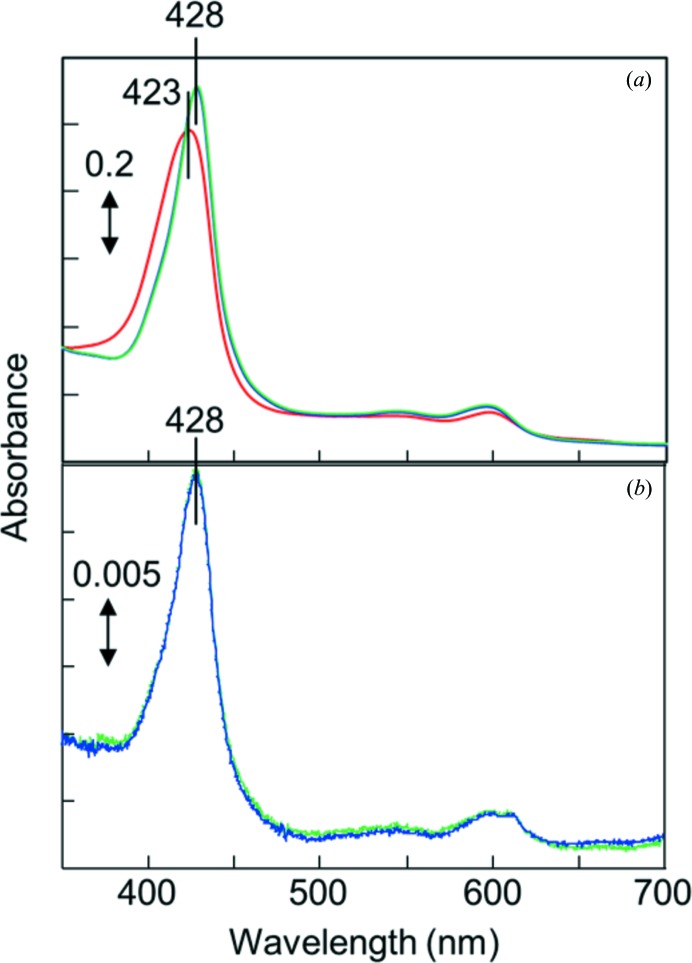
CcO in the fully oxidized state was purified from bovine heart mitochondria and crystallized as described previously (Tsukihara et al., 1995 ▶). The oxidized CcO crystals were stabilized at 277 K in 40 mM MES–Tris buffer pH 5.8 containing 0.2%(w/v) n-decyl β-d-maltoside and 1%(w/v) polyethylene glycol 4000 (Merck) (solution A). The CN−-bound oxidized CcO was prepared by equilibrating the crystals in 10 mM potassium cyanide solution as follows. The solution containing CcO in the fully oxidized resting state exhibited a Soret band peak at 423 nm. The peak position was shifted to 428 nm by the addition of cyanide ion (Fig. 1 ▶ a). The CN−-bound oxidized crystals, washed with solution A as described above and dissolved in 100 mM sodium phosphate buffer pH 7.4 containing 0.2%(w/v) n-decyl β-d-maltoside without potassium cyanide, provided a typical CN−-bound oxidized CcO spectrum showing the Soret band peak at 428 nm. The peak position was not shifted by the addition of cyanide ion (Fig. 1 ▶ b), confirming complete binding of CN− to the fully oxidized crystal. For the X-ray diffraction measurements, crystals were cooled in a cryo-nitrogen stream at 100 K in the presence of 45% ethylene glycol as a cryoprotectant.
Figure 1.
Absorption spectra of CcO. (a) Spectra of a solution containing 10 µM CcO in the fully oxidized resting state in 100 mM sodium phosphate buffer pH 7.4 containing 0.2%(w/v) n-decyl β-d-maltoside (red line) and 2 h (blue line) or 1 d (green line) after the addition of 5 mM potassium cyanide. (b) Spectra of a solution containing dissolved CcO crystals after CN− treatment in 100 mM sodium phosphate buffer pH 7.4 and 0.2%(w/v) n-decyl β-d-maltoside (blue line) and 2 h after the addition of 5 mM potassium cyanide (green line).
2.2. X-ray structure determination
X-ray diffraction intensity was recorded with an image-plate detector on BL44XU at SPring-8, Japan. Experimental conditions are given in Table 1 ▶. To decrease the radiation damage in the diffraction images, the irradiation position of the crystal was translated by a distance of 50 µm every 30 shots. Data processing and scaling were carried out using DENZO and SCALEPACK, respectively (Otwinowski & Minor, 1997 ▶). To increase the redundancy of the reflections, diffraction data measured from two crystals were merged. The structure amplitudes (|F o|) were calculated using the CCP4 program TRUNCATE (French & Wilson, 1978 ▶). The initial phases at 4 Å resolution were obtained by the molecular-replacement (MR) method using the previously determined structure of the fully oxidized protein (Shinzawa-Itoh et al., 2007 ▶; PDB entry 2dyr) as a model. Phase extension to 2.0 Å resolution was carried out by density modification in conjunction with noncrystallographic symmetry averaging using the CCP4 program DM (Cowtan, 1994 ▶). The resultant phase angles (αMR/DM) were used to calculate the electron-density map (MR/DM map) with Fourier coefficients |F o|exp(iαMR/DM).
Table 1. X-ray diffraction data and refinement statistics for CN-bound oxidized CcO.
| Crystal 1 | Crystal 2 | |
|---|---|---|
| Experimental conditions | ||
| Beamline | BL44XU, SPring-8 | |
| Wavelength () | 0.90 | |
| Detector | DIP6040 | |
| Oscillation angle () | 0.50 | |
| Exposure period (s) | 10.0 | |
| Temperature (K) | 100 | |
| No. of crystals used | 2 | |
| Beam size (m) | 50 80 | 50 70 |
| Crystal-to-detector distance (mm) | 310 | 360 |
| No. of diffraction images | 180 | 200 |
| Total rotation range () | 90 | 100 |
| Diffraction data | ||
| Space group | P212121 | |
| Unit-cell parameters () | a = 183.7, b = 206.7, c = 178.2 | |
| Resolution () | 200.02.00 (2.022.00) | |
| Cutoff | 3.0 | |
| Observed reflections | 3385108 (76493) | |
| Independent reflections | 451915 (11249) | |
| Averaged redundancy† | 7.5 (6.8) | |
| I/(I)‡ | 18.5 (1.2) | |
| Completeness§ | 0.999 (0.999) | |
| R merge ¶ | 0.125 (2.038) | |
| R p.i.m. †† | 0.049 (0.832) | |
| CC1/2 ‡‡ | 0.997 (0.513) | |
| Refinement | ||
| Resolution () | 40.02.00 (2.022.00) | |
| R §§ | 0.187 (0.304) | |
| R free ¶¶ | 0.214 (0.326) | |
| R.m.s.d.†††, bonds () | 0.0287 | |
| R.m.s.d.†††, angles () | 2.3 | |
| Averaged B factors (2) | ||
| Protein atoms | 42.4 | |
| Heavy metals | 30.4 | |
| Lipids and detergents | 83.1 | |
| Waters | 48.2 | |
| All atoms | 45.0 | |
Redundancy is the number of observed reflections for each independent reflection.
I/(I) is the average of the intensity signal-to-noise ratio.
Completeness is the ratio of independent reflections to observed reflections.
R
merge = 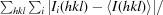
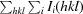 , where Ii(hkl) is the intensity value of the ith measurement of hkl and I(hkl) is the corresponding mean value of Ii(hkl) for all i measurements. The summation is over reflections with I/(I) larger than 3.0.
, where Ii(hkl) is the intensity value of the ith measurement of hkl and I(hkl) is the corresponding mean value of Ii(hkl) for all i measurements. The summation is over reflections with I/(I) larger than 3.0.
R
p.i.m. = 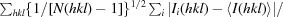
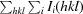 , where N(hkl) is the multiplicity of reflection hkl (Weiss, 2001 ▶).
, where N(hkl) is the multiplicity of reflection hkl (Weiss, 2001 ▶).
CC1/2 is the correlation coefficient between random half data sets calculated using AIMLESS (Evans Murshudov, 2013 ▶).
R is the conventional crystallographic R factor, R = 
 , where F
obs and F
calc are the observed and calculated structure factors, respectively.
, where F
obs and F
calc are the observed and calculated structure factors, respectively.
R free is the free R factor for 5% of the reflections that were excluded from refinement.
Root-mean-square deviation.
The atomic coordinates of fully oxidized CcO (PDB entry 2dyr) were used to build an initial model in the MR/DM map. Structure refinement was initiated with X-PLOR (Brünger et al., 1987 ▶) followed by the CCP4 program REFMAC5 (Murshudov et al., 2011 ▶). The atomic model was rebuilt by iterative cycles of refinement calculations and visual inspection of the (F o − F c) electron-density map calculated with Fourier coefficients (|F o| − |F c|)exp(iαc), as well as the MR/DM map in Coot (Emsley et al., 2010 ▶), where |F c| and αc are the structure amplitudes and phase angles, respectively, calculated from the refined model. Bulk-solvent correction, anisotropic scaling of the observed and calculated structure amplitudes. and TLS parameters were incorporated into the refinement calculation. Since two monomers in the asymmetric unit show different bulk motions from each other (Tomizaki et al., 1999 ▶), each monomer was defined as a separate TLS group in the refinement calculation. The individual anisotropic B factors were refined for all of the Fe, Cu and Zn atoms. During the refinement, the C—N bond length was fixed at 1.15 Å, whereas the Fea 3—C and CuB—N distances and the Fea 3—C—N and C—N—CuB angles were not geometrically restrained. The progress of refinement was assessed by the decrease in the R and R free values (Brünger, 1992 ▶) calculated at each step of the refinement. Two water molecules, one between two propionates of haem a 3 and the other forming a hydrogen bond to Oγ1 of Thr316 and the hydroxide group of the hydroxyl farnesylethyl group of haem a 3, were removed from the structure-factor calculations during the refinement. These two water sites were used as monitors to estimate the occupancy of a water molecule in the vicinity of Tyr244. In the final stage of the refinement, the occupancy of the water molecule located near Tyr244 was fixed to the value estimated by comparing its peak height with those of the monitors, and the water molecules used as monitors were included in the structure-factor calculations.
3. Results and discussion
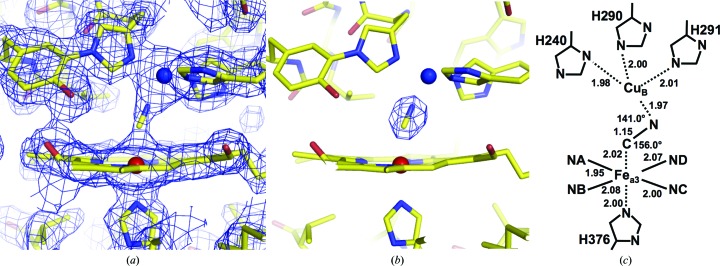
Statistics for the intensity data set and the structure refinement at 2.0 Å resolution are summarized in Table 1 ▶. The MR/DM and (F o − F c) electron-density maps, calculated as described in the previous section, clearly revealed electron density in the Fea 3–CuB site (Figs. 2 ▶ a and 2 ▶ b). The (F o − F c) electron density of the fully oxidized form calculated at 2.1 Å resolution reveals an elongated peak distinct from that of the CN− ion at the dioxygen-reduction site, which is 1.55 times higher than the average peak height of the reference water molecules (Fig. 3 ▶ a; Aoyama et al., 2009 ▶). The peak in (F o − F c) electron density between the metal centres was 1.56 times higher than the average peak height of the same reference water molecules as those in the fully oxidized form. The peak height ratio of 1.56 is consistent with the ratio of the number of electrons of the CN− ion and the water molecule (14.0/10.0 = 1.40). The electron-density hump between CuB and Fea 3 was assignable as a CN− ion. At the current resolution, however, it is difficult to distinguish C and N in the electron-density map. C was assigned on the Fea 3 side according to spectroscopic investigations (Hirota et al., 1996 ▶). The CN model was incorporated into the electron density and refinement was continued. Two crystallographically independent molecules are present in each asymmetric unit. One of the two molecules has a B factor that is lower than that of the other by about 8 Å2; structural descriptions are given for the molecule with the lower B factor. The refined structure showed that the Fea 3–C and CuB–N distances were 2.02 and 1.97 Å, respectively, and the Fea 3—C—N and CuB—N—C angles were 156.0 and 141.0°, respectively (Fig. 2 ▶ c). These structures indicate that the CN− ion bridges between the two metal centres. Within experimental error, the distance between the two metal ions (4.88 Å) is almost identical to the corresponding distances in the fully oxidized state (4.97 Å; PDB entry 2dyr) and the fully reduced CN−-bound form (4.99 Å; PDB entry 3ag4) and significantly shorter than that of 5.14 Å in the fully reduced enzyme (Muramoto et al., 2007 ▶; PDB entry 2eij), which has no bridging ligand between CuB and Fea 3.
Figure 2.
Structure of the oxygen-reduction centre containing CN−. (a) MR/DM electron-density map drawn at the 1.5σ level. (b) (F o − F c) electron-density map drawn at the 9.0σ level. (c) Schematic description of the CN−-bound metal sites, giving distances (in Å) and angles.
Figure 3.
Structure of the cavity containing the water molecule that forms a hydrogen bond to the OH of Tyr244. The cavity is surrounded by His240, Tyr244, Val287, His290, Thr309 and Ile312. The side-chain structure of Ile312 exhibits multiple conformations, the major and minor components of which are coloured yellow and green, respectively. The distance between the O atom of the water and Cδ1 of the major component of Ile312 is 2.02 Å, whereas that between the O atom and Cδ1 of the minor component of Ile312 is 3.93 Å.
The peak height of the water molecule that forms a hydrogen bond to the OH of Tyr244 was 37% of the average peak height of the monitor waters in the (F o − F c) map. Thus, the occupancy of the water O atom was estimated to be 0.37. The electron density at this site in the fully oxidized form increases in proportion to the X-ray exposure period (Aoyama et al., 2009 ▶). Because CN− completely removes the bound peroxide, X-ray reduction of heme a 3 of the CN−-bound enzyme is unlikely to generate any water molecules. The water peak in the CN−-bound enzyme must be owing to spontaneously introduced exogenous water molecules. Before X-ray irradiation, the fully oxidized enzyme must have the same amount of spontaneously introduced water molecules in the vicinity of the OH of Tyr244 as the CN−-bound enzyme. Thus, the electron density near the OH of Tyr244 in the fully oxidized enzyme, determined by X-ray diffraction at SPring-8, is owing to spontaneously introduced exogenous water molecules as well as waters induced by X-ray reduction. The water molecule in the cavity is surrounded by His240, Tyr244, Val287, His290, Thr309 and Ile312 (Fig. 3 ▶). Ile312 has two conformations: the main structure of Ile312 clashes with the water molecule, whereas the minor structure accommodates the water molecule in the cavity. Structure refinement with occupancies of 0.63 and 0.37 for the major and the minor conformation of Ile312, respectively, converged well, with similar average B factors of 26.3 and 28.0 Å2 for the major and the minor components of Ile312, respectively.
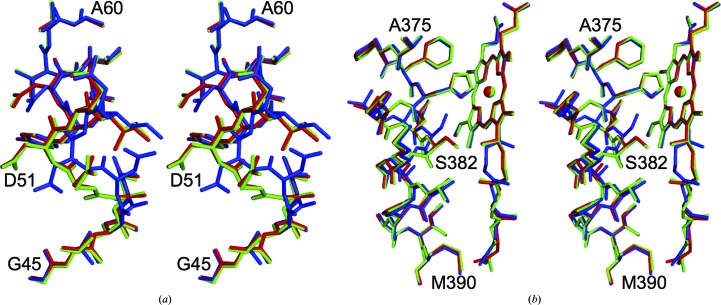
Comparison of the structures of the oxidized and reduced enzymes revealed significant conformational differences in residues 49–55 of subunit I (Yoshikawa et al., 1998 ▶), in residues 380–386 of helix X (371–400) and in the hydroxyfarnesylethyl group of haem a (Tsukihara et al., 2003 ▶). Structures showing the redox-coupled changes were compared with those of the CN−-bound oxidized enzyme, as shown in Fig. 4 ▶. These structures of the CN−-bound oxidized enzyme were identical to those of the oxidized enzyme. These observations strongly suggest that the structural effect of water production induced by X-ray reduction of the fully oxidized enzyme is restricted to the region near the OH of Tyr244.
Figure 4.
Comparison of the structure of CN−-bound oxidized CcO with those of the reduced (PDB entry 2eij) and oxidized (PDB entry 2dyr) enzymes. The structures of CN−-bound oxidized CcO and reduced CcO were superposed on that of oxidized CcO by a least-squares method using Coot (Emsley et al., 2010 ▶). The CN−-bound oxidized CcO, the reduced CcO and the oxidized CcO are coloured green, blue and red, respectively. The three structures for (a) residues 45–60 and (b) residues 375–390 of subunit I and haem a are shown as stereoscopic pairs.
Supplementary Material
PDB reference: bovine heart cytochrome c oxidase, 3x2q
Acknowledgments
This work was supported by Grants-in-Aid for the Global Center of Excellence Program and for Scientific Research (A) 2247012 and (B) 26234567 provided by the Japanese Ministry of Education, Culture, Sports, Science and Technology (SY), JST/CREST (TT) and JSPS Kakenhi Grants 11780475 and 22370060 (KM). SY is a Senior Visiting Scientist at the RIKEN Harima Institute.
References
- Aoyama, H., Muramoto, K., Shinzawa-Itoh, K., Hirata, K., Yamashita, E., Tsukihara, T., Ogura, T. & Yoshikawa, S. (2009). Proc. Natl Acad. Sci. USA, 106, 2165–2169. [DOI] [PMC free article] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Brünger, A. T., Kuriyan, J. & Karplus, M. (1987). Science, 235, 458–460. [DOI] [PubMed]
- Cowtan, K. (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 31, 34–38.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Ferguson-Miller, S. & Babcock, G. T. (1996). Chem. Rev. 96, 2889–2908. [DOI] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Hirota, S., Ogura, T., Shinzawa-Itoh, K., Yoshikawa, S. & Kitagawa, T. (1996). J. Phys. Chem. 100, 15274–15279.
- Muramoto, K., Hirata, K., Shinzawa-Itoh, K., Yoko-o, S., Yamashita, E., Aoyama, H., Tsukihara, T. & Yoshikawa, S. (2007). Proc. Natl Acad. Sci. USA, 104, 7881–7886. [DOI] [PMC free article] [PubMed]
- Muramoto, K., Ohta, K., Shinzawa-Itoh, K., Kanda, K., Taniguchi, M., Nabekura, H., Yamashita, E., Tsukihara, T. & Yoshikawa, S. (2010). Proc. Natl Acad. Sci. USA, 107, 7740–7745. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Shinzawa-Itoh, K., Aoyama, H., Muramoto, K., Terada, H., Kurauchi, T., Tadehara, Y., Yamasaki, A., Sugimura, T., Kurono, S., Tsujimoto, K., Mizushima, T., Yamashita, E., Tsukihara, T. & Yoshikawa, S. (2007). EMBO J. 26, 1713–1725. [DOI] [PMC free article] [PubMed]
- Tomizaki, T., Yamashita, E., Yamaguchi, H., Aoyama, H., Tsukihara, T., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1999). Acta Cryst. D55, 31–45. [DOI] [PubMed]
- Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1995). Science, 269, 1069–1074. [DOI] [PubMed]
- Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1996). Science, 272, 1136–1144. [DOI] [PubMed]
- Tsukihara, T., Shimokata, K., Katayama, Y., Shimada, H., Muramoto, K., Aoyama, H., Mochizuki, M., Shinzawa-Itoh, K., Yamashita, E., Yao, M., Ishimura, Y. & Yoshikawa, S. (2003). Proc. Natl Acad. Sci. USA, 100, 15304–15309. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst. 34, 130–135.
- Yoshikawa, S., Shinzawa-Itoh, K., Nakashima, R., Yaono, R., Yamashita, E., Inoue, N., Yao, M., Fei, M. J., Libeu, C. P., Mizushima, T., Yamaguchi, H., Tomizaki, T. & Tsukihara, T. (1998). Science, 280, 1723–1729. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: bovine heart cytochrome c oxidase, 3x2q