Abstract
Human metapneumovirus (hMPV) is a newly discovered pathogen associated with respiratory tract illness, primarily in young children, immunocompromised individuals, and the elderly. The genomic sequence of the prototype hMPV isolate NL/1/00 without the terminal leader and trailer sequences has been reported previously. Here we describe the leader and trailer sequences of two hMPV isolates, NL/1/00 and NL/1/99, representing the two main genetic lineages of hMPV. Minigenome constructs in which the green fluorescent protein or chloramphenicol acetyltransferase genes are flanked by the viral genomic ends derived from both hMPV lineages and transcribed using a T7 RNA polymerase promoter-terminator cassette were generated. Cotransfection of minigenome constructs with plasmids expressing the polymerase complex components L, P, N, and M2.1 in 293T or baby hamster kidney cells resulted in expression of the reporter genes. When the minigenome was replaced by a sense or antisense full-length cDNA copy of the NL/1/00 or NL/1/99 viral genomes, recombinant virus was recovered from transfected cells. Viral titers up to 107.2 and 105.7 50% tissue culture infective dose/ml were achieved with the sense and antisense plasmids, respectively. The recombinant viruses replicated with kinetics similar to those of the parental viruses in Vero cells. This reverse genetics system provides an important new tool for applied and fundamental research.
The human metapneumovirus (hMPV) was recently identified in respiratory specimens obtained from children in The Netherlands suffering from respiratory tract illness (RTI) (38). Subsequently, hMPV has been detected around the world, in clinical samples collected from children, immunocompromised individuals, and elderly individuals suffering from RTI (3, 6, 8, 13-15, 20, 26, 28, 34, 36, 41). Clinical signs and symptoms associated with hMPV infection are similar to those caused by respiratory syncytial virus (RSV), ranging from mild respiratory problems to severe cough, bronchiolitis, and pneumonia, often accompanied by high fever, myalgia, and vomiting (5, 14, 15, 23, 26, 27, 34, 39, 40). The genomic organization of hMPV resembles that of avian pneumovirus (APV), and hMPV has therefore been classified as the first mammalian member of the family Paramyxoviridae, subfamily Pneumovirinae, genus Metapneumovirus (37). Two main genetic lineages of hMPV were first described for the Dutch isolates (38), and the presence of these genotypes has been confirmed worldwide (2, 4, 6, 26, 28, 41). Recently, the two main lineages were found to represent two serotypes of hMPV, A and B, each of which can be further divided in two genetic sublineages, A1, A2, B1, and B2 (40).
For APV, the other member of the Metapneumovirus genus, a minigenome system has been established (30), but a recombinant virus rescue system is not yet available. Such reverse genetics systems have been reported for other paramyxoviruses, including RSV, measles virus, parainfluenza viruses, rinderpest virus, and canine distemper virus (1, 10, 17, 19, 29). The systems for the recovery of recombinant paramyxovirus use a cDNA plasmid encoding the full-length viral RNA in sense or antisense orientation flanked by sequences to ensure the transcription of genome-length RNA such as a T7 RNA polymerase promoter-terminator cassette and a hepatitis delta virus (HDV) ribozyme sequence. Upon transcription of the full-length hMPV cDNA by T7 RNA polymerase expressed either from plasmids, modified vaccinia virus Ankara, or recombinant fowl pox virus and coexpression of the viral polymerase protein complex (N, P, L, and M2.1), a recombinant virus can be produced.
Reverse genetics systems provide a powerful tool for fundamental virus research and for the generation of vaccine candidates, including live-attenuated vaccines, because point mutations, deletions, and insertions can be engineered to suit specific needs. Foreign genes can be introduced in order to attenuate viruses or to create chimeric vaccines. By using reverse genetics, one can envisage creating chimeric viruses between APV and hMPV as potential vaccine candidates in humans and poultry or chimeric vaccines between hMPV and other human respiratory pathogens (RSV, parainfluenza virus) to combat RTI in humans.
Here, we describe the sequence analysis of the genomic termini of NL/1/00 and NL/1/99, representative strains for hMPV serotypes A (lineage A1) and B (lineage B1), respectively. We successfully used this information for the design of minigenome replication systems using chloramphenicol acetyltransferase (CAT) or green fluorescent protein (GFP) as reporter genes. Subsequently, the reporter genes of the minireplicons were replaced with full-length cDNA copies of the NL/1/00 (serotype A) or NL/1/99 (serotype B) genomes, and recombinant hMPVs were recovered.
MATERIALS AND METHODS
Cells and viruses.
Virus isolates hMPV NL/1/00 and NL/1/99 were propagated in tertiary monkey kidney cells as described previously (38). 293T cells were grown in Dulbecco's modified eagle medium (BioWhittaker, Verviers, Belgium), supplemented with 10% fetal calf serum (FCS), nonessential amino acids, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM glutamine. BSR-T7 cells, a kind gift of K. K. Conzelmann, were grown in the same media without nonessential amino acids and supplemented with 0.5 mg of G418 (Life Technologies, Breda, The Netherlands)/ml. Vero cells were grown in Iscove's modified Dulbecco's medium (BioWhittaker) supplemented with 10% FCS, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM glutamine. For hMPV replication, Vero cells were grown in Dulbecco's modified Eagle medium supplemented with 3% FCS, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, 2 mM glutamine, and 0.25 mg of trypsin/ml. For virus titration or plaque assays, Vero cells were grown in Iscove's modified Dulbecco's medium supplemented with 4% bovine serum albumin fraction V (Invitrogen, Breda, The Netherlands), 100 IU of penicillin/ml, 100 μg of streptomycin/ml, 2 mM glutamine, and 3.75 μg of trypsin/ml.
293T cells were transfected using the CaPO4 precipitation method (25), and BSR-T7 cells were transfected using Lipofectamine 2000 (Invitrogen), according to the instructions of the manufacturer.
Identification of leader and trailer sequences of hMPV.
Viral RNA was isolated from concentrated NL/1/00 and NL/1/99 virus stocks by using RNAzolB according to instructions from the manufacturer (Campro Scientific, Veenendaal, The Netherlands). The viral RNA was circularized using T4 RNA ligase (New England Biolabs, Frankfurt am Main, Germany) for 1 h at 37°C, and cDNA was synthesized using random hexamer primers (Promega, Leiden, The Netherlands) and RNase H-free superscript II reverse transcriptase (Invitrogen) for 50 min at 42°C. The ligated junction of the leader and trailer was PCR amplified using primers in the 5′ end of the N open reading frame (ORF) and the 3′ end of the L ORF. The PCR products were sequenced upon cloning in pCR2.1 (Invitrogen).
A second approach was used to identify the terminal nucleotides of the leader of NL/1/00. Viral RNA was isolated using a QIAamp viral RNA mini kit (QIAGEN, Valencia, Calif.), according to the instructions from the manufacturer. This viral RNA was polyadenylated by incubation with poly(A) polymerase (Ambion, Austin, Tex.) at 37°C for 1 h and purified using a NucAway spin column (Ambion). The viral RNA was reverse transcribed using a primer complementary to the poly(A) tail region and superscript I reverse transcriptase (Invitrogen). PCRs were carried out and sequenced upon cloning in plasmid pCR2.1 (Invitrogen).
Plasmid construction. (i) Minigenome constructs.
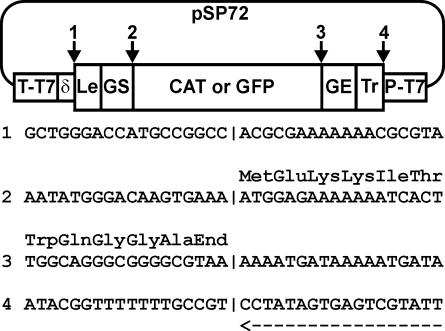
An NdeI (nucleotide [nt] 2379)-to-HpaI (nt 136) fragment was removed from plasmid pSP72 (Promega) and replaced by a synthetic T7 promoter extended with two or three G residues, two BsmBI sites, the HDV ribozyme, and a T7 terminator to yield pSP72-PT7-δ-TT7. The hMPV genomic leader sequence with the gene start of N and the genomic trailer sequence with the gene end of L were amplified by PCR and ligated and were separated by two BsmBI sites. This fragment was ligated in the BsmBI site of pSP72-PT7-δ-TT7 to yield pSP72-PT7-Tr-Le-δ-TT7. The ORFs of GFP or CAT were amplified by PCR using primers with type II restriction sites (BbsI, BsaI, or BsmBI) and cloned in the BsmBI sites between the N gene start and L gene end signals of hMPV to yield pSP72-PT7-Tr-CAT-Le-δ-TT7 and pSP72-PT7-Tr-GFP-Le-δ-TT7. The junctions between the elements of the minigenome plasmids are shown in Fig. 1.
FIG. 1.
Schematic representation of a minigenome construct. A CAT or GFP reporter gene is flanked by the viral leader sequence (Le) and gene start signal (GS) of the nucleocapsid gene on one end and the gene end signal (GE) of the polymerase gene and the viral trailer (Tr) on the other end. This minigenome is cloned in the context of a T7 RNA polymerase promoter-terminator (P-T7, T-T7) cassette. The authenticity of the transcribed ends of the negative-sense minigenome are determined by the position of the T7 promoter sequence and a HDV ribozyme sequence (δ). The important junctions, depicted with numbers 1 to 4, are shown in detail below the map, with the translated sequence of the CAT gene as the example.
(ii) Polymerase constructs.
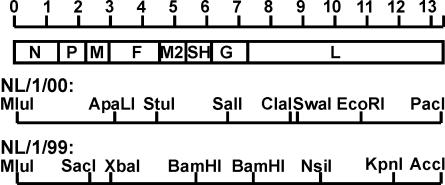
The N, P, and M2.1 ORFs of NL/1/00 and NL/1/99 were amplified by PCR using primers spanning the start and stop codons and flanked by NcoI and XhoI sites, respectively, and were cloned in the multiple cloning site of pCITE (Novagen) to yield plasmids pCITE-N, pCITE-P, and pCITE-M2.1. Constructs encoding the L gene of NL/1/00 or NL/1/99 were assembled from overlapping PCR fragments using restriction sites in the L gene (Fig. 2) and were cloned in pCITE. The restriction sites used were NcoI (introduced at nt 7180 before the start codon of L), SwaI (nt 8806), EcoRI (nt 10998), and SmaI (introduced after the PacI site at nt 13283 in the trailer) for NL/1/00, and BamHI (nt 7127), NsiI (nt 9565), KpnI (nt 11753), and AccI (nt 13276 in the trailer) for NL/1/99.
FIG. 2.
Restriction maps of hMPV NL/1/00 and NL/1/99. The genomic organization of hMPV is shown, roughly drawn to scale. The restriction sites that were used to generate the full-length cDNA constructs and the polymerase expression plasmids are shown for both virus isolates.
(iii) Full-length cDNA vectors.
The minigenome plasmids (pSP72-PT7-Tr-Le-δ-TT7) of NL/1/00 and NL/1/99 were used to construct full-length cDNA vectors. The cDNA encoding the genome of NL/1/00 was assembled from overlapping PCR fragments using restriction sites MluI (nt 12), ApaLI (nt 3293), StuI (nt 4480), SalI (nt 6649), ClaI (nt 8678), EcoRI (nt 10998), and PacI (nt 13283) (Fig. 2). The cDNA encoding the genome of NL/1/99 was assembled from overlapping PCR fragments using restriction sites MluI (nt 12), SacI (nt 2034), XbaI (nt 2979), BamHI (nt 5500 and 7127), NsiI (nt 9565), KpnI (nt 11753), and AccI (nt 13276) (Fig. 2). In the genome of both NL/1/00 and NL/1/99, a genetic marker, an AflII restriction enzyme site, was introduced at amino acid (aa) position 8 in the N ORF of the hMPV genome by using a QuickChange site-directed mutagenesis kit (Stratagene). Also, for NL/1/99, a negative-sense full-length clone was generated with the extra AflII restriction enzyme site, to compare the functionality of rescue with positive- or negative-sense plasmids.
All plasmid inserts were sequenced to ensure the absence of mutations. All primer sequences used for plasmid construction are available upon request.
Minigenome assays.
Minigenome plasmids were cotransfected with plasmid pAR3126, expressing a T7 RNA polymerase, and pCITE-L, pCITE-N, pCITE-P, and pCITE-M2.1 in six-well plates containing 3 × 105 to 5 × 105 293T cells per well (25). Transfections were done overnight and medium was refreshed the next day. In a second approach, transfections were done using BSR-T7 cells, a baby hamster kidney cell line stably expressing T7 RNA polymerase (7). Minigenomes were cotransfected with pCITE-L, pCITE-N, pCITE-P, and pCITE-M2.1 in six-well plates when cell monolayers were 80 to 95% confluent. Cells were analyzed 3 days after transfection by using a CAT enzyme-linked immunosorbent assay (Roche Diagnostics, Almere, The Netherlands) according to the instructions from the manufacturer or a flow cytometer equipped with an argon laser emitting at 488 nm (Becton Dickinson, Erembodegem, Belgium).
Recovery of recombinant hMPV.
Both 293T cells and BSR-T7 cells were used for rescue of recombinant hMPV. 293T cells were transfected overnight using the CaPO4 method with the four pCITE polymerase complex plasmids, pAR3126, and the full-length hMPV cDNA plasmid. BSR-T7 cells were transfected for 5 h with Lipofectamine 2000 (Invitrogen) and the same plasmids without pAR3126. After transfection, the media was replaced with fresh media supplemented with trypsin and incubated for 2 to 4 days. Cells were either used to prepare lysates or for direct cocultivation with Vero cells. To prepare lysates, cells were scraped with a rubber cell scraper, pooled with the supernatant, and subjected to one −70°C freeze-thaw cycle. The virus preparation was cleared from cellular debris by centrifugation and used to inoculate 60 to 70% confluent Vero cells in 100-mm dishes. After 3 or 4 days, half of the medium was replaced with fresh medium, and after 6 or 7 days, the supernatant was collected and the infected Vero cells were immunostained using a guinea pig polyclonal antiserum raised against hMPV and a fluorescein isothiocyanate-labeled rabbit anti-guinea pig serum (DakoCytomation, Heverlee, Belgium). The cells were analyzed using a flow cytometer, and the supernatant was used for virus titrations, reverse transcription (RT)-PCR, and restriction digests to confirm rescue of recombinant virus.
RT-PCR to identify genetic marker.
In both NL/1/00 and NL/1/99, a silent mutation was introduced in the codon that encodes leucine at aa position 8 in the N ORF of the full-length genome. At this position, CCTAAG (NL/1/99) or CCTGAG (NL/1/00) were changed to CTTAAG in order to introduce an AflII restriction site. After harvesting the rescued virus and RNA isolation with a High Pure RNA isolation kit (Roche Diagnostics), RT-PCR was performed with specific primers to amplify the region in which the mutation was introduced. After amplification, a control digestion was done with AflII to confirm that the recovered virus was recombinant in nature.
Virus titration.
Titrations were performed with 10-fold serial dilutions in 96-well plates. Briefly, confluent monolayers of Vero cells were spin-inoculated (15 min, 2000 × g) with 100 μl of 10-fold serial dilutions of each sample and incubated at 37°C. Fresh media (100 μl) was added to each well after 3 days. Seven days after inoculation, infected wells were identified by immunofluorescence assays with hMPV-specific polyclonal antiserum raised in guinea pigs. Titers expressed as 50% tissue culture infectious doses (TCID50) were calculated as described by Reed and Muench (31).
Plaque assay.
Twenty-four-well plates containing 95% confluent monolayers of Vero cells were inoculated with 10-fold serial virus dilutions for 1 h at 37°C, after which the media was replaced by 0.5 ml of fresh media and 0.5 ml of 2% methyl cellulose (MSD, Haarlem, The Netherlands) and cells were incubated at 37°C for 4 days. Methyl cellulose overlays were removed and cells were fixed with 100% methanol. Blocking was performed for 1 h at 37°C with 5% (wt/vol) nonfat dry milk in phosphate-buffered saline, and cells were subsequently incubated with hMPV-specific polyclonal antiserum for 1 h at 37°C, followed by incubation with horseradish peroxidase-labeled rabbit anti-guinea pig antibodies (DakoCytomation). Positive plaques were counted after incubation with AEC substrate chromogen (DakoCytomation) to determine viral titers.
Growth curve.
To generate growth curves, 25-cm2 flasks containing confluent Vero cells were inoculated at 37°C for 2 h with wild-type and recombinant hMPV virus strains at a multiplicity of infection of 0.1. After adsorption of the virus to the cells, the inoculum was removed and cells were washed two times with media before addition of 7 ml of fresh media and incubation at 37°C. Every 2 days, 1 ml of the supernatant was collected and replaced by fresh media. Plaque assays or end-point titrations were performed to determine viral titers in PFU per milliliter or TCID50 per milliliter, respectively. There is a general correspondence between viral titers determined by these two methods, but the end-point titrations yielded approximately 10-fold-higher titers; thus, 10 TCID50/ml corresponds with approximately 1 PFU/ml.
Nucleotide sequence accession numbers. The updated and new sequences discussed here were submitted to GenBank under accession no. AF371337 and AY525843, respectively.
RESULTS
Identification of leader and trailer sequences.
In order to determine the authentic nucleotide sequences of the hMPV genomic RNA leader and trailer, viral RNA isolated from concentrated virus stocks was circularized using T4 RNA ligase. cDNA was synthesized from this RNA, and the region containing the ligated junction of the genomic termini was amplified by PCR and cloned in plasmid pCR2.1. For NL/1/00 and NL/1/99, 26 and 35 clones were sequenced, respectively, and the position of the junction was determined after comparing all sequences. All but two clones showed the same trailer cDNA sequence, 5′-TACGGTTTTTTTGCCGT-3′. In the two deviant clones, the terminal T nucleotide was missing. The majority of the leader cDNA sequences were 5′-GCGAAAAAAACGCGTAT-3′, two nucleotides shorter than we had anticipated based on the leader sequences published for APV and RSV (21, 30). All other clones displayed three, four, five, or even longer nucleotide deletions at the leader end (Table 1). We next performed 3′ rapid amplification of cDNA ends (RACE) analysis as an alternative strategy to determine the 3′ end of the hMPV genome. To this end, fresh viral genomic RNA of NL/1/00 was isolated, which was polyadenylated and subsequently reverse transcribed using a primer complementary to the poly(A) tail region. The cDNA was amplified by PCR and cloned in plasmid pCR2.1. Sequence analysis revealed that 15 out of 79 clones (19%) contained the hMPV leader sequence 5′ ACGCGAAAAAAACGCGTAT 3′, which included the AC terminal dinucleotide observed for other paramyxovirus leader termini, but were lacking in the constructs derived upon RT-PCR of ligated viral RNA. The other 81% of the sequences revealed the absence of 3′-proximal nucleotides. From these data, we conclude that the correct leader cDNA sequence starts with AC and that the shorter leader sequences resulted from nuclease activity present in the virus preparations. It is of interest that the hMPV leader and trailer sequences are less complementary than the termini of related paramyxoviruses (Fig. 3).
TABLE 1.
Determination of leader sequences of hMPV NL/1/99 and NL/1/00 obtained by RT-PCR following ligation of viral RNA and 3′ RACEa
| Leader sequence positive strand | NL/1/99 RT-PCR circles (n = 26) | NL/1/00 RT-PCR circles (n = 35) | NL/1/00 poly(A) tailing and 3′ RACE (n = 79) |
|---|---|---|---|
| ACGCGAAAAAAACGCG | 0 (0) | 0 (0) | 15 (19) |
| CGCGAAAAAAACGCG | 0 (0) | 0 (0) | 6 (8) |
| GCGAAAAAAACGCG | 15 (58) | 30 (85) | 5 (6) |
| CGAAAAAAACGCG | 6 (23) | 2 (6) | 23 (29) |
| GAAAAAAACGCG | 2 (8) | 1 (3) | 0 (0) |
| AAAAAAACGCG | 2 (8) | 0 (0) | 0 (0) |
| Larger deletion | 1 (4) | 2 (6) | 30 (38) |
All data given as number (%).
FIG. 3.
Complementarity of leader and trailer sequences of hMPV, APV, and RSV. Viral leader (Le) and trailer (Tr) sequences were aligned, and the complementarity is indicated with connection lines, showing that the terminal nucleotides of hMPV were less complementary than those of RSV and APV. The 3′-terminal U residue in the APV genome has not been detected experimentally but was added in analogy to other paramyxoviruses (30).
hMPV minigenome assays.
Minigenome assays were performed to test whether the sequences of the genomic termini were correct and the plasmids encoding the viral polymerase complex components were functional. For this purpose, we used minigenome reporter constructs containing GFP or CAT ORFs in antisense orientation and flanked by the genomic termini of hMPV. A T7 phage RNA polymerase promoter was added upstream of the last genomic hMPV nucleotide of the trailer, and a HDV ribozyme sequence was linked to the hMPV leader to ensure authentic ends of the viral RNAs (Fig. 1). The minigenome constructs were cotransfected overnight in 293T cells with pAR3126 expressing T7 RNA polymerase and pCITE plasmids containing the N, P, L, and M2.1 ORFs. In the first experiment, 1.5 μg of minigenome, 1.5 μg of T7 RNA polymerase, and 1.8 μg of polymerase expression plasmids (0.6 μg of N, 0.6 μg of P, 0.3 μg of L, and 0.3 μg of M2.1) were cotransfected, yielding 1.3 ng of CAT/ml after 24 h and 11 ng of CAT/ml after 48 h (data not shown). Constructs with and without the terminal AC dinucleotide in the leader were both functional, but the construct without the terminal AC nucleotides gave rise to lower (32% reduced) levels of CAT expression (data not shown). Subsequently, we optimized the amounts of total plasmid DNA for transfection and the relative amounts of each individual plasmid using CAT expression as the readout. The optimal amounts of plasmids were 1 μg of minigenome, 1.5 μg of pAR3126, 0.8 μg of pCITE-N, 0.4 μg of pCITE-P, 0.4 μg of pCITE-M2.1, and 0.4 μg of pCITE-L. By using these optimized amounts, we achieved levels of CAT expression between 20 and 80 ng/ml for both the NL/1/00 and NL/1/99 minigenome systems. When the vector expressing the M2.1 protein is excluded from the experiments, the reporter gene is expressed at lower levels, indicating that the M2.1 protein is dispensable for minireplicon replication, whereas each of the other components is indispensable (Fig. 4). In BSR-T7 cells transfected with 1.5 μg of minigenome, 1.2 μg of pCITE-N, 0.6 μg of pCITE-P, 0.6 μg of pCITE-M2.1, and 0.6 μg of pCITE-L, CAT expression levels up to 140 ng/ml were obtained 48 h after transfection (data not shown).
FIG. 4.
CAT expression in minigenome replication assays in 293T cells. 293T cells were transfected with a T7 RNA polymerase-expressing plasmid, plasmids pCITE-L, pCITE-N, pCITE-P, and pCITE-M2.1, and the minigenome vector of either NL/1/00 (left panel) or NL/1/99 (right panel). Each of the individual plasmids was omitted from the transfection mixture to test the requirement of each individual component. Cell lysates were harvested 48 h after transfection, and levels of CAT were determined by enzyme-linked immunosorbent assay.
We next tested the GFP minigenome constructs in 293T cells by using the optimal amounts of plasmids determined with the CAT minigenome constructs. Forty-eight hours after cotransfection, up to 13% of the cells revealed high fluorescence by flow cytometry when all constructs were transfected, whereas such GFP expression was absent when the pCITE expression vectors were omitted (data not shown).
In BSR-T7 cells transfected with the GFP minigenome and pCITE expression vectors, 67.4% of the cells were positive by flow cytometry and 1.1% by fluorescence microscopy. This difference in the proportion of GFP-positive cells likely reflected the detection limits of the two assays; only the proportion of cells expressing very high levels of GFP as determined by flow cytometry can be detected under a normal fluorescence microscope.
Assembly of hMPV cDNA and recovery of recombinant hMPV.
Full-length hMPV cDNA clones were generated for both NL/1/00 and NL/1/99 by assembling DNA fragments of up to 3 kb that were generated by RT-PCR. This was done by using unique restriction enzyme sites present in the viral genome (Fig. 2). The full-length genomes were cloned in the minigenome vector (Fig. 1), by using restriction sites present in the leader and trailer. In order to demonstrate that the virus recovered by reverse genetics was derived from plasmid DNA, a restriction enzyme site (AflII) was added at aa position 8 in the N ORF of the full-length cDNA construct. The NL/1/00 cDNA construct contained three nucleotide substitutions and a five-nucleotide insertion compared to the GenBank sequence (AF371337). Upon subsequent sequence analysis of the original virus stock, the three nucleotide substitutions were found to be incorrect in the GenBank sequence entry (nt positions 3838, 8350, and 12218 in the GenBank sequence were changed to A, T, and T, respectively). A five-nucleotide insertion, TAAAA, was found in the intergenic region between F and M at position 4709 compared to the GenBank sequence. This nucleotide insertion was absent in the virus stock from which the GenBank sequence was derived but present in all other passages of virus stocks of NL/1/00. The plasmid encoding the full-length genome of NL/1/99 did not show any mutations compared to the wild-type sequence (accession no. AY525843).
Various approaches were followed for virus rescue from plasmids and were found to be successful. Initially, recombinant virus was rescued from 293T cells transfected overnight with 5 μg of the full-length cDNA plasmid, 1.5 μg of pAR3126, 0.8 μg of pCITE-N, 0.4 μg of pCITE-L, 0.4 μg of pCITE-P, and 0.4 μg of pCITE-M2.1. Recombinant virus was also rescued from 293T cells infected with fowlpox-T7 and subsequently transfected with plasmids. However, the most efficient virus rescue was observed in BSR-T7 cells transfected for 5 h with 3 μg of the full-length cDNA plasmid, 0.3 μg of pCITE-N, 0.3 μg of pCITE-P, 0.15 μg of pCITE-L, and 0.24 μg of pCITE-M2.1. The cells and supernatant were harvested 48 h after transfection, subjected to one freeze-thaw cycle, and used to inoculate Vero cells. Cytopathic effects were observed after 6 days in culture, at which time the majority of cells expressed hMPV proteins as determined by staining with an hMPV polyclonal antiserum and flow cytometry (data not shown). Virus titers in the supernatants collected at day 6 ranged from 105.7 to 107.2 TCID50/ml.
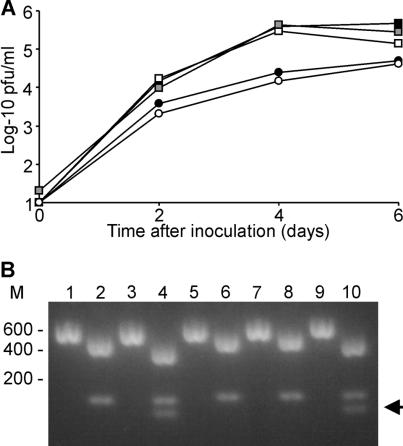
The replication kinetics of the wild-type and recombinant hMPV strains are shown in Fig. 5A, revealing that they were indistinguishable. Both wild-type and recombinant NL/1/00 appeared to replicate less efficiently than NL/1/99 in Vero cells. This difference in replication kinetics was observed in all assays that we performed with the NL/1/00 and NL/1/99 viruses. Probably the Vero cell line used in these experiments is more permissive for NL/1/99 than for NL/1/00.
FIG. 5.
Replication kinetics of wild-type and recombinant hMPV in Vero cells and verification of the presence of recombinant genomes. Vero cells, infected at a multiplicity of infection of 0.1 with wild-type and recombinant hMPV NL/1/00 (circles) and NL/1/99 (squares), were washed and incubated for 6 days. Samples were collected every 2 days, and virus titers were determined by plaque assay (A). Closed symbols represent wild-type virus, open symbols represent recombinant virus with an additional AflII site, and the gray symbol represents a NL/1/99 recombinant without the additional AflII site. The presence of the AflII site was confirmed by RT-PCR of viral RNA and AflII digestion (B). Lanes 1, 3, 5, 7, and 9, undigested DNA; lanes 2, 4, 6, 8, and 10, AflII-digested DNA. Lanes 1 and 2, wild-type NL/1/00; lanes 3 and 4, recombinant NL/1/00; lanes 5 and 6, wild-type NL/1/99; lanes 7 and 8, recombinant NL/1/99 without the AflII site; lanes 9 and 10, recombinant NL/1/99 with the AflII site. When the wild-type 603-bp N fragment is digested with AflII, fragments of 469 and 134 bp are generated. Due to the introduction of the additional AflII site, the recombinant 469-bp fragment is further digested to yield 402- and 67-bp fragments (the latter fragment is indicated with an arrowhead).
For NL/1/99, recombinant viruses were rescued using plasmids in which the cDNA genome was either cloned in the plus or minus sense orientation. Virus rescue from positive-sense cDNA resulted in virus titers higher than those for negative-sense cDNA (107 and 105.7 TCID50, respectively). Virus rescue in the presence and absence of the pCITE vector expressing M2.1 was equally efficient (data not shown).
The AflII restriction site in the NL/1/99 genome did not affect virus replication in Vero cells (Fig. 5A) or the efficiency of virus rescue (cDNA constructs with and without AflII sites gave rise to virus titers of 107 and 107.2, respectively). From these virus stocks, RNA was isolated and subjected to RT-PCR analysis and AflII restriction digestion, confirming that both NL/1/00 and NL/1/99 were recovered from plasmid DNA (Fig. 5B).
DISCUSSION
Here we describe the recovery of recombinant hMPV solely from cloned cDNAs. We selected two virus isolates, NL/1/00 and NL/1/99, representing each of the two main genetic lineages or serotypes of hMPV (38). These isolates were selected because they were found to replicate efficiently in tertiary monkey kidney cells and Vero cells.
The extreme termini of the viral genomes of NL/1/00 and NL/1/99 were determined by circularization of viral RNA with T4 RNA ligase, RT-PCR amplification, and sequencing. Whereas the trailer sequences were of the expected length, the leader sequences were two or more nucleotides shorter than expected from the leader sequence of RSV (21) according to this approach. A 3′ RACE method was used as an alternative strategy to determine the cDNA leader sequence of hMPV, revealing the presence of an additional AC dinucleotide in a proportion of the sequenced clones. In analogy with other paramyxoviruses, we assumed that the leader sequence of hMPV is 3′ UGCGCUUUUUUUGCG 5′ and that the truncated versions of this leader sequence that were detected were due to the presence of nuclease activity in the virus preparations. It is interesting that Randhawa et al. observed truncation of the leader sequence of APV with a single nucleotide in their experiments, but when a hypothetical U residue was added to their minigenome constructs in analogy with other paramyxoviruses, this minigenome was replicated efficiently (30). The leader and trailer of hMPV appear to be less complementary than those of RSV and APV (Fig. 3); only three of the five terminal residues of hMPV are complementary, compared to five of five and four of five for APV and RSV, respectively. With the completion of the full-length genomic sequences described here and those described recently by Biacchesi et al., full-length genomic sequences for each of the two main genetic lineages that represent hMPV serotypes A and B and each of the sublineages (A1, A2, B1, and B2) are now available from public databases (4, 40).
The sequences of the genomic termini were used to generate minigenome constructs, containing CAT or GFP as reporter genes. To increase transcription from the T7 promoter, either two or three G residues were placed between the T7 promoter and the end of the trailer sequence (24). Neither the addition of these two or three G residues at the trailer end nor the absence or presence of the terminal UG residues in the leader significantly affected CAT expression from minigenome vectors, suggesting that the authenticity of the sequence was not critical in minigenome assays. In minigenome assays, the N, P, and L proteins were found to be indispensable, whereas the M2.1 protein was not. For RSV, the M2.1 protein was shown to enhance the processivity of the viral polymerase, which is important for the efficient synthesis of full-length mRNA (10) and the read-through of intergenic regions (16, 18). The hMPV M2.1 protein may have the same function, but this cannot be concluded yet from our experiments. Although we were able to rescue virus in the absence of the M2.1 expression vector, it was shown for RSV that the simple expedient of omitting an expression plasmid is invalid for evaluating recovery requirements (9). Thus, the functional analysis of M2.1 requires further study.
When the CAT ORF of the minigenome construct was replaced by a full-length cDNA copy of the NL/1/00 or NL/1/99 genome, we could recover infectious virus from transfected cells. An additional AflII restriction site introduced in the N gene was used to confirm that the rescued viruses were recombinants. The AflII restriction site did not affect the efficiency of virus replication in vitro, since all recombinant viruses produced replicated in Vero cells with similar kinetics as their wild-type counterparts. Although transfections in both 293T cells and BSR-T7 cells resulted in the rescue of recombinant virus, the BSR-T7 cells yielded higher virus titers shortly after transfection, perhaps due to higher expression of T7 RNA polymerase in all cells. When we used a fowlpox virus expressing the T7 RNA polymerase gene as an alternative way for T7 delivery in 293T cells, we also successfully recovered recombinant hMPV NL/1/00. Since the efficiency of virus rescue using fowlpox-T7 was not significantly higher than rescue using plasmid-derived T7 RNA polymerase, the latter system was preferred since only plasmid DNAs are used without the need for infectious viruses. We were able to rescue recombinant NL/1/99 by using both positive- and negative-sense full-length cDNA plasmids, revealing that the efficiency with negative-sense cDNA resulted in lower titers than for positive-sense cDNA (105.7 and 107.2 TCID50, respectively). This is in agreement with the general belief that for rescue of negative-sense nonsegmented viruses, the simultaneous presence of viral mRNAs and naked negative-sense genomic RNA will result in hybridization, preventing the assembly of the genome into the RNP.
A plethora of live-attenuated viruses and chimeric viruses have been generated using reverse genetics (11, 12, 22, 32, 33, 35), and the same can now be envisaged for hMPV. For instance, recombinant hMPV strains harboring the surface glycoproteins of both serotype A and B isolates of hMPV can be generated that may induce a broad antibody response in infected hosts. Chimeric live-attenuated vaccines based on the hMPV genome, in which genes of RSV and/or parainfluenza viruses are inserted, may be useful as multivalent vaccine candidates. Recently, a chimeric bovine/human parainfluenza virus type 3 vector expressing the hMPV F protein was described and was found to induce protective hMPV antibody titers in a hamster model (35). In addition to its use for the generation of vaccine candidates, the reverse genetics system described here will be useful for fundamental and applied metapneumovirus research.
Acknowledgments
We thank J. Dunn, Brookhaven National Laboratory, for plasmid pAR3126, A. Garcia-Sastre for plasmid pPolI-CAT-RT, and K. Conzelmann for BSR-T7 cells. We thank Monique Spronken, Emmie de Wit, Roel van Eijk, Fiona Fernandes, Leenas Bicha, and Jackie Zhao for excellent technical assistance and James Simon for critically reading the manuscript.
R.F. is a fellow of the Royal Dutch Academy of Arts and Sciences. This work was sponsored in part by the framework five grant “Hammocs” from the European Union.
REFERENCES
- 1.Baron, M. D., and T. Barrett. 1997. Rescue of rinderpest virus from cloned cDNA. J. Virol. 71:1265-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastien, N., D. Ward, P. Van Caeseele, K. Brandt, S. H. Lee, G. McNabb, B. Klisko, E. Chan, and Y. Li. 2003. Human metapneumovirus infection in the Canadian population. J. Clin. Microbiol. 41:4642-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 6.Boivin, G., G. De Serres, S. Cote, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Dery. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, A., S. A. Nordbo, S. Jeansson, and S. Slordahl. 2003. Lower respiratory tract infection caused by human metapneumovirus in two children: the first report of human metapneumovirus infection in Norway. Scand. J. Infect. Dis. 35:772-774. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. L., E. Camargo, and M. G. Hill. 1999. Support plasmids and support proteins required for recovery of recombinant respiratory syncytial virus. Virology 259:251-255. [DOI] [PubMed] [Google Scholar]
- 10.Collins, P. L., M. G. Hill, E. Camargo, H. Grosfeld, R. M. Chanock, and B. R. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, P. L., S. S. Whitehead, A. Bukreyev, R. Fearns, M. N. Teng, K. Juhasz, R. M. Chanock, and B. R. Murphy. 1999. Rational design of live-attenuated recombinant vaccine virus for human respiratory syncytial virus by reverse genetics. Adv. Virus Res. 54:423-451. [DOI] [PubMed] [Google Scholar]
- 12.Durbin, A. P., M. H. Skiadopoulos, J. M. McAuliffe, J. M. Riggs, S. R. Surman, P. L. Collins, and B. R. Murphy. 2000. Human parainfluenza virus type 3 (PIV3) expressing the hemagglutinin protein of measles virus provides a potential method for immunization against measles virus and PIV3 in early infancy. J. Virol. 74:6821-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, M. Yoshioka, X. Ma, and K. Kobayashi. 2003. Seroprevalence of human metapneumovirus in Japan. J. Med. Virol. 70:281-283. [DOI] [PubMed] [Google Scholar]
- 14.Esper, F., D. Boucher, C. Weibel, R. A. Martinello, and J. S. Kahn. 2003. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111:1407-1410. [DOI] [PubMed] [Google Scholar]
- 15.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 16.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gassen, U., F. M. Collins, W. P. Duprex, and B. K. Rima. 2000. Establishment of a rescue system for canine distemper virus. J. Virol. 74:10737-10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman, M. A., and A. K. Banerjee. 1997. An infectious clone of human parainfluenza virus type 3. J. Virol. 71:4272-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maggi, F., M. Pifferi, M. Vatteroni, C. Fornai, E. Tempestini, S. Anzilotti, L. Lanini, E. Andreoli, V. Ragazzo, M. Pistello, S. Specter, and M. Bendinelli. 2003. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 41:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mink, M. A., D. S. Stec, and P. L. Collins. 1991. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology 185:615-624. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, B. R., and P. L. Collins. 2002. Live-attenuated virus vaccines for respiratory syncytial and parainfluenza viruses: applications of reverse genetics. J. Clin. Investig. 110:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterhaus, A., and R. Fouchier. 2003. Human metapneumovirus in the community. Lancet 361:890-891. [DOI] [PubMed] [Google Scholar]
- 24.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69:1011-1020. [DOI] [PubMed] [Google Scholar]
- 25.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peiris, J. S., W. H. Tang, K. H. Chan, P. L. Khong, Y. Guan, Y. L. Lau, and S. S. Chiu. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg. Infect. Dis. 9:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier, G., P. Dery, Y. Abed, and G. Boivin. 2002. Respiratory tract reinfections by the new human Metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 8:976-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randhawa, J. S., A. C. Marriott, C. R. Pringle, and A. J. Easton. 1997. Rescue of synthetic minireplicons establishes the absence of the NS1 and NS2 genes from avian pneumovirus. J. Virol. 71:9849-9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. J. Hyg. 27:493-497. [Google Scholar]
- 32.Schmidt, A. C., J. M. McAuliffe, B. R. Murphy, and P. L. Collins. 2001. Recombinant bovine/human parainfluenza virus type 3 (B/HPIV3) expressing the respiratory syncytial virus (RSV) G and F proteins can be used to achieve simultaneous mucosal immunization against RSV and HPIV3. J. Virol. 75:4594-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skiadopoulos, M. H., S. R. Surman, J. M. Riggs, P. L. Collins, and B. R. Murphy. 2001. A chimeric human-bovine parainfluenza virus type 3 expressing measles virus hemagglutinin is attenuated for replication but is still immunogenic in rhesus monkeys. J. Virol. 75:10498-10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takao, S., H. Shimozono, H. Kashiwa, Y. Shimazu, S. Fukuda, M. Kuwayama, and K. Miyazaki. 2003. Clinical study of pediatric cases of acute respiratory diseases associated with human metapneumovirus in Japan. Jpn. J. Infect. Dis. 56:127-129. [PubMed] [Google Scholar]
- 35.Tang, R. S., J. H. Schickli, M. MacPhail, F. Fernandes, L. Bicha, J. Spaete, R. A. Fouchier, A. D. Osterhaus, R. Spaete, and A. A. Haller. 2003. Effects of human metapneumovirus and respiratory syncytial virus antigen insertion in two 3′ proximal genome positions of bovine/human parainfluenza virus type 3 on virus replication and immunogenicity. J. Virol. 77:10819-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanasugarn, W., R. Samransamruajkit, P. Vanapongtipagorn, N. Prapphal, B. Van den Hoogen, A. D. Osterhaus, and Y. Poovorawan. 2003. Human metapneumovirus infection in Thai children. Scand. J. Infect. Dis. 35:754-756. [DOI] [PubMed] [Google Scholar]
- 37.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 38.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Hoogen, B. G., G. J. J. van Doornum, J. C. Fockens, J. J. Cornelissen, W. E. P. Beyer, R. de Groot, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 188:1571-1577. [DOI] [PubMed] [Google Scholar]
- 40.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10:658-666. [DOI] [PMC free article] [PubMed]
- 41.Viazov, S., F. Ratjen, R. Scheidhauer, M. Fiedler, and M. Roggendorf. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 41:3043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]