Abstract
Mx proteins form a family of interferon (IFN)-induced GTPases with potent antiviral activity against various single-stranded RNA viruses in mammals and chickens. In fish, alpha/beta IFN has been reported to inhibit the replication of infectious pancreatic necrosis virus (IPNV), but the mode of action has not been elucidated. A correlation between the inhibition of IPNV and Mx protein expression has, however, been observed. To examine whether Atlantic salmon Mx1 protein (ASMx1) possesses antiviral activity against IPNV, CHSE-214 cells constitutively expressing ASMx1 were established. ASMx1 appeared to be localized in the cytoplasm. The ASMx1-expressing clone selected showed a severely reduced IPNV-induced cytopathic effect, which was confirmed by a 500-fold reduction in virus yield. The antiviral activity against IPNV was further confirmed by the inhibition of virus protein synthesis and the reduced accumulation of virus transcripts. The present work further adds to the body of evidence which suggests that antiviral activity is a major functional role of vertebrate Mx proteins. Moreover, the list of viruses inhibited by Mx proteins is extended to include double-stranded RNA viruses.
Alpha/beta interferon (IFN) induces antiviral activity in cells and forms an important early line of defense against virus infection in vertebrates. In mammals, it is well documented that antiviral activity is mediated by several IFN-inducible proteins, among which the Mx proteins are some of the most studied (10). Mx proteins belong to the dynamin superfamily of large GTPases and were originally identified as the single dominant determinant of influenza virus A resistance in a strain of mice (15, 27). Antiviral activity due to IFN has been demonstrated in a number of fish species in vitro and in vivo (8). Recently, alpha/beta IFNs were cloned from zebra fish (1), Atlantic salmon (35), and channel catfish (28). Atlantic salmon IFN has been shown to have an antiviral effect against infectious pancreatic necrosis virus (IPNV) in salmon cells (35), but the mode of action has not been elucidated. A correlation between the inhibition of IPNV and Mx protein expression has, however, been observed in IFN-stimulated salmon cells (16, 31). Mx cDNAs encoding two different Mx proteins have been cloned from Atlantic salmon (36). Salmon Mx proteins have been shown to be induced by poly(I-C) and macrophage-derived IFN supernatants (17) and recently also by recombinant Atlantic salmon IFN (35).
The importance of Mx proteins in the IFN response of vertebrates is suggested by the presence and conservation of Mx genes in mammals (14), birds (4, 5), and teleost fish (18, 24, 33, 36, 42, 43). Until recently, however, antiviral activity had been established only for Mx proteins of mice (2), rats (30), and humans (12). In mice and rats, both nuclear and cytoplasmic Mx protein forms exist, and the antiviral specificity correlates with their subcellular locations (25). Human MxA, located in the cytoplasm, has a broader antiviral spectrum and inhibits viruses replicating both in the cytoplasm and in the nucleus. This group includes viruses with both negative and positive single-stranded RNA genomes and a virus with a DNA genome (11, 12). The mechanism by which MxA can inhibit such a variety of viruses is still unknown, but several reports suggest a direct interaction of Mx proteins and viral targets (20, 22). Mx proteins from chickens and birds first appeared to be devoid of antiviral activity (4, 5). However, a closer examination of different chicken breeds revealed polymorphisms of the Mx gene and confirmed the antiviral activity of Mx proteins from some breeds against influenza virus and vesicular stomatitis virus (VSV) (19). Recently, it was also found that pig Mx1 confers resistance to VSV (3). The antiviral effects of fish Mx proteins have been uncertain. Rainbow trout Mx proteins expressed by transient transfection of trout cells had no apparent inhibitory effect against replication of the rhabdovirus infectious hematopoietic necrosis virus (IHNV) (41). On the other hand, a recent study reported that the replication of fish rhabdoviruses was reduced in a fish cell line transfected with Japanese flounder Mx proteins (6).
IPNV is strongly inhibited in salmon cells expressing high levels of Mx proteins after treatment with IFN or poly(I-C) (16, 17, 31, 35). This makes it an interesting candidate virus for testing the antiviral activity of Atlantic salmon Mx proteins. IPNV is a naked bisegmented double-stranded RNA (dsRNA) virus belonging to the family Birnaviridae (7). Aquatic birnavirus has a worldwide distribution and can infect a range of species of fish and shellfish. IPNV is one of the most economically important viral pathogens of Atlantic salmon, causing problems in young fish and in smolts after transfer from freshwater to seawater. A better understanding of the IFN system and its mode of action against viruses might be useful in developing new strategies to control IPNV.
In the present work, the antiviral activity of Atlantic salmon Mx1 protein (ASMx1) against IPNV was studied by establishing Chinook salmon embryo (CHSE-214) cells expressing ASMx1 constitutively. ASMx1 expression was found to inhibit the virus-induced cytopathic effect (CPE) and viral protein synthesis and to reduce the transcription of viral RNA. These findings support the notion that fish Mx proteins have an important role in innate antiviral immunity. Moreover, the list of viruses inhibited by Mx proteins is extended to include dsRNA viruses.
MATERIALS AND METHODS
Cell cultures and virus.
CHSE-214 cells were grown in Eagle's MEM (EMEM) containing 100 μg of streptomycin per ml, 100 U of penicillin per ml, 2 mM l-glutamine, 1% nonessential amino acids, and 7.5% fetal bovine serum (FBS; EuroClone, Milan, Italy) in 5% CO2 at 20°C. Transfected cells were maintained in culture medium containing 500 μg of phleomycin D1 (Zeocin; Invitrogen, Groningen, The Netherlands) per ml. IPNV strain Sp, kindly provided by the Norwegian Institute of Fisheries and Aquaculture (Tromsø, Norway), was propagated in CHSE-214 cells at 15°C as described previously (16). Virus titers were calculated by the 50% tissue culture infective dose (TCID50) method (34).
Antibodies.
A polyclonal rabbit antibody (anti-RBTMx3) generated against a fragment of the rainbow trout Mx3 protein prepared as described previously (41) was used for the detection of Mx protein. The plasmid used to express the rainbow trout Mx3 protein fragment was a gift from Jo-Ann C. Leong (Department of Microbiology, Oregon State University, Corvallis). For the detection of IPNV proteins, Karen Elina Christie (Intervet Norbio, Bergen, Norway) kindly provided a polyclonal rabbit anti-IPNV serum raised against purified IPNV. The antibody gives a higher intensity for VP3 than for VP2 in Western blots, indicating that VP3 mainly contains linear epitopes while VP2 contains more conformation-dependent epitopes. The antiserum was diluted (1:3) in phosphate-buffered saline (PBS) and then incubated with a monolayer of Atlantic salmon TO cells for 2 h at 15°C. It was further purified through a HiTrap affinity column (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's instructions.
Generation of an ASMx1-expressing cell clone.
The ASMx1 gene (36) was cloned between the EcoRI and NotI sites of the eukaryotic expression vector pcDNA3.1/Zeo(+) (Invitrogen) so that it was under the control of the cytomegalovirus promoter. The gene for green fluorescent protein (GFP) was cloned into the same expression vector as a negative control. To obtain cells permanently expressing ASMx1 or GFP, monolayers of CHSE-214 cells seeded in six-well culture plates (5 × 105 cells per well) were transfected with 1 μg of plasmid pcDNA3.1/Zeo-ASMx1 or pcDNA3.1/Zeo-GFP together with 3 μl of Fugene (Roche, Indianapolis, Ind.) diluted in 30 μl of serum-free EMEM for 24 h. Transfected cells were selected in culture medium containing 500 μg of phleomycin D1 per ml and grown in monolayers. Resistant colonies were examined for Mx protein expression by Western blot analysis, and colonies positive for Mx protein expression were subjected to subcloning by limiting dilution. GFP-transfected cells were examined for GFP expression by using a fluorescence microscope (Nikon Diaphot TMD) with a fluorescein isothiocyanate filter.
IFN stimulation and transfection with poly(I-C).
As positive controls for antiviral activity and Mx protein expression, CHSE-214 cells grown in monolayers were incubated for 24 h with 1:200 dilutions of supernatants from HEK293 cells transfected with the Atlantic salmon IFN-α1 gene as described previously (35). As a positive control for reverse transcription (RT)-PCR analyses, CHSE-214 cells were transfected with poly(I-C) (Amersham Pharmacia Biotech), which is a potent inducer of alpha/beta IFN (16). Poly(I-C) was dissolved in 0.9% NaCl at a concentration of 0.5 mg per ml. CHSE-214 cells seeded in six-well culture plates (5 × 105 cells per well) were transfected with 0.1 μg of poly(I-C) together with 3 μl of Fugene dissolved in 30 μl of serum-free EMEM for 24 h. Fresh culture medium was added, and the cells were grown for 24 h before being harvested or infected.
Indirect immunofluorescence staining of Mx proteins.
Cells seeded in 96-well culture plates (2.5 × 104 cells per well) and grown for 3 days were fixed with 3% paraformaldehyde solution in PBS for 20 min. After being washed with PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min. The cells were washed once in PBS and blocked with 5% dry milk in PBS for 30 min before being incubated for 1 h with primary antibody dissolved in 0.5% dry milk. Anti-RBTMx3 was used to detect Mx protein expression. The cells were washed for 10 min with PBS and incubated with secondary antibody for 30 min. Alexa 594- goat anti-rabbit immunoglobulin G (Molecular Probes Europe BV, Leiden, The Netherlands) diluted 1:200 in PBS was used as the secondary antibody. Finally, the cells were washed for 10 min with PBS and examined by using a fluorescence microscope (Nikon Diaphot TMD) with a Texas red filter.
Cell growth curves.
Growth curves for the ASMx1-expressing clone as well as for GFP-expressing cells and untreated control CHSE-214 cells were obtained. Cells were seeded in 96-well culture plates (2 × 104 cells per well) in quadruplicate, and nuclei were counted daily in a Bürker chamber after the addition of lysis buffer (0.1 M citric acid, 1% Tween 20, 0.05% crystal violet). The experiment was done twice.
Infection with IPNV.
Cells grown in monolayers at 20°C for 3 days were washed once with EMEM, and IPNV diluted in EMEM without serum was added at a multiplicity of infection (MOI) of 0.1. Virus was allowed to adsorb for 1.5 h at 15°C. For cells grown in 96-well or 24-well culture plates, culture medium with 4% FBS was added to give a final concentration of 2% FBS. For cells grown in six-well culture plates, virus was removed before culture medium with 2% FBS was added. Infected cells were incubated in 5% CO2 at 15°C.
Analysis of antiviral activity.
A cytopathic effect (CPE) reduction assay (29) and a virus yield reduction assay were used to study the antiviral activity of the ASMx1-expressing clone. The CPE reduction assay measures the protection of cells against virus-induced lysis by calculating the percentage of surviving cells during virus infection. Cells were seeded in 96-well culture plates (2.5 × 104 cells per well); 12 parallel wells were infected with IPNV, while 4 parallel wells were kept as uninfected controls. At 3 days postinfection, when CPE was evident in untreated, infected control cells, the cells were washed once with PBS and fixed and stained for 10 min with 1% crystal violet in 20% ethanol. The cells were washed three times with distilled water before the stain was dissolved in 100 μl of 50% ethanol containing 0.05 M sodium citrate and 0.05 M citric acid. Dye absorbance was measured at 550 nm with a SpectraMax 190 (Molecular Devices). The results are presented as the percentage of surviving cells, whereby 100% represents the dye absorbance of uninfected cells treated in the same way as infected cells. For the virus yield reduction assay, culture supernatants from infected wells were pooled when CPE was evident and stored at −70°C until titration. Titration was done with CHSE-214 cells by the TCID50 method (34). Since viruses were not removed during infection, the amount of infectious units added was subtracted from the final titers to obtain the virus yield.
Western blot analyses of Mx and IPNV proteins.
Cells seeded in 24-well culture plates (2 × 105 cells per well) were washed once with PBS, lysed with 50 μl of sodium dodecyl sulfate (SDS) gel sample buffer (0.1 M Tris-HCl [pH 6.8], 0.2 M dithiothreitol, 2% SDS, 20% glycerol, 0.1% bromophenol blue), boiled for 5 min, and centrifuged for 5 min at 20,800 x g. Cell extract (15 μl) was applied to each lane and subjected to SDS- 10% polyacrylamide gel electrophoresis in a Bio-Rad II electrophoresis cell. MagicMark Western Standard (Invitrogen) was included on each gel for molecular mass estimation. Blotting, blocking, and antibody incubation were performed as described previously (17). Polyclonal rabbit anti-RBTMx3 antibody and polyclonal rabbit anti-IPNV antibody were used for the detection of Mx and IPNV proteins, respectively. Horseradish peroxidase-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) diluted 1:10,000 was used as the secondary antibody. Detection was performed by using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.). To ensure equal protein loading, membranes were stripped and immunostained for actin by using polyclonal antiactin antibody (Sigma, St. Louis, Mo.) diluted 1:500 as the primary antibody (17). To compare Mx protein expression levels, the densities of Mx protein bands and actin bands were measured by using NHI Image 1.62/ppc software. The values for the Mx protein bands were normalized to the actin band values.
RT-PCR assay for detecting IFN, Mx protein, and VP2 expression.
Cells were seeded in six-well culture plates (5 × 105 cells per well) in two parallel rows, and total RNA was isolated at selected times by using Trizol reagent (Invitrogen) in accordance with the manufacturer's instructions. RNA (1 μg) from each sample was reverse transcribed into cDNA at 42°C for 60 min. Synthesis was performed with 20 μl of buffer containing 50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, and 10 mM dithiothreitol (pH 8.3) and supplemented with 0.5 μg of random hexamers, 0.5 mM deoxynucleoside triphosphates, 20 U of RNasin, and 200 U of Moloney murine leukemia virus reverse transcriptase (all from Promega, Mannheim, Germany). Five percent of the resulting cDNA was used as a template in each PCR. A 220-bp fragment was amplified by PCR with Atlantic salmon IFN-specific primers (5′-ATAGATGATGGCGAGGTTGAGGAC-3′ and 5′-TCAACTTCTTGAAGTAGCGTTTCAG-3′) (35). The following conditions were applied: 96°C for 2 min and 34 cycles of 96°C for 20 s, 57°C for 20 s, and 72°C for 20 s. The identity of the PCR product was confirmed by DNA sequencing. Sequence analysis revealed 97% identity between the Chinook salmon and Atlantic salmon IFN genes in this region.
For analysis of the transcription of IPNV genes, primers with specificity for the VP2 segment (5′-GCGAATTCTGATTGGTCTGAGCACGC-3′ and 5′-ATGAATTCGAACCCCAGGACAAAGT-3′) gave rise to a 624-bp fragment under the following PCR conditions: 96°C for 2 min and 23 cycles of 96°C for 20 s, 57°C for 20 s, and 72°C for 30 s.
Samples were analyzed for Mx protein expression by amplification of a 222-bp product with primers which amplify both trout and Atlantic salmon Mx cDNAs (5′-TCGAGCTGAAGATGAAGAGGAAGAAAGA-3′ and 5′-CTGGCAGGTCGATGAGTGTGAGGT-3′). PCR was performed under the following conditions: 96°C for 2 min and 28 cycles of 96°C for 20 s, 65°C for 20 s, and 72°C for 20 s.
To confirm that all of the samples contained the same amount of cDNA, PCR was also performed with actin-specific primers (5′-CACTCAACCCCAAAGCCAACAGG-3′ and 5′-AAAGTCCAGCGCCACGTAGCACAG-3′) under the following conditions: 96°C for 2 min and 20 cycles of 96°C for 20 s, 68°C for 20 s, and 72°C for 20 s.
RESULTS
Generation of CHSE-214 cells constitutively expressing ASMx1.
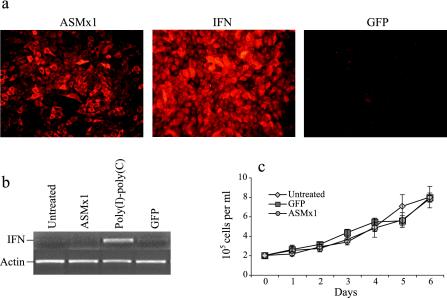
In order to investigate the antiviral effect of ASMx1, we wanted to establish a salmonid cell line expressing ASMx1 constitutively. This was achieved by transfecting CHSE-214 cells with a plasmid containing the phleomycin D1 resistance gene and the ASMx1 gene under the control of the cytomegalovirus promoter and subsequently selecting for phleomycin D1-resistant cells. CHSE-214 cells were chosen because they can be grown in single colonies and because they showed higher transfection rates than Atlantic salmon cell lines. Individual colonies were isolated, grown, and tested for the expression of Mx protein by Western blotting (data not shown). Positive colonies were subjected to subcloning by limiting dilution, and one clone was selected for further studies. CHSE-214 cells permanently transfected with GFP were generated as a negative control. CHSE-214 cells stimulated with recombinant Atlantic salmon alpha/beta IFN (35) were used as a positive control. Control cells and the ASMx1-transfected clone were analyzed for Mx protein expression by indirect immunofluorescence. As shown in Fig. 1a, the ASMx1-transfected clone consisted of a heterogeneous population of cells with variable Mx protein expression levels. As expected, the majority of CHSE-214 cells stimulated with recombinant IFN showed a relatively high level of expression of Mx protein, while GFP-transfected cells showed no expression of Mx protein. Both IFN-treated cells and cells transfected with ASMx1 appeared to express Mx protein in the cytoplasm (Fig. 1a).
FIG. 1.
Analysis of the ASMx1-expressing clone. (a) Mx protein expression was detected by an indirect immunofluorescence assay with a polyclonal antibody generated against rainbow trout Mx3 protein. The cells shown are CHSE-214 cells permanently transfected with ASMx1, CHSE-214 cells incubated with recombinant IFN for 24 h, and CHSE-214 cells permanently transfected with GFP. All cells were monolayers, and the percentage of ASMx1-expressing cells was calculated to be about 65%. (b) Expression of IFN, as determined by RT-PCR, in untreated CHSE-214 cells, the ASMx1-expressing clone, CHSE-214 cells transfected with poly(I-C) for 48 h, and CHSE-214 cells permanently transfected with GFP. Actin bands confirmed the presence of equal amounts of cDNA in all samples. (c) Growth curves for the ASMx1-expressing clone, GFP-expressing CHSE-214 cells, and untreated CHSE-214 cells. Cells seeded in quadruplicate at a confluence of 2 × 104 cells per well in 96-well plates were lysed and counted daily. Bars show standard errors. Passage 2 of the ASMx1-expressing clone was used in all experiments.
To exclude the possibility of IFN production by ASMx1-expressing cells, transcription of IFN was assessed by RT-PCR. CHSE-214 cells transfected with poly(I-C) served as a positive control for the induction of IFN, while GFP-expressing cells and untreated cells served as negative controls. Poly(I-C)-transfected cells gave rise to one PCR product (Fig. 1b), which was confirmed to originate from IFN transcripts by cloning and sequencing. In contrast, no IFN transcripts were detected in the ASMx1-expressing clone or the GFP-transfected cells and untreated cells. These results reveal that IFN is not induced in the ASMx1-expressing CHSE-214 clone. The influence of ASMx1 and GFP on cell growth was assessed (Fig. 1c), but no significant difference in the growth rates of transfected cells and untreated control cells was seen.
Protection against IPNV-induced CPE in the ASMx1-expressing clone.
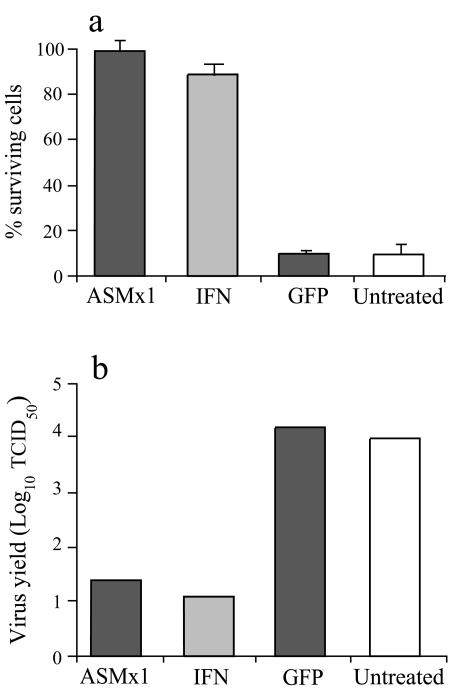
The CPE reduction assay (29) and the virus yield reduction assay were used to measure the antiviral activity of the ASMx1-expressing clone. CHSE-214 cells stimulated with recombinant IFN served as a positive control, while GFP-expressing cells and untreated CHSE-214 cells were included as negative controls. The ASMx1-expressing clone and control cells grown in 96-well culture plates were infected with IPNV. When untreated control cells showed distinct CPE, at about 3 days postinfection, supernatants were harvested and cells were fixed and stained with crystal violet. The stain was dissolved, and the OD550 was measured. The ASMx1-expressing clone and IFN-stimulated cells showed survival rates of 99 and 88%, respectively (Fig. 2a). GFP-expressing cells and untreated cells, on the other hand, showed a survival rate of only about 10%. The experiment was repeated three times, and small variations in protection were observed; however, the ASMx1-expressing clone always showed considerably higher survival rates than negative control cells, as did IFN-stimulated cells.
FIG. 2.
Antiviral activity of the ASMx1-expressing clone against IPNV. The ASMx1-expressing clone (passage 2), CHSE-214 cells treated with recombinant IFN for 24 h, CHSE-214 cells permanently transfected with GFP, and untreated CHSE-214 cells were seeded in 96-well plates and infected with IPNV (MOI, 0.1). (a) For the CPE reduction assay, the cells were fixed and stained with crystal violet in ethanol at 72 h postinfection, and the OD550 was measured. Results are presented as the percentage of surviving cells, calculated as the OD550 of infected cells (n = 12) divided by the OD550 of uninfected cells (n = 4). Bars show standard deviations. (b) The virus yield of the supernatants at 72 h postinfection was determined by the TCID50 method with CHSE-214 cells.
These results were confirmed by the virus yield reduction assay. Cell supernatants from infected cells were harvested before fixation, and the TCID50 was measured. As shown in Fig. 2b, virus yield was about 500-fold lower in the ASMx1-expressing clone and IFN-stimulated cells than in GFP-expressing and untreated control cells. These results indicate that Mx proteins have an antiviral effect against IPNV in vitro.
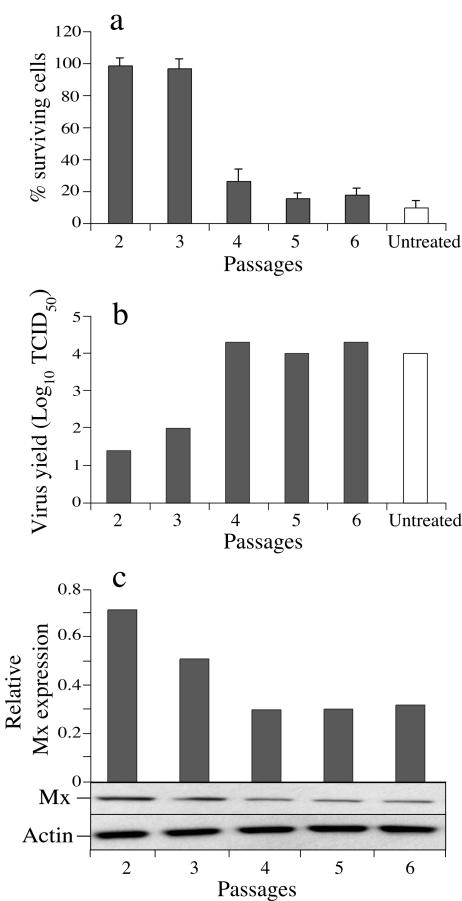
Unfortunately, the antiviral activity of the ASMx1-expressing clone was lost after a few passages in selective medium. As shown in Fig. 3a, passages 2 and 3 showed almost full protection against virus-induced CPE; in contrast, in passages 4 to 6, less than 30% of cells survived. These results were confirmed by more than a 100-fold-higher virus yield for passages 4 to 6 than for passages 2 and 3 (Fig. 3b). The virus yield for passages 4 to 6 was similar to that for untreated control cells. To investigate whether the loss of the antiviral effect was due to a reduction in Mx protein expression, the Mx protein level in each passage was assessed by Western blotting. ASMx1 expression in passages 4 to 6 was twofold lower than that in passage 2 (Fig. 3c). These experiments were repeated three times. The passage at which antiviral activity disappeared varied between the experiments but was always associated with decreased Mx protein expression. In contrast, no decrease in GFP expression was apparent during the passaging of GFP-transfected cells. Immunostaining of cells from later passages indicated reduced expression of ASMx1 in all cells rather than a high level of expression in a few cells (data not shown).
FIG. 3.
Reduction in antiviral activity and progressive loss of Mx protein expression in the ASMx1-expressing clone during selective growth. (a) For each passage (2 to 6), the ASMx1-expressing clone was infected with IPNV (MOI, 0.1). Untreated CHSE-214 cells were included as a control. For the CPE reduction assay, cells were fixed and stained with crystal violet in ethanol at 72 h postinfection, and the OD550 was measured. Results are presented as the percentage of surviving cells, calculated as the OD550 of infected cells (n = 12) divided by the OD550 of uninfected cells (n = 4). Bars show standard deviations. (b) The virus yield of the supernatants from infected cells was determined by the TCID50 method with CHSE-214 cells. (c) Cell extracts from the ASMx1-expressing clone grown in 24-well culture plates for 3 days were harvested for each passage (2 to 6) and analyzed for Mx protein and actin expression by Western blotting. Histograms express the density values of Mx protein bands normalized to the density values of the corresponding actin bands.
Decreased synthesis of virus proteins and viral RNA in the ASMx1-expressing clone infected with IPNV.
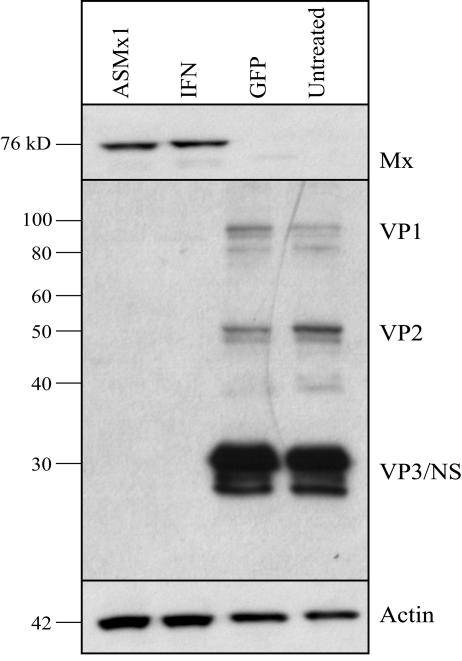
The inhibition of viral replication in the ASMx1-expressing clone was further confirmed by analysis of viral protein synthesis. Monolayers of the ASMx1-expressing clone in 24-well plates were infected with IPNV, and cell extracts were harvested at 48 h postinfection. CHSE-214 cells stimulated with recombinant IFN served as a positive control, and GFP-expressing cells and untreated CHSE-214 cells were included as negative controls. Western blotting of IPNV-infected cells showed the synthesis of viral proteins in both untreated cells and GFP-expressing cells (Fig. 4). In contrast, the synthesis of viral proteins was not detectable in the ASMx1-expressing clone or in IFN-stimulated cells at 48 h postinfection. These results indicated that IPNV protein synthesis was inhibited in the presence of ASMx1.
FIG. 4.
Inhibition of IPNV protein synthesis in the ASMx1-expressing clone. The ASMx1-expressing clone (passage 2), CHSE-214 cells treated with recombinant IFN for 24 h, CHSE-214 cells permanently transfected with GFP, and untreated CHSE-214 cells were seeded in 24 wells and infected with IPNV (MOI, 0.1). Cell extracts were harvested at 48 h postinfection and analyzed for the expression of IPNV proteins (VP1, VP2, VP3, and nonstructural protein [NS]) by immunoblotting. To show the expression of Mx protein and actin, the blot was stripped and incubated with anti-Mx protein antibody and with antiactin antibody, respectively. The experiment was done twice.
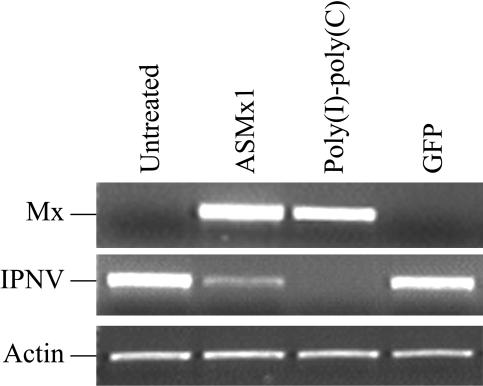
The influence of ASMx1 on viral transcription was examined by RT-PCR with primers specific for the VP2 segment of the IPNV genome. Total RNA was harvested from monolayers of the ASMx1-expressing clone at 24 h after infection with IPNV. Poly(I-C)-transfected cells were used as a positive control, and GFP-transfected cells and untreated CHSE-214 cells were used as negative controls. The expression of VP2 transcripts was markedly weaker in the ASMx1-expressing clone than in GFP-expressing cells and untreated cells at 24 h postinfection (Fig. 5). Poly(I-C)-transfected control cells, on the other hand, showed no VP2 band. An examination of Mx protein transcripts revealed the presence of Mx proteins in both the ASMx1-expressing clone and the positive control but not in the two negative controls. Reduced transcription of VP2 in the ASMx1-expressing clone was also observed at an MOI of 1 (data not shown). These results showed that the transcription of VP2 was reduced in the presence of ASMx1. The results are representative of three independent experiments.
FIG. 5.
Decrease in IPNV transcription in the ASMx1-expressing clone. Untreated CHSE-214 cells, the ASMx1-expressing clone (passage 2), CHSE-214 cells transfected with poly(I-C) for 24 h, and CHSE-214 cells permanently transfected with GFP, all grown as monolayers in six-well plates, were infected with IPNV (MOI, 0.1). Total RNA was harvested at 24 h postinfection, and the expression of VP2 transcripts of IPNV and Mx protein transcripts was analyzed by semiquantitative RT-PCR. The amount of cDNA in each sample was verified by actin-specific RT-PCR.
DISCUSSION
The anti-IPNV activity of Atlantic salmon alpha/beta IFN is well established (17, 31, 35), but the mechanisms involved have not been studied earlier. A correlation between inhibition of IPNV and Mx protein expression in IFN-stimulated salmon cells has, however, been observed (16, 31). In the present work, CHSE-214 cells constitutively expressing ASMx1 were used to examine the antiviral properties against IPNV. Several lines of evidence confirm the antiviral activity of ASMx1 against IPNV: (i) protection against virus-induced lysis shown by the CPE reduction assay, (ii) reduction in virus yield, (iii) inhibition of expression of viral proteins, and (iv) reduction in transcripts of the virus capsid protein VP2. Since IFN transcripts were not detectable in ASMx1-transfected CHSE-214 cells, endogenous IFN is unlikely to contribute to the protective effect of ASMx1.
The antiviral activity of fish Mx proteins has been uncertain because cells transfected with three rainbow trout Mx genes were found not to inhibit replication of the rhabdovirus IHNV (41). However, a recent study reported that replication of fish rhabdoviruses was reduced in a fish cell line transfected with Japanese flounder Mx protein (6). The present work further adds to the body of evidence which suggests that antiviral activity is a major functional role of vertebrate Mx proteins.
ASMx1 was found to be expressed in the cytoplasm (Fig. 1a). This finding is in accordance with the antiviral effect induced against IPNV, which replicates in the cytoplasm (7). Endogenously expressed Atlantic salmon Mx proteins were also located in the cytoplasm and not in the nucleus (unpublished data). This finding indicates that Atlantic salmon express Mx proteins only in the cytoplasm, as is the case for chickens. Human MxA is expressed in the cytoplasm and has a wide antiviral spectrum against different viruses, irrespective of their intracellular replication site (12). Chicken Mx protein was recently found to inhibit both influenza virus and VSV replicating in the nucleus and cytoplasm (19). In rodents, on the other hand, both nuclear and cytoplasmic Mx protein forms exist, and their antiviral specificity correlates with their subcellular location. Mx1 is located in the nucleus and inhibits orthomyxoviruses, which replicate in the nucleus, while Mx2 accumulates in the cytoplasm and inhibits rhabdovirus and bunyavirus, which replicate in the cytoplasm (25). To further investigate the antiviral spectrum of ASMx1, it would be of interest to examine antiviral properties against fish rhabdoviruses such as viral hemorrhagic septicemia virus and IHNV and fish orthomyxoviruses such as infectious salmon anemia virus.
The mechanism by which ASMx1 inhibits IPNV proliferation is still not known although viral transcripts were reduced in the ASMx1-expressing clone. Mammalian Mx proteins appear to interfere with viral replication at different stages depending on host cell and virus combinations. Mouse Mx1 protein interferes with primary transcription of orthomyxviruses, probably by interaction with the polymerase (32, 40). Human MxA protein appears to interfere with different stages of replication, depending of the virus species and the site of replication. The rhabdovirus VSV and measles virus are inhibited at the level of primary transcription (37-39). In the case of the orthomyxovirus Thogoto virus, MxA binds to the nucleoprotein, preventing the infecting particle from entering the nucleus and thereby blocking transcription of the viral genome (20, 21). MxA is also found to interact with the nucleoprotein of La Crosse virus and other bunyaviruses, causing the depletion of nucleoprotein from the replication site near the Golgi apparatus (22). Through physical interactions with viral nucleoproteins, MxA may thus direct viral components to other locations, where they become unavailable for the generation of new virus particles (13). For hepatitis B virus, no interaction between nucleocapsids and MxA was, however, detected. MxA was instead found to inhibit the export of viral RNAs from the nucleus (11). Certain viruses, such as Semliki Forest virus and measles virus, are inhibited by MxA in a cell type-specific manner, suggesting that unknown cellular factors may influence antiviral activity (23, 37). In future studies, it will be of particular interest to study whether ASMx1 binds to any IPNV proteins.
The antiviral effect of ASMx1-transfected CHSE-214 cells was lost after several passages in selective medium (Fig. 3a and b), raising the question as to whether this was due to a decrease in Mx protein expression. A progressive decrease in ASMx1 expression with passaging was indeed documented (Fig. 3c). The possibility that high-level expression of ASMx1 interferes with cell growth was excluded by the demonstration of similar growth rates in ASMx1- and GFP-expressing cells and untreated control cells. There is, however, a possibility that ASMx1 might have disturbing effects on cell functions, causing Mx gene expression to be suppressed by silencing or other mechanisms. This hypothesis is supported by the fact that the GFP gene and the gene for phleomycin D1 resistance remained expressed after several passages. The fact that ASMx1 was expressed at different levels in the permanently transfected cells adds to the possibility that cells with low levels of expression are selected for during passaging. Incompatibility between high levels of constitutive Mx protein expression and cell function has been reported in several other cases. Permanently transfected chicken cell lines expressing high levels of chicken Mx protein could not be established, suggesting low metabolic stability of chicken Mx protein or incompatibility with cell proliferation (5). Moreover, attempts to establish transgenic mice expressing Mx protein showed that constitutive expression might be deleterious, eliminating high-level expression of the Mx gene (2). In addition, overexpression of MxA in patients with Fanconi anemia induces apoptosis, indicating that a loss of control of Mx protein expression is deleterious for the organism (26).
The fact that cell passages with no antiviral activity still expressed ASMx1 (Fig. 3), although in smaller amounts, suggests that Mx protein expression must be at a certain level for full inhibition of IPNV replication to occur. Also, for human MxA it has been shown that the extent of viral inhibition correlates with the amount of MxA protein in infected cells (9, 23). Inhibition of IPNV transcription was complete in poly(I-C)-transfected cells, while the ASMx1-expressing clone showed only reduced viral transcription (Fig. 5). This finding may be explained by a more uniform expression of Mx protein in poly(I-C)-stimulated cells. In addition, IFN-inducible genes other than the Mx gene are likely to participate in the antiviral activity of poly(I-C)- and IFN-stimulated cells.
In conclusion, the present work demonstrates that the antiviral activity of Mx protein is not restricted to mammals and birds but also includes fish. We display evidence for antiviral activity of ASMx1 against IPNV, providing an explanation for the protective effect induced against IPNV by IFN in salmonid cells. Additional studies are, however, required to further define the mechanism(s) of action of ASMx1, and the antiviral spectrum of ASMx1 has yet to be explored. Moreover, the list of viruses inhibited by Mx proteins is extended to include viruses with a dsRNA genome.
REFERENCES
- 1.Altmann, S. M., M. T. Mellon, D. L. Distel, and C. H. Kim. 2003. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 77:1992-2002. [DOI] [PMC free article] [PubMed]
- 2.Arnheiter, H., M. Frese, R. Kambadur, E. Meier, and O. Haller. 1996. Mx transgenic mice—animal models of health. Curr. Top. Microbiol Immunol. 206:119-147. [DOI] [PubMed] [Google Scholar]
- 3.Asano, A., J. H. Ko, T. Morozumi, N. Hamashima, and T. Watanabe. 2002. Polymorphisms and the antiviral property of porcine Mx1 protein. J. Vet. Med. Sci. 64:1085-1089. [DOI] [PubMed] [Google Scholar]
- 4.Bazzigher, L., A. Schwarz, and P. Staeheli. 1993. No enhanced influenza virus resistance of murine and avian cells expressing cloned duck Mx protein. Virology 195:100-112. [DOI] [PubMed] [Google Scholar]
- 5.Bernasconi, D., U. Schultz, and P. Staeheli. 1995. The interferon-induced Mx protein of chickens lacks antiviral activity. J. Interferon Cytokine Res. 15:47-53. [DOI] [PubMed] [Google Scholar]
- 6.Caipang, C. M., I. Hirono, and T. Aoki. 2003. In vitro inhibition of fish rhabdoviruses by Japanese flounder, Paralichthys olivaceus Mx. Virology 317:373-382. [DOI] [PubMed] [Google Scholar]
- 7.Dobos, P. 1995. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 5:25-54. [Google Scholar]
- 8.Ellis, A. E. 2001. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 25:827-839. [DOI] [PubMed] [Google Scholar]
- 9.Frese, M., G. Kochs, U. Meier-Dieter, J. Siebler, and O. Haller. 1995. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J. Virol. 69:3904-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 11.Gordien, E., O. Rosmorduc, C. Peltekian, F. Garreau, C. Brechot, and D. Kremsdorf. 2001. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 75:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Technol. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 13.Haller, O., and G. Kochs. 2002. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 14.Horisberger, M. A., and M. C. Gunst. 1991. Interferon-induced proteins: identification of Mx proteins in various mammalian species. Virology 180:185-190. [DOI] [PubMed] [Google Scholar]
- 15.Horisberger, M. A., P. Staeheli, and O. Haller. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc. Natl. Acad. Sci. USA 80:1910-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, I., R. Larsen, and B. Robertsen. 2002. An antiviral state induced in Chinook salmon embryo cells (CHSE-214) by transfection with the double-stranded RNA poly I:C. Fish Shellfish Immunol. 13:367-378. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, I., and B. Robertsen. 2002. Effect of double-stranded RNA and interferon on the antiviral activity of Atlantic salmon cells against infectious salmon anemia virus and infectious pancreatic necrosis virus. Fish Shellfish Immunol. 13:221-241. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, V., and B. Robertsen. 2000. Cloning of an Mx cDNA from Atlantic halibut (Hippoglossus hippoglossus) and characterization of Mx mRNA expression in response to double-stranded RNA or infectious pancreatic necrosis virus. J. Interferon Cytokine Res. 20:701-710. [DOI] [PubMed] [Google Scholar]
- 19.Ko, J. H., H. K. Jin, A. Asano, A. Takada, A. Ninomiya, H. Kida, H. Hokiyama, M. Ohara, M. Tsuzuki, M. Nishibori, M. Mizutani, and T. Watanabe. 2002. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 12:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kochs, G., and O. Haller. 1999. GTP-bound human MxA protein interacts with the nucleocapsids of Thogoto virus (Orthomyxoviridae). J. Biol. Chem. 274:4370-4376. [DOI] [PubMed] [Google Scholar]
- 21.Kochs, G., and O. Haller. 1999. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl. Acad. Sci. USA 96:2082-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochs, G., C. Janzen, H. Hohenberg, and O. Haller. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc. Natl. Acad. Sci. USA 99:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landis, H., A. Simon-Jodicke, A. Kloti, C. Di Paolo, J. J. Schnorr, S. Schneider-Schaulies, H. P. Hefti, and J. Pavlovic. 1998. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J. Virol. 72:1516-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. Y., I. Hirono, and T. Aoki. 2000. Cloning and analysis of expression of Mx cDNA in Japanese flounder, Paralichthys olivaceus. Dev. Comp. Immunol. 24:407-415. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. H., and S. M. Vidal. 2002. Functional diversity of Mx proteins: variations on a theme of host resistance to infection. Genome Res. 12:527-530. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., and H. Youssoufian. 1997. MxA overexpression reveals a common genetic link in four Fanconi anemia complementation groups. J. Clin. Investig. 100:2873-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenmann, J. 1962. Resistance of mice to mouse adapted influenza A virus. Virology 16:203-204. [DOI] [PubMed] [Google Scholar]
- 28.Long, S., M. Wilson, E. Bengten, L. Bryan, L. W. Clem, N. W. Miller, and V. G. Chinchar. 2004. Identification of a cDNA encoding channel catfish interferon. Dev. Comp. Immunol. 28:97-111. [DOI] [PubMed] [Google Scholar]
- 29.Meager, A. 1987. Quantification of interferons by antiviral-assay and their standarization, p. 129-147. In M. J. Clemens, A. G. Morris, and A. J. H. Gearing (ed.), Lymphokines and interferons: a practical approach. IRL Press, Oxford, England.
- 30.Meier, E., G. Kunz, O. Haller, and H. Arnheiter. 1990. Activity of rat Mx proteins against a rhabdovirus. J. Virol. 64:6263-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nygaard, R., S. Husgard, A. I. Sommer, J. A. Leong, and B. Robertsen. 2000. Induction of Mx protein by interferon and double-stranded RNA in salmonid cells. Fish Shellfish Immunol. 10:435-450. [DOI] [PubMed] [Google Scholar]
- 32.Pavlovic, J., O. Haller, and P. Staeheli. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 66:2564-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plant, K. P., and R. L. Thune. 2004. Cloning and characterisation of a channel catfish (Ictalurus punctatus) Mx gene. Fish Shellfish Immunol. 16:391-405. [DOI] [PubMed] [Google Scholar]
- 34.Reed, L., and H. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 35.Robertsen, B., V. Bergan, T. Rokenes, R. Larsen, and A. Albuquerque. 2003. Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 23:601-612. [DOI] [PubMed] [Google Scholar]
- 36.Robertsen, B., G. Trobridge, and J. A. Leong. 1997. Molecular cloning of double-stranded RNA inducible Mx genes from Atlantic salmon (Salmo salar L.). Dev. Comp. Immunol. 21:397-412. [DOI] [PubMed] [Google Scholar]
- 37.Schneider-Schaulies, S., J. Schneider-Schaulies, A. Schuster, M. Bayer, J. Pavlovic, and V. ter Meulen. 1994. Cell type-specific MxA-mediated inhibition of measles virus transcription in human brain cells. J. Virol. 68:6910-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwemmle, M., K. C. Weining, M. F. Richter, B. Schumacher, and P. Staeheli. 1995. Vesicular stomatitis virus transcription inhibited by purified MxA protein. Virology 206:545-554. [DOI] [PubMed] [Google Scholar]
- 39.Staeheli, P., and J. Pavlovic. 1991. Inhibition of vesicular stomatitis virus mRNA synthesis by human MxA protein. J. Virol. 65:4498-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stranden, A. M., P. Staeheli, and J. Pavlovic. 1993. Function of the mouse Mx1 protein is inhibited by overexpression of the PB2 protein of influenza virus. Virology 197:642-651. [DOI] [PubMed] [Google Scholar]
- 41.Trobridge, G. D., P. P. Chiou, and J. A. Leong. 1997. Cloning of the rainbow trout (Oncorhynchus mykiss) Mx2 and Mx3 cDNAs and characterization of trout Mx protein expression in salmon cells. J. Virol. 71:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trobridge, G. D., and J. A. Leong. 1995. Characterization of a rainbow trout Mx gene. J. Interferon Cytokine Res. 15:691-702. [DOI] [PubMed] [Google Scholar]
- 43.Yap, W. H., A. Tay, S. Brenner, and B. Venkatesh. 2003. Molecular cloning of the pufferfish (Takifugu rubripes) Mx gene and functional characterization of its promoter. Immunogenetics 54:705-713. [DOI] [PubMed] [Google Scholar]