Abstract
Bluetongue virus is a large and structurally complex virus composed of three concentric capsid layers that surround 10 segments of a double-stranded RNA genome. X-ray crystallographic analysis of the particles without the outer capsid layer has provided atomic structural details of VP3 and VP7, which form the inner two layers. However, limited structural information is available on the other five proteins in the virion—two of which are important for receptor recognition, hemagglutination, and membrane interaction—are in the outer layer, and the others, important for endogenous transcriptase activity are internal. Here we report the electron cryomicroscopy (cryo-EM) reconstruction of the mature particle, which shows that the outer layer has a unique non-T = 13 icosahedral organization consisting of two distinct triskelion and globular motifs interacting extensively with the underlying T = 13 layer. Comparative cryo-EM analysis of the recombinant corelike particles has shown that VP1 (viral polymerase) and VP4 (capping enzyme) together form a flower-shaped structure attached to the underside of VP3, directly beneath the fivefold axis. The structural data have been substantiated by biochemical studies demonstrating the interactions between the individual outer and inner capsid proteins.
Bluetongue virus (BTV) is an economically important member of the Orbivirus genus in the family Reoviridae. This is a large family and contains important viruses isolated from vertebrates (including humans), invertebrates, and plants. BTV infects mainly ruminants and is transmitted by insect vectors of the Culicoides species.
BTV is a large (∼850-Å-diameter) and structurally complex virus composed of three concentric capsid layers that surround 10 segments of a double-stranded RNA (dsRNA) genome. The outer layer, composed of VP2 (111 kDa) and VP5 (∼59 kDa), is removed during the initial stages of the viral life cycle revealing the transcriptionally competent inner capsid termed the “core” particle. The outer layer of the core particle is composed of 260 trimers of VP7 (∼38 kDa) organized on a T = 13 icosahedral lattice. This VP7 layer interacts with the underlying innermost layer made from 120 copies of VP3 (∼103 kDa) arranged as 60 dimers on a T = 1 icosahedral lattice (10). Such a unique icosahedral organization indeed appears to be a common feature of the dsRNA viruses (26). The VP3 layer houses the segmented genome as well as three minor structural proteins: VP1, an RNA-dependent RNA polymerase (1); VP4, a methyl transferase (28, 30); and VP6, a helicase (18, 33). The transcriptionally competent core particles have been extensively studied both by electron cryomicroscopy (cryo-EM) and X-ray crystallographic techniques (9-11). Although these studies provided detailed structural description of the VP7 and the VP3 layers, little information was gleaned about the endogenous transcription enzyme complex of the virus.
In contrast to the core particles, structural studies on the intact virions are more difficult as a result of the labile nature of the outer capsid. Both intact BTV virions and the recombinant virus-like particles (VLPs) formed by the coexpression of outer layer proteins VP5 and VP2 along with the core proteins VP3 and VP7 have been studied at a very low resolution (∼40 Å) by cryo-EM techniques (14-16). These have revealed that the outer capsid is composed of 120 globular regions and 60 triskelion structures. However, at the resolution of ∼40 Å, interpretation of the precise juxtaposition and molecular interactions between the inner and outer layers was necessarily limited. It has been previously proposed that VP2 protein form the triskelion motifs of the virus while VP5 has the globular configuration (15). It is noteworthy that VP2 possesses the virus hemagglutination and neutralization activity, and is the cellular receptor binding protein (12). VP2 is the most variable protein of the seven BTV capsid proteins across the 24 BTV serotypes and thus is likely one of the most accessible proteins of BTV (22). In contrast to VP2, VP5 is reasonably well conserved and has recently been shown to be the membrane penetration protein and possesses membrane fusion-like activity (6, 13).
In this report, we have used cryo-EM techniques to obtain higher resolution structure (∼24 Å) of the intact virus to delineate the structural features of the outer capsid proteins and their interactions with the underlying VP7 layer by docking the X-ray structure of the VP7. With the availability of the X-ray structure of the BTV core particle, the present cryo-EM study improves upon previous reports considerably and reveals previously unseen details in the structure of the BTV particle. It was previously thought that there were very few connections between the outer and the inner layers, as the outer capsid is so easily removed. However, our studies show that the outer layer proteins interact extensively with the VP7 layer. To identify the locations of the two of the enzymes that are required for the endogenous transcription activity of the core particles, we have expressed recombinant core-like particles (CLPs) (formed of VP3 and VP7) with and without the minor proteins VP1 and VP4. The cryo-EM analysis of these particles has allowed us to unambiguously identify the locations of these internal proteins and examine their interactions with the inner VP3 shell.
MATERIALS AND METHODS
Virion preparation.
U.S. prototype BTV serotype 10 was produced in BHK 21 cell monolayers using previously described methods (23).
Synthesis and purification of recombinant CLPs.
Spodoptera frugiperda (Sf) cells were grown in suspension or monolayer cultures at 28°C in SF900II serum-free medium (GibcoBRL). For the synthesis of CLPs, Sf 9 cell cultures were infected with recombinant baculovirus expressing VP3 and VP7, in combination with recombinant baculoviruses expressing VP1 and VP4 using a multiplicity of infection of 5 PFU per cell. CLPs were recovered from infected cells and purified from cell debris by CsCl gradient centrifugation as described previously (7). The presence of each protein was analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10%PAGE) and Western blotting using anti-BTV-10 polyclonal antibodies as well as by EM.
Radiolabeling of baculovirus-infected insect cells and immunoprecipitation.
Sf 9 cells infected with recombinant baculoviruses at a multiplicity of infection of 5 were incubated in SF900 serum-free media at 28°C. After 30 h postinfection cells were pulsed with 100 μCi of [35S]Met/Cys (NEN) for 2 h. Cells were then harvested, washed and lysed with radio-immunoprecipitation (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM EDTA), incubated on ice for 15 min, and centrifuged at 13,000 × g. Cell lysates were incubated separately with monoclonal or monospecific polyclonal antibodies to individual BTV proteins as indicated and the complexes were pulled down with protein A-agarose beads (Pierce Biotechnology, Rockford, Ill.), washed with RIPA buffer, analyzed by SDS-PAGE and autoradiographed.
In vitro transcription-translation.
In vitro proteins complexes were generated using a TNT T7 Quick Coupled Transcription/Translation System (Promega Corporation, Madison, Wis.) following the manufacturer's procedure. In brief, plasmid DNA constructs in which BTV VP2, VP5, and VP7 genes are under the control of T7 promoter were used as DNA templates to generate the protein complexes. For each plasmid 0.5 μg was used in the reaction mix containing 20 μl of TNT T7 Quick Master Mix, 2 μl of [35S]methionine (10 mCi/ml), and 3 μl of nuclease-free water. The amount of each protein synthesized in vitro was first verified by analyzing an aliquot of each reaction on a SDS-10% PAGE. Each reaction mix was incubated for 1 h at 30°C and 10 μl of the each reaction mix were incubated with protein A beads already conjugated with BTV antibodies for the immunoprecipitation assay.
Conventional electron microscopy.
Purified CLPs and intact virions were first examined for concentration and particle integrity using conventional negative staining techniques. The 5-μl volume of particles, resuspended in water, was absorbed onto carbon-coated copper 400-mesh EM grids for 15 min, washed with water, and stained with 1% (wt/vol) uranyl acetate. Grids were examined in a Hitachi H-7000 electron microscope at 75 kV.
Cryo-EM.
Specimen preparation for cryo-EM was carried out using standard procedures (4). Images of the specimen embedded in vitreous ice were recorded in a JEOL 1200 electron cryomicroscope, using a 100-kV electron beam at a dose of 5e−/Å2 and at a magnification of ×30,000. From each region, a focal pair was recorded with intended defocus values of 1.2 and 2.4 μm. The BTV cores were imaged with a 400 kV JEOL 4000 electron cryomicroscope at a magnification of ×30,000 using spot scan procedures as described previously (11).
Three-dimensional structural analysis.
Micrographs, selected based on particle concentration, quality of ice, and appropriate defocus, were digitized on a Zeiss SCAI microdensitometer (Carl Zeiss, Inc., Englewood, Colo.), using a 7-μm step size. The pixels were then averaged to give a 14-μm step size that corresponded to 4.67 Å/pixel in the object. Virions, cores and baculovirus expressed particles were boxed with a pixel area of 256 by 256. Determination of the orientational parameters (3), their refinement (8), and the three-dimensional reconstructions (3) were carried out using the ICOS Toolkit software suite (21). The reconstructions of the various particles were computed to a resolution within the first zero of the contrast transfer function (CTF) of the corresponding micrograph. The defocus values as determined from CTF ring positions in the sum of particle Fourier transforms ranged from 1.2 to 1.48 μm. The reconstructions were corrected for the effects of the CTF using procedures described earlier (39). The final resolution for each reconstruction was determined by Fourier ring correlation analysis (35). Contour levels in each reconstruction were chosen to represent equal volume between the radii ∼234 and ∼270 Å (which contains mass common to all the reconstructions).
Fitting the X-ray coordinates of VP7 into the native virion.
The VP7 from the X-ray crystallographic structure of core (10) was docked into the cryo-EM map of the whole virion using the same procedures as described earlier (11) and visualized using the program O (17).
RESULTS
Cryo-EM.
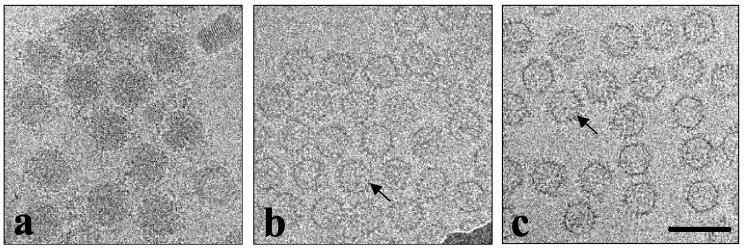
Micrographs of unstained, frozen hydrated BTV (Fig. 1a), and baculovirus expressed CLPs (Fig. 1b and c) are shown. The intact virions have a diameter of ∼820 Å and have a diffuse periphery as observed previously (15, 23, 36). As in the images of core particles, the CLPs exhibit a similar bristly appearance due to projections of VP7 trimers emanating from the inner shell (5, 7, 34). However, in contrast to core particle, CLPs clearly appear empty because of the absence of the genome. The CLPs have been shown to be very similar in size and morphology to native cores (7, 14). Observations of the cryo-electron micrographs show no visible differences between the CLPs containing VP1 and/or VP4 (Fig. 1c) with those that contain neither of these proteins (Fig. 1b).
FIG. 1.
Cryo-EM images of BTV virions (a) and baculovirus-expressed CLPs (VP3/7 [b] and VP3/7/1/4 [c]) embedded in vitreous ice. Arrow indicates the knobby protrusions of VP7. Bar, 1,000 Å.
Image processing.
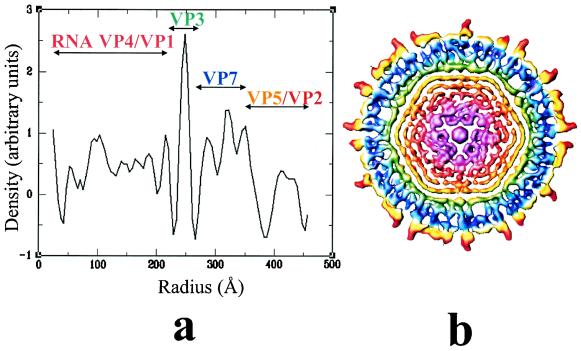
The digitized micrographs for each preparation were used for image reconstructions. Three-dimensional reconstruction of the native virions to ∼24-Å resolutions was computed using 67 particles from a single micrograph. The cores were computed to the same resolution using 91 particles. The baculovirus-expressed CLPs were each reconstructed to a resolution of ∼26 Å from 112 particles (VP1, VP3, and VP7), 105 particles (VP3, VP4, and VP7), and 117 particles (VP1, VP3, VP4, and VP7). In each case, the orientations for the virions, core particles and baculovirus expressed particles, adequately represented the icosahedral asymmetric unit. It was found that 95% of the inverse Eigen values were below 0.1, further indicating sufficient sampling of the orientations in the reconstructions at the specified resolution. The radial density profile (Fig. 2a) computed from the native reconstruction, except for the peak between the radii 380 and 450 Å, is very similar to that seen in core reconstruction (see Fig. 3 in Grimes et al. [11]). Thus, it is clear that the outer layer extends from a radius of 380 Å to ∼440 Å. The intact virion has a major peak at a radius of ∼248 Å. Based on the previous structural studies on the cores, this peak between the radii 230 and 270 Å is due to the VP3 layer, and peaks between the radii 270 and 350 Å are due to the VP7 layer. Immediately inside the VP3 shell of all the particle types, except for CLPs, is a series of peaks spaced at ∼26 Å, corresponding to the internal concentric layers of density (Fig. 2b).
FIG. 2.
Radial distribution of density in the virion reconstruction. (a) Radial density profile computed from the virion reconstruction. (b) Central cross section from the virion reconstruction, perpendicular to icosahedral threefold axes, color-coded as indicated in panel a.
FIG. 3.
Virion and core structures. (a) Surface-shaded representation of the virion reconstruction viewed along the icosahedral threefold axis. The structure is radially color-coded as in Fig. 1b. (b) Close-up view of the virion structure showing the two structural motifs those constitute the outer layer. The triskelion propellers are shown in red, and the globular domains are shown in yellow. (c) Surface representation of the core reconstruction. Three of the icosahedral fivefold axes that define a facet, locations of type II and III channels, and five types of quasi-equivalent VP7 trimers (P, Q, R, S, and T) are indicated. (d) Virion reconstruction, at approximately the same outer radius of the core structure, showing the triangle-shaped feature at the type II and type III channels. A set of three fivefold axes as in panel c is also indicated.
Unique organization of the outer capsid of BTV.
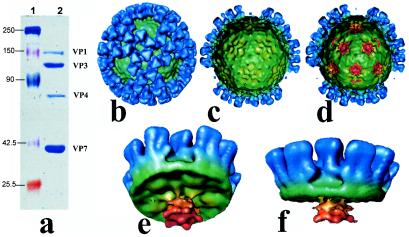
The outer capsid of the BTV virion exhibits a unique organization made from two distinct motifs, a triskelion and a globular motif. Sixty triskelion densities and 120 globular densities form the outer layer between 380- and 440-Å radii (Fig. 3a and b). Thus, unlike other notable members of the Reoviridae family like rotaviruses and reoviruses, the outer capsid of this virus does not conform to a T = 13 icosahedral symmetry. The most external part of the outer capsid is the propeller-shaped triskelion motif. Each blade of the propeller is ∼75 Å in length and 28 Å wide. At the terminal tip of each blade, the molecule broadens out to 60 Å wide and at this point, bends upwards perpendicular to the plane of the virus. These bent tips give the entire virion a diameter of ∼880 Å and extend from the main body of the particle by 30 Å. Interspersed between the triskelions and lying more internally between 380 and 420 Å radii are the globular densities of ∼60 Å in diameter. These are also entirely exposed in the virion.
Interactions between the outer capsid layer and the VP7 layer.
The structural organization of the outer layer represents a drastic mismatch with the underlying VP7 layer. In the core reconstruction, as in the X-ray structure, the VP7 trimers, organized on a T = 13 icosahedral lattice, form pentameric and hexametric rings defining three types of channels (Fig. 3c). These trimers are distinct with a triangular shaped top portion, and at lower radii, they are connected together by densities at the local and strict 2-fold axes of the T = 13 lattice. Based on their quasi-equivalent locations on the T = 13 lattice, the VP7 trimers are classified into five types (Fig. 3c).
With respect to the T = 13 VP7 layer, the triskelion densities of the outer layer sit right above the Q type VP7 trimer, whereas the globular densities sit right above the type II and type III channels (Fig. 3a, c, and d). The triskelion densities are attached to the inner capsid layer by their underside in four places. The base of the triskelion interacts with the Q type VP7, whereas the propeller-like arms make three other connections with the P, R, and S VP7 trimers (Fig. 3b). Thus, each triskelion motif essentially interacts with four VP7 trimers. All the VP7 molecules of the core are covered by the connections that are made with the tips of the propeller except the VP7 at the icosahedral threefold axis (T type). The top of the VP7 trimer at this position is thus clearly exposed to the exterior.
Examination of the virion reconstruction at the radii that correspond to the VP7 layer in the core and CLPs clearly showed differences. In the virion reconstruction, in addition to the densities that correspond to the VP7 trimers, clearly there are additional densities. The channels at the two local sixfold axes (type II and III) are filled with triangle-shaped densities that are connected to the globular morphological units seen in the outer layer (Fig. 3d). Each triangle-shaped density interacts with five out of the six surrounding VP7 trimers. Thus, there are three distinct density features, which can be attributed to the outer layer proteins: triskelion and the globular features that are apparent from the outside and the inner triangle-shaped features that fill the channels in the VP7 layer.
VP7 residues that interact with the outer layer proteins.
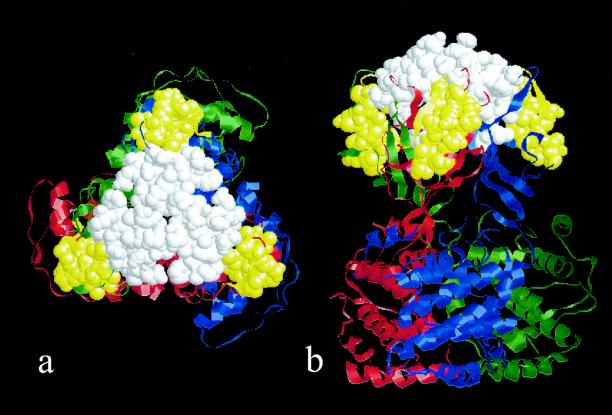
To further understand which residues in the VP7 are involved in the interactions with outer layer, we docked the X-ray of the VP7 trimer in to the virion reconstruction. The real space correlation coefficient after the fitting was about 0.76. At the precise radius (approximated by the pixel size) that corresponds to the top of VP7 trimers in the core (or CLP) reconstruction, the densities in the virion reconstruction due to the VP7 trimers and the outer layer proteins coalesce, making delineation of the boundaries between VP7 and outer layer proteins difficult. However, at a radius one pixel lower, the VP7 densities are clearly identifiable. From the virion reconstructions it is clear that the outer layer proteins make extensive contacts with both the top and the sides of the VP7 trimers. The triskelion density of the outer capsid interacts exclusively with the upper flattish surface of the VP7 trimer, which includes amino acid residues 141 to 143, 164 to 166, 195 to 205, 238 to 241 of the VP7 (Fig. 4). The globular unit makes contacts with the sides of VP7 trimers, which face type II or III channels. The interactions extend from a radius of 330 to 350 Å and include residues 168 to 173, 210 to 215, and 226 to 234 of the distal β-barrel domain of VP7 (Fig. 4). The inner triangle-shaped density interacts mainly with the lower portion of the β-barrel domain.
FIG. 4.
VP7 residues involved in interactions with outer layer proteins. (a) Top head-on view. (b) A side view. The X-ray structure of the VP7 trimer is shown in RIBBON representation. The trimeric subunits are colored in red, blue, and green. The locations on the VP7 where the triskelion motifs interact are shown in white and where the globular domains interact is shown in yellow.
Biochemical characterization of the interactions between outer layer proteins and VP7.
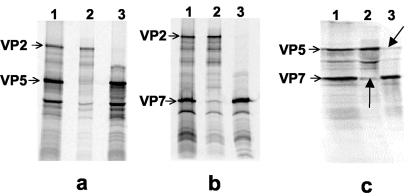
To confirm the structural observations described above, we coexpressed different combinations of VP2, VP5, and VP7, such as VP2-VP5, VP5-VP7, and VP2-VP7, by an in vitro transcription-translation as described in Materials and Methods. The in vitro generated proteins were subsequently used in an immunoprecipitation assay where complexes were pulled down using different antibodies raised against BTV VP2, VP5, and VP7 and analyzed by SDS-10% PAGE. Both VP2 and VP5 showed interactions with VP7 (Fig. 5b and c), and VP2-VP7 or VP5-VP7 complexes could easily be pulled down by either VP7 monoclonal antibody or by VP2 or VP5 polyclonal antibodies (Fig. 5b and c). However, only a very small amount of the proteins (VP2 or VP7) could be identified by heterologous antibodies in comparison to homologous antibody in the case of VP2-VP7 complexes (Fig. 5b). The data suggest that interaction between VP2 and VP7 is weaker than that of VP5 and VP7 (Fig. 5c). In contrast to VP7-VP5, it was not possible to pull down the VP2-VP5 complexes by using this assay system. Neither anti-VP2 monoclonal nor anti-VP5 polyclonal antibody could precipitate the heterologous proteins (Fig. 5a). Thus, it appears that there is no strong interaction between the two outer capsid proteins. These results confirm the structural data that revealed minimal interactions between the two distinct morphological features of the outer layer, representing VP5 and VP2, but extensive interactions individually with the underlying VP7 trimers. Alternatively, it is also possible that VP2 is easily removed from the complex due to its weak interaction with VP5.
FIG. 5.
Immunoprecipitation of the in vitro synthesized protein complexes. Plasmids expressing VP2, VP5, or VP7 proteins were used as templates for in vitro transcription-translation assays and the synthesized protein complexes were pulled down using different BTV antibodies. Reactions containing VP2-VP5, VP2-VP7, or VP5-VP7 templates were set up as controls to verify the correct size of each protein. (a) VP2/VP5 complex precipitated either using, both anti-VP2 and anti-VP5 (lane 1) as control, or using only anti-VP2 (lane 2) or only anti-VP5 (lane 3) antibodies. (b) VP2/VP7 complex precipitated either using both anti-VP2 and anti-VP7 antisera (lane 1) as control, or using anti-VP2 (lane 2) or anti-VP7 antisera (lane 3). (c) VP5/VP7 complex precipitated either using both anti-VP5 and anti-VP7 antisera (lane 1) as control, or using only anti-VP5 (lane 2) or only anti-VP7 antisera (lane 3).
Visualization of the transcription complex.
Since atomic details of the structural organization of the VP7 and VP3 was provided by the X-ray structure of the BTV core, our next focus was to obtain a better understanding of the locations of the various internal proteins. In both the virion and the core reconstructions there is a significant amount of density internal to the VP3 layer. Concentric layers of density with spacing of ∼26Å, consistent with the X-ray structure of the core, are observed in these reconstructions (11). Much of this density has been attributed to the packed layers of dsRNA genome (9). Similar concentric shells of internal densities have been found in several other members of the Reoviridae family that have been studied structurally (25, 31, 38).
To localize the density that is due to the internal proteins, we examined the structures of the CLPs with and without the internal proteins, in particular, VP1 and VP4 generated by recombinant baculoviruses (Fig. 6a) as described previously (7). Both proteins when incorporated within the CLPs either individually or together the molar ratios of each remained the same and were identical to that of virions (data not shown). We have shown previously that the largest minor protein VP1 is the RNA polymerase while VP4 possesses both guanylyltransferase and transmethylase activities and together they form the transcription complex (1, 28, 30). In all CLP reconstructions, the VP7 and the VP3 layers exhibit identical features. These features are similar to those found in the native core structure. However, the trimers that surround the aqueous channel at the fivefold axes, the P type, are missing in the CLP reconstructions (Fig. 6b). As a result, in CLPs, there are only 200 VP7 trimers instead of the expected 260 trimers. Compared to the virion and the core reconstructions, the CLPs have expectedly less internal density and are essentially empty particles (Fig. 6b and c). Virtually no internal density is seen in the CLPs with only VP3 and VP7. This is also true in the reconstructions of CLPs obtained by the coexpression of VP3 and VP7 with either VP1 or VP4. However, in the particles obtained by the coexpression of VP3, VP7 along with VP1 and VP4, a flower-shaped density directly beneath the icosahedral fivefold axes and attached to the underside of the VP3 layer is clearly observed (Fig. 6d to f). Similar feature has been observed in the recombinant rotavirus particles containing homologous proteins (27).
FIG. 6.
Organization of internal proteins in CLPs. (a) SDS-10% PAGE of purified CLPs (lane 2) containing VP1, VP3, VP4, and VP7. Insect cells were coinfected with two recombinant baculoviruses, one expressing VP3 and VP7 and the other expressing VP1 and VP4. CLPs were subsequently purified by CsCl gradient from cell lysates as described in Materials and Methods. Note that the four proteins are in same molar ratios as the virion core. Molecular markers are shown in lane 1. (b) Cryo-EM structures of the recombinant CLPs. Surface features of the CLP with and without the internal proteins (VP1 and VP4) are indistinguishable. Surface representation of the CLP reconstruction with VP3 and VP7 along the icosahedral threefold axes is shown as an example. The coloring of the VP7 and VP3 layers is same as in Fig. 2 and 3. The peripentonal VP7 are missing in all the CLP reconstructions. (c) A view from inside the VP3-VP7 CLP. Cryo-EM reconstructions of the VP3-VP7-VP1 and VP3-VP7-VP4 CLPs also look the same without any internal features. (d) Inside view of the CLP reconstruction with VP3, VP7, VP1, and VP4. A flower-shaped density features (red) attached to the inside surface of VP3 (green) at all the fivefold axes are clearly seen. (e and f) Conical cutaway from the reconstruction shown in panel c, providing close-up views of the flower-shaped structure and its interaction with the VP3 layer.
VP3 interacts directly with VP1 and VP4 complex.
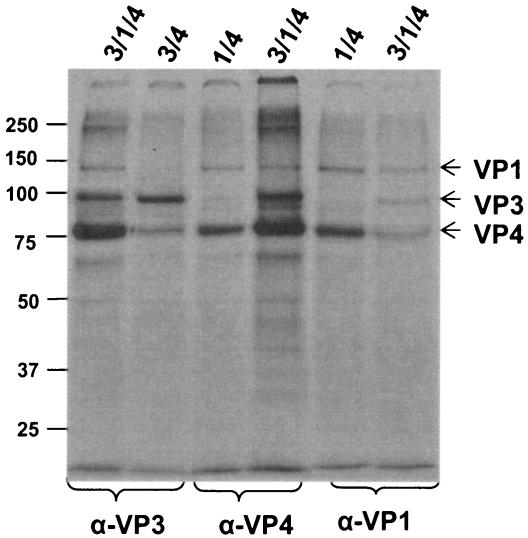
To examine if VP1 and VP4 interact with each other and if they individually also interact with VP3 we carried out immunoprecipitation experiments with radiolabeled recombinant proteins. Hi5 cells were infected with recombinant baculoviruses expressing VP1, VP3 and VP4 or only VP1 and VP4. The proteins were radiolabeled by adding the [35S]Met/Cys in the infected cell for 2 h and recovered by lysing the cells as described in Materials and Methods. The concentration of each protein was estimated and for each pull down assay same amount of protein was used. The BTV protein complexes were pulled down with different monospecific antibodies raised against BTV VP1, VP3 and VP4 protein and each antibody has been extensively used in previous published reports (1, 7, 28). Each immunoprecipitated sample was subsequently analyzed by SDS-PAGE and autoradiographed (Fig. 7). The data clearly show that anti-VP3 antibody could pull down both VP1 and VP4 indicating that indeed VP3 is closely associated with these minor proteins (Fig. 7, first lane). However, an interaction between VP1 and VP3 was less obvious when anti-VP1 antibody was used to pull down the complex (Fig. 7, last lane). This is in contrary to the interactions between VP3 and VP4 as either VP3 or VP4 antibody could pull down the complexes very efficiently (Fig. 7, lanes 2 and 4). As expected, VP1 and VP4 formed strong complex with each other and the resulting complex was pulled down either by VP4 or VP1 antibody and thus indicating that bonding between these two proteins are quite stable (Fig. 7).
FIG. 7.
Autoradiogram showing the interaction between VP3, VP1, and VP4 after immunoprecipitation. Hi5 cells infected either with recombinant baculoviruses expressing VP1, VP3, and VP4 or only with VP3 and VP4 or with VP1 and VP4. The infected cells were radiolabeled (30 h postinfection) for 2 h with [35S]methionine as described in Materials and Methods. Each complex was immunoprecipitated separately (as indicated in each lane) using anti-BTV VP3, VP1, and VP4 polyclonal antibody as indicated at the bottom of each lane. The molecular weights of each protein match exactly with those of VP1, VP4, and VP3, and the protein levels in each lane are comparable; note that there is no other major nonspecific or breakdown product band. Standard molecular weight markers were also run to estimate the molecular weights of the proteins.
DISCUSSION
Outer layer exhibits unique organization with a symmetry mismatch.
Like other members in the family Reoviridae, such as Rotavirus, Reovirus, and Aquareovirus (19), BTV has three concentric capsid layers enclosing multiple segments of dsRNA. In all these viruses, the innermost layer which forms the scaffold for the assembly of the outer two layers, exhibits a unique T = 1 icosahedral organization with two molecules in the icosahedral asymmetric unit. The middle layer built upon this T = 1 layer has a T = 13 icosahedral symmetry. In reoviruses and rotaviruses, the outer layer closely follows the same symmetric organization of the middle layer. In BTV, however, structural organization of the outer layer is quite unique and significantly deviates from the underlying T = 13 organization, thereby creating an organizational mismatch. This mismatch between the outer layer and the middle T = 13 layer is to some extent reminiscent of what is seen between the T = 13 layer and the innermost T = 1 layer. One difference, however, is that the innermost layer of BTV is contiguous, composed of one structural protein VP3, whereas the outer layer interrupted by two distinct motifs, globular and triskelion, is not contiguous. Although quite distinct from the other members of Reoviridae, the organization of the outer layer among the members of Orbivirus genus appears to be conserved. Similar organization of the outer layer with a triskelion and globular structural elements is also seen in a 23 Å cryo-EM structure of Broadhaven virus (32).
The 60 triskelion motifs appear to selectively interact with underlying VP7 trimers that surround the type I channels located at the icosahedral fivefold axes of the T = 13 lattice. The globular motifs selectively sit on the 120 of the 132 channels which are located at the quasi-sixfold axes of the underlying T = 13 VP7 layer. Based on their selective interactions with the VP7 layer and given that these two motifs exhibit minimal interactions with other, it is likely that the two structural motifs assemble independently on the VP7 layer. One possibility is that the assembly of globular structures first precludes the assembly of the triskelions on the VP7 trimers except the ones around the fivefold axes.
Composition of the triskelion and globular motifs.
From the biochemical data it is clear that the outer layer of BTV is composed of two proteins, VP5 and VP2. Earlier low-resolution studies had suggested that the VP2 forms the triskelion motifs and VP5 forms the globular motifs of the outer layer. Such an assignment making the VP2 protein most accessible part of the virion is consistent with the receptor binding and hemagglutinating activity exhibited by VP2. VP2 is also suggested to confer serotype specificity, as this protein exhibits more variability between the BTV serotypes. Based on the shape, it is likely that each triskelion is a trimer of VP2. The volume calculation from our BTV structure, assuming protein mass for VP2 as 111 kDa and a protein density of 1.30 g/cm3, agrees with each triskelion being composed of three VP2 molecules.
The assignment of VP2 to the triskelions implies that the globular motif is composed of the other outer layer protein VP5. Unlike in the case of the triskelion motif, it is difficult to unequivocally argue based on the shape as to how many VP5 molecules compose each globular motif. However, it is noteworthy that VP5 forms trimers readily in solution (13). Based on the volume calculations from the cryo-EM map, with a mass of VP5 as 59 kDa, and keeping in mind the uncertainties arising from the resolution and the close interactions between the globular motifs and VP7 trimers, it appears that each globular motif may be composed of two to three molecules of VP5. The triangular-shaped internal density of the globular motif observed inside the channels of the VP7 layer appears to be suggestive of three VP5 molecules constituting each globular motif.
Recent studies have indicated that VP5 has membrane fusion and destabilization activity (6). Analysis of the VP5 sequence using secondary structure predictions algorithms indicates that this protein is predominantly α-helical, with an amphipathic helical domain at the N terminus followed by a coiled coil domain. Such α-helical motifs are commonly seen in membrane fusion proteins of envelope viruses such as gp41 of human immunodeficiency virus and hemagglutinin of influenza virus. This amphipathic helical domain is functional causing the cell death, i.e., the membrane destabilization. It is tempting to speculate that the internal triangular shaped density represents these N-terminal regions of VP5. It is possible that this domain is protected and externalized only during receptor interactions facilitating membrane penetration.
Interactions between the outer layer proteins and the VP7 layer.
Our studies show that both VP2 triskelions and VP5 globular motifs interact extensively with the VP7 layer. While the VP2 triskelion motifs interact only with the top flat portion of the VP7 trimers, the VP5 globular motifs interact along side the VP7 trimers surrounding two of the three channels. Each triskelion motif predominantly interacts with just one of the trimer (P type), whereas the each globular motif interacts with five of the six VP7 trimers surrounding a channel. Stronger and more extensive interaction of VP5 with VP7 is consistent with our immunoprecipitation results and also the observation that VP2 comes off rather easily by high salt treatment, while VP5 remain attached to core. The latter observation supports our hypothesis in the previous section that assembly of VP5 precedes that of VP2. It surprising that despite the extensive interactions between the outer and VP7 layers, as observed in our reconstruction, the outer layer is so labile in BTV. The observed organizational mismatch between the outer layer proteins and the VP7 layer perhaps suggests that the interactions between these proteins are not highly specific leading to rather a weak association of the two capsid layers.
Location of the VP1 and VP4 inside the core.
Internal to the VP3 layer, the features in the cryo-EM reconstruction of either the mature BTV or the core particle are difficult to interpret in terms of internal proteins and the genomic RNA. Our comparative cryo-EM analysis of the authentic cores and the CLPs obtained by coexpressing VP3 and VP7 along with VP1 and/or VP4 has shown that these two proteins, most likely as a heterodimer, are attached to inside surface of the VP3 layer at each of the icosahedral fivefold vertex. Similar results are obtained for recombinant rotavirus particles (27). In these rotavirus particles the extra density was visualized only in the reconstruction of the recombinant particles that were obtained by coexpressing the polymerase and the capping enzyme together with VP2 and VP6 (homologues of VP3 and VP7 in BTV, respectively). It is noteworthy that the helicase protein, VP6 of BTV, unlike the VP1 and VP4 are not readily incorporated within the CLPs, and therefore VP6 was not included in our studies. It is likely that VP6 needs to be associated with BTV RNAs in order to be encapsidated. Our structural results are also strongly supported by the coimmunoprecipitation studies, which indicate VP1 and VP4 not only interact with one another strongly but each interacts with VP3. Such close interactions between the polymerase and the capping enzyme may indeed be required for capping the nascent mRNA as they emerging from the polymerase. In rotavirus particles it has been shown that transcripts as short as 7 nucleotides are capped, indicating that the capping site is only 7 nucleotides away from the enzymatic site in the polymerase (20).
It is difficult to partition the individual contributions from VP1 and VP4 in the flower-shaped density observed in the reconstruction of recombinant VP3/7/4/1 CLPs as we could not see any density in the CLPs, which contained only one of these proteins. Based on the recent results on reovirus (37), it is likely that central mass is predominantly due to the polymerase and the some of the sideward projecting density could be due to the VP4. We have to express caution in such an interpretation, as the density that we have attributed to VP4 and VP1 is fivefold averaged. From the volume of this fivefold averaged flower-shaped mass, the most likely interpretation is that each flower-shaped complex contains one molecule of each of VP1 and VP4. Biochemical analysis suggests that VP4 exists as a dimer (29). From our structural analysis, an unequivocal determination of the precise stoichiometry and oligomerization state of VP4 is difficult. The location of the polymerase complex is consistent with the model proposed for genome organization in BTV from X-ray crystallographic studies (9). Although it is only in BTV and rotavirus particles that the location of the polymerase complex is unequivocally demonstrated using recombinant particles, similar densities are seen in the reconstructions of several other dsRNA viruses, including viruses from outside the Reoviridae family. Thus, in all these viruses, the overall structural mechanism of endogenous transcription is likely to be very similar. The unique icosahedral organization of the innermost layer, found in majority of the dsRNA viruses including those of bacterial (2) and fungal origin (24), may be a critical requirement for the appropriate positioning of the transcription enzyme complex and genome organization. In viruses with multiple dsRNA segments a consensus model consistent with the available structural and biochemical data is that each segment is associated with a transcription complex at the fivefold axis to allow for its independent and simultaneous transcription.
Acknowledgments
This work was supported partly by an NIH grant and partly by a Wellcome Trust grant (United Kingdom).
We thank Ana Oliveria for expert technical assistance.
REFERENCES
- 1.Boyce, M., J. Wehrfritz, R. Noad, and P. Roy. 2004. Purified recombinant Bluetongue virus VP1 exhibits RNA replicase activity. J. Virol. 78:3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butcher, S. J., T. Dokland, P. M. Ojala, D. H. Bamford, and S. D. Fuller. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 16:4477-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos. Trans. R Soc. Lond. B Biol. Sci. 261:221-230. [DOI] [PubMed] [Google Scholar]
- 4.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 5.Els, H. J., and D. W. Verwoerd. 1969. Morphology of bluetongue virus. Virology 38:213-219. [DOI] [PubMed] [Google Scholar]
- 6.Forzan, M., C. Wirblich, and P. Roy. 2004. A capsid protein of a non-enveloped Bluetongue virus exhibits membrane fusion activity. Proc. Natl. Acad. Sci. USA 101:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French, T. J., and P. Roy. 1990. Synthesis of bluetongue virus (BTV) corelike particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J. Virol. 64:1530-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller, S. D. 1987. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell 48:923-934. [DOI] [PubMed] [Google Scholar]
- 9.Gouet, P., J. M. Diprose, J. M. Grimes, R. Malby, J. N. Burroughs, S. Zientara, D. I. Stuart, and P. P. Mertens. 1999. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97:481-490. [DOI] [PubMed] [Google Scholar]
- 10.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 11.Grimes, J. M., J. Jakana, M. Ghosh, A. K. Basak, P. Roy, W. Chiu, D. I. Stuart, and B. V. V. Prasad. 1997. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure 5:885-893. [DOI] [PubMed] [Google Scholar]
- 12.Hassan, S. H., and P. Roy. 1999. Expression and functional characterization of bluetongue virus VP2 protein: role in cell entry. J. Virol. 73:9832-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan, S. H., C. Wirblich, M. Forzan, and P. Roy. 2001. Expression and functional characterization of bluetongue virus VP5 protein: role in cellular permeabilization. J. Virol. 75:8356-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewat, E. A., T. F. Booth, P. T. Loudon, and P. Roy. 1992. Three-dimensional reconstruction of baculovirus expressed bluetongue virus core-like particles by cryo-electron microscopy. Virology 189:10-20. [DOI] [PubMed] [Google Scholar]
- 15.Hewat, E. A., T. F. Booth, and P. Roy. 1992. Structure of bluetongue virus particles by cryoelectron microscopy. J. Struct. Biol. 109:61-69. [DOI] [PubMed] [Google Scholar]
- 16.Hewat, E. A., T. F. Booth, and P. Roy. 1994. Structure of correctly self-assembled bluetongue virus-like particles. J. Struct. Biol. 112:183-191. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Kar, A. K., and P. Roy. 2003. Defining the structure-function relationships of bluetongue virus helicase protein VP6. J. Virol. 77:11347-11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawton, J., M. Estes, and B. V. V. Prasad. 2000. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 55:185-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawton, J. A., M. K. Estes, and B. V. V. Prasad. 2001. Identification and characterization of a transcription pause site in rotavirus. J. Virol. 75:1632-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawton, J. A., and B. V. V. Prasad. 1996. Automated software package for icosahedral virus reconstruction. J. Struct. Biol. 116:209-215. [DOI] [PubMed] [Google Scholar]
- 22.Marshall, J. J., and P. Roy. 1990. High level expression of the two outer capsid proteins of bluetongue virus serotype 10: their relationship with the neutralization of virus infection. Virus Res. 15:189-195. [DOI] [PubMed] [Google Scholar]
- 23.Mertens, P. P., J. N. Burroughs, and J. Anderson. 1987. Purification and properties of virus particles, infectious subviral particles, and cores of bluetongue virus serotypes 1 and 4. Virology 157:375-386. [DOI] [PubMed] [Google Scholar]
- 24.Naitow, H., J. Tang, M. Canady, R. B. Wickner, and J. E. Johnson. 2002. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9:725-728. [DOI] [PubMed] [Google Scholar]
- 25.Pesavento, J. B., J. A. Lawton, M. E. Estes, and B. V. Venkataram Prasad. 2001. The reversible condensation and expansion of the rotavirus genome. Proc. Natl. Acad. Sci. USA 98:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad, B. V. V., and P. E. Prevelige, Jr. 2003. Viral genome organization. Adv. Protein Chem. 64:219-258. [DOI] [PubMed] [Google Scholar]
- 27.Prasad, B. V. V., R. Rothnagel, C. Q. Zeng, J. Jakana, J. A. Lawton, W. Chiu, and M. K. Estes. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471-473. [DOI] [PubMed] [Google Scholar]
- 28.Ramadevi, N., J. N. Burroughs, P. P. C. Mertens, I. M. Jones, and P. Roy. 1998. Capping and methylation of mRNA by purified recombinant VP4 protein of Bluetongue virus. Proc. Natl. Acad. Sci. USA 95:13537-13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramadevi, N., J. Rodriguez, and P. Roy. 1998. A leucine zipper-like domain is essential for dimerization and encapsidation of bluetongue virus nucleocapsid protein VP4. J. Virol. 72:2983-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramadevi, N., and P. Roy. 1998. Bluetongue virus core protein VP4 has nucleoside triphosphate phosphohydrolase activity. J. Gen. Virol. 79:2475-2480. [DOI] [PubMed] [Google Scholar]
- 31.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 A resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 32.Schoehn, G., S. R. Moss, P. A. Nuttall, and E. A. Hewat. 1997. Structure of Broadhaven virus by cryoelectron microscopy: correlation of structural and antigenic properties of Broadhaven virus and bluetongue virus outer capsid proteins. Virology 235:191-200. [DOI] [PubMed] [Google Scholar]
- 33.Stauber, N., J. Martinez-Costas, G. Sutton, K. Monastyrskaya, and P. Roy. 1997. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol. 71:7220-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studdert, M. J., J. Pangborn, and R. B. Addison. 1966. Bluetongue virus structure. Virology 29:509-511. [DOI] [PubMed] [Google Scholar]
- 35.van Heel, M. 1987. Similarity measures between images. Ultramicroscopy 21:95-100. [Google Scholar]
- 36.Verwoerd, D. W., H. J. Els, E. M. De Villiers, and H. Huismans. 1972. Structure of the bluetongue virus capsid. J. Virol. 10:783-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, X., S. B. Walker, P. R. Chipman, M. L. Nibert, and T. S. Baker. 2003. Reovirus polymerase lambda3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat. Struct. Biol. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, Z. H., M. L. Baker, W. Jiang, M. Dougherty, J. Jakana, G. Dong, G. Lu, and W. Chiu. 2001. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat. Struct. Biol. 8:868-873. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, Z. H., B. V. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]