PREFACE
Protein synthesis is principally regulated at the initiation stage (rather than during elongation or termination), allowing rapid, reversible and spatial control over gene expression. Progress over recent years in determining the structures and activities of initiation factors, and in mapping their interactions within ribosomal initiation complexes, has significantly advanced our understanding of the complex translation initiation process. These developments have provided a solid foundation for studies of regulation of initiation by mechanisms that include modulation of the activity of initiation factors (which affects almost all scanning-dependent initiation), or via sequence-specific RNA-binding proteins and microRNAs (which thus impact individual mRNAs).
INTRODUCTION
Translation initiation is the process of assembly of elongation-competent 80S ribosomes, in which the initiation codon is base-paired with initiator tRNA in the ribosomal P-site1. It requires at least 9 eukaryotic initiation factors (eIFs; Table 1) and comprises two steps: first, formation of 48S initiation complexes with established codon-anticodon base-pairing in the P-site of the 40S ribosomal subunits, and second, joining of 48S complexes with 60S subunits. On most mRNAs, 48S complexes form by the “scanning” mechanism: a 43S preinitiation complex (comprising a 40S subunit, the eIF2–GTP–Met-tRNAMeti ternary complex (eIF2-TC), eIF3, eIF1, eIF1A and likely eIF5) attaches to the capped 5'-proximal region of mRNA in a step that involves unwinding of the mRNA’s 5’ terminal secondary structure by eIF4A, eIF4B and eIF4F. The 43S complex then scans the 5’-untranslated region (5’-UTR) in the 5’→3’ direction to the initiation codon (Fig.1). After initiation codon recognition and 48S complex formation, eIF5 and eIF5B promote hydrolysis of eIF2-bound GTP, displacement of eIFs and joining of a 60S subunit. Although most mRNAs use the scanning mechanism, initiation on a few is mediated by IRESs (Box 1).
Table 1.
Eukaryotic initiation factors
| Name | Subunits Mol. wt. (kDa) | Function |
|---|---|---|
| Core Initiation Factors | ||
| eIF2 | 3 (36.1, 38.4, 51.1) | Forms an eIF2/GTP/Met-tRNAi ternary complex that binds to the 40S subunit, thus mediating ribosomal recruitment of Met-tRNAi |
| eIF3 | 13 (800 total) | Binds 40S subunits, eIFs 1, 4G, 5; stimulates binding of eIF2/GTP/Met-tRNAi to 40S subunits; promotes attachment of 43S complexes to mRNA and subsequent scanning; possesses ribosome dissociation and anti-association activities preventing joining of 40S and 60S subunits |
| eIF1 | 1 (12.7) | Ensures the fidelity of initiation codon selection; promotes ribosomal scanning; stimulates binding of eIF2/GTP/Met-tRNAi to 40S subunits; prevents premature eIF5-induced hydrolysis of eIF2-bound GTP and Pi release |
| eIF1A | 1 (16.5) | Stimulates binding of eIF2/GTP/Met-tRNAi to 40S subunits; cooperates with eIF1 in promoting ribosomal scanning and initiation codon selection |
| eIF4E | 1 (24.5) | Binds to the m7GpppG 5’-terminal ‘cap’ structure of mRNA |
| eIF4A* | 1 (46.1) | DEAD-box ATPase and ATP-dependent RNA helicase |
| eIF4G** | 1 (175.5) | Binds eIF4E, eIF4A, eIF3, PABP, SLIP1 and mRNA (Fig. 3A); enhances eIF4A’s helicase activity |
| eIF4F | 3 (246.1 total) | Cap-binding complex, comprising eIF4E, eIF4A and eIF4G; unwinds 5’-proximal region of mRNA and mediates attachment to it of 43S complexes; assist ribosomal complexes during scanning |
| eIF4B | 1 (69.3) | RNA-binding protein; enhances eIF4A’s helicase activity |
| eIF4H | 1 (27.4) | RNA-binding protein; enhances eIF4A’s helicase activity; homologous to a fragment of eIF4B |
| eIF5 | 1 (49.2) | GTPase-activating protein specific for eIF2•GTP that induces hydrolysis of eIF2-bound GTP upon recognition of the initiation codon |
| eIF5B | 1 (138.9) | Ribosome-dependent GTPase; mediates ribosomal subunit joining |
| eIF2B | 5 (33.7, 39.0, 50.2, 59.7, 80.3) | Guanosine nucleoside exchange factor that promotes GDP/GTP exchange on eIF2 |
| Auxiliary factors | ||
| DHX29 | 1 (155.3) | A DExH-box-containing protein; binds 40S subunit; promotes ribosomal scanning on mRNAs with long, highly-structured 5’UTRs |
| Ded1p | 1 (65.6) | DEAD-box NTPase/RNA helicase; potentially promotes scanning in S. cerevisiae |
| eIF6 | 1 (26.6) | Binds 60S subunits; anti-association factor that prevents joining of 40S subunits to 60S subunits |
| p97 | 1 (102.4) | Closely related to the carboxy-terminal two-thirds of eIF4GI; binds eIF4A and eIF3; promotes initiation in a potentially mRNA-specific manner |
| PABP | 1 (70.7) | Binds to the mRNA 3’-poly(A) tail, eIF4G and eRF3; enhances binding of eIF4F to the cap; may facilitate recruitment of recycled post-termination 40S subunits back to the 5’-end of mRNA. |
Two paralogues (eIF4AI and eIF4AII), encoded by different genes, are functionally indistinguishable, but “eIF4AIII” has no activity as an eIF.
Two paralogues (eIF4GI and eIF4GII), encoded by different genes, are functionally similar but show some selectivity towards different mRNAs. eIF4GI is generally the more abundant.
Figure 1. Model of the canonical pathway of eukaryotic translation initiation.
This pathway is divided into eight stages (2–9), which follow (1) recycling of post-termination complexes to yield separated 40S and 60S ribosomal subunits, and result in formation of an 80S ribosomal initiation complex in which Met-tRNAMeti is base-paired with the initiation codon in the ribosomal P-site and which is competent to start the elongation stage of translation. These stages are: (2) formation of the eIF2•GTP/Met-tRNAMeti ternary complex; (3) formation of a 43S preinitiation complex comprising a 40S subunit, eIF1, eIF1A, eIF3, eIF2•GTP/Met-tRNAMeti and probably eIF5; (4) mRNA activation, during which the mRNA cap-proximal region is unwound in an ATP-dependent manner by eIF4F with eIF4B; (5) attachment of the 43S complex to this mRNA region; (6) scanning of the 5’UTR in a 5’→3’direction by 43S complexes; (7) recognition of the initiation codon and 48S initiation complex formation, which switches the scanning complex to a ‘closed’ conformation and leads to displacement of eIF1, permitting eIF5-mediated hydrolysis of eIF2-bound GTP and Pi release; (8) joining of 60S subunits to 48S complexes and concomitant displacement of eIF2•GDP and other factors (eIF1, eIF3, eIF4B, eIF4F and eIF5) mediated by eIF5B; and (9) GTP hydrolysis by eIF5B and release of eIF1A and eIF5B•GDP from assembled elongation-competent 80S ribosomes. Translation is a cyclical process in which termination follows elongation, and leads to recycling (1) which generates separated ribosomal subunits. The model omits potential ‘closed-loop’ interactions involving poly(A) binding protein (PABP), eukaryotic release facto 3 (eRF3) and eIF4F during recycling (Supplementary information S5), and recycling of eIF2•GDP by eIF2B. Whether eRF3 is still present on ribosomes at the recycling stage is unknown.
Box 1.
IRES-mediated translation initiation
Here, we will summarize the current state of knowledge concerning the mechanism of initiation in vertebrates and discuss the principles underlying its regulation, focusing on examples where the regulatory mechanism is well understood and/or the biological significance is particularly high, and introducing evidence from lower eukaryotes only when it significantly enhances understanding of mechanisms in vertebrates.
MECHANISM OF 5’-END-DEPENDENT INITIATION
The canonical mechanism of translation initiation can be divided into several stages (Fig.1), as described below.
Formation of 43S preinitiation complexes
Translation initiation requires a pool of separated ribosomal subunits. Translation is a cyclical process, and ribosomal subunits that participate in initiation are derived by recycling of post-termination ribosomal complexes (post-TCs), which comprise and 80S ribosome still bound to mRNA, P-site deacylated tRNA and at least one release factor, eukaryotic release factor 1 (eRF1). Post-TCs are recycled by releasing these ligands and dissociating ribosomes into subunits. At a relatively low free (nucleotide-unbound) Mg2+ concentration (1 mM), recycling can be mediated by eIFs2. eIF3, in cooperation with its loosely-associated eIF3j subunit, eIF1 and eIF1A, dissociates post-TCs into free 60S subunits and mRNA- and tRNA-bound 40S subunits. Subsequently eIF1 promotes release of tRNA, and then eIF3j, which binds 40S subunits with negative cooperativity with mRNA3,4, mediates mRNA dissociation. eIF3, and likely eIF1 and eIF1A, remain associated with recycled 40S subunits, preventing their re-association with 60S subunits. Recycling at even slightly elevated Mg2+ concentrations (which stabilize ribosomal subunit association) also requires ATP-binding cassette subfamily E member 1 (ABCE1) (A.V. Pisarev, M.A. Skabkin, V.P. Pisareva, O.V. Skabkina, A. Rakotondrafara, M.W. Hentze, C.U.T.H. and T.V.P., unpublished observations), an essential ATP-binding cassette protein5. ABCE1 splits post-TCs into free 60S subunits and tRNA- and mRNA-bound 40S subunits, and subsequent release of P-site tRNA and mRNA from these 40S subunits also requires eIF3, eIF1 and eIF1A. Thus, eIF3, eIF1 and eIF1A are recruited to 40S subunits during recycling, whereas eIF2–GTP–Met-tRNAMeti subsequently attaches to recycled 40S subunits, bound simultaneously to eIF3, eIF1 and eIF1A, to form 43S complexes. Another protein that can prevent ribosomal subunit re-association by binding to 60S subunits is eIF6, but its status as an initiation factor is uncertain (Supplementary information S1).
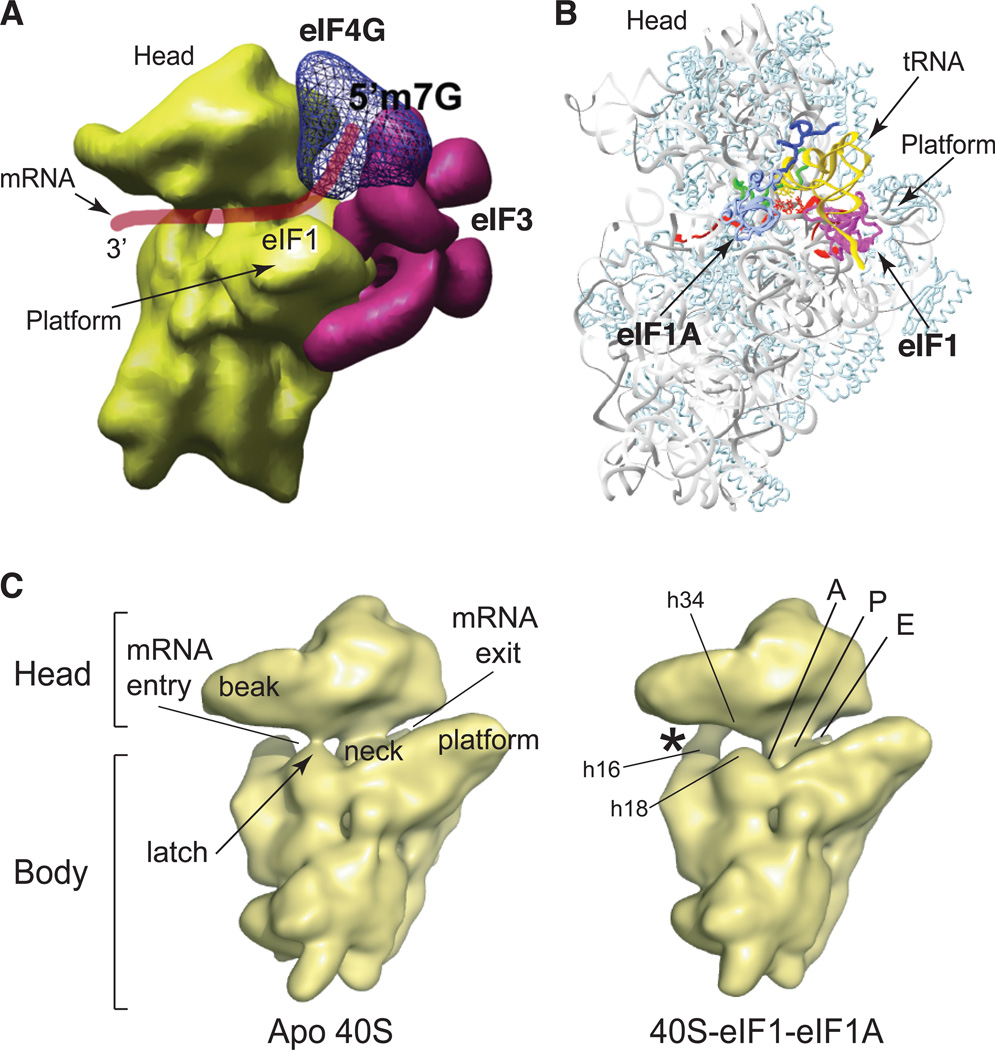
Recent studies have yielded insights into the architecture of 43S complexes. Eukaryotic and prokaryotic small ribosomal subunits share a common structural core that includes the decoding center, whereas additional eukaryotic ribosomal proteins (rp) and 18S rRNA expansion segments (rapidly evolving regions interspersed throughout the conserved rRNA core that might function in eukaryote-specific aspects of translation) are situated peripherally6. The highest-resolution structure of eukaryotic ribosomes (7.3Å) has been determined by cryoelectronmicroscopy (EM)7, but the structural homology between eukaryotic and prokaryotic ribosomes allows high-resolution crystal structures of prokaryotic ribosomes to be used to model 40S–eIF interactions based on biochemical data. The 40S subunit consists of the head, platform and body with the mRNA-binding channel wrapping around the neck (Fig.2). The bulk of the five-lobed eIF3 molecule binds to the 40S subunit side facing the solvent8 (Fig.2A), whereas the eIF3j C-terminal domai localizes in the mRNA-binding channel in the A-site area on the intersubunit side4. Although eIF2’s structure is available9, the position of eIF2–GTP–Met-tRNAMeti on 40S subunits has not been determined. However, in 43S complexes, the Met-tRNAiMet anticodon loop is likely inserted less deeply into the P-site than in ribosomal complexes with established codon-anticodon base-pairing (as shown in Fig.2B), and its acceptor-end might be rotated towards the E-site10–12. eIF1 binds to the interface between the platform and Met-tRNAMeti (Ref. 10), eIF1A’s structured domain resides in the A-site, forming a bridge over the mRNA channel, whereas its N- and C-terminal tails extend into the P-site13 (Fig.2B). Importantly, binding of eIF1 and eIF1A to 40S subunits induces conformational changes14, which involve opening of the mRNA entry channel 'latch' formed by helix (h) 18 in the body and h34 and rpS3 in the neck, and establishment of a new head–body connection on the solvent side between h16 and rpS3 (Fig.2C).
Figure 2. Architecture of ribosomal initiation complexes.
(A) Model of a 40S subunit with eIF3 on its exterior (solvent) surface and eIF4G bound to eIF3 near the E-site, based on cryoelectron microscopy analysis, and showing positions of mRNA (red line) and eIF1 on the subunit interface. Binding of eIF3 to the solvent surface is compatible with its potential partial retention on ribosomes during translation of short upstream open reading frames (uORFs). Adapted with permission from Ref. 8. (B) Positions of eIF1 (magenta) and eIF1A (with its structured domain in light blue , its carboxy-terminal tail in dark blue and its amino-terminal tail in green) on the 40S subunit relative to mRNA (red) and P-site tRNA (yellow), based on directed hydroxyl radical probing data10, 13 and modeled using T. thermophilus 30S subunit crystal structures (PDB codes 1JGO and 1JGP). (C) Cryoelectron microscopy reconstructions of yeast apo 40S subunits (left panel) and 40S–eIF1–eIF1A complexes (right panel), labelled to indicate the A-, P- and E-sites in the mRNA-binding channel, and the positions of rRNA helices h16, h18 and h34, which are involved in forming the mRNA entry-channel (h18-h34) and the eIF1- and eIF1A–induced head–shoulder connection (h16-rpS3) (indicated by an asterisk). Adapted from Ref. 14 with permission.
Attachment of 43S complexes to mRNA
Although 43S complexes are intrinsically capable of 5’-end-dependent attachment to model mRNAs with completely unstructured 5’-UTRs15, natural 5’-UTRs possess sufficient secondary structure that loading of 43S complexes onto them requires the cooperative action of eIF4F and eIF4B or eIF4H, which unwind the 5’ cap-proximal region of mRNA to prepare it for ribosomal attachment. eIF4F comprises the cap-binding protein eIF4E, the DEAD-box RNA helicase eIF4A and eIF4G, which functions as a “scaffold” that binds eIF4E, eIF4A, the poly(A)-binding protein (PABP) and eIF3 (Fig.3A). eIF4B and eIF4H enhance eIF4A’s helicase activity, contain RRM domains and are homologous over the entire length of eIF4H1. The ‘cap’ stacks between two tryptophan residues on eIF4E’s concave surface; additional contacts with the cap-proximal nucleotide stabilize eIF4E’s binding to capped mRNA16. A segment of eIF4G wraps around eIF4E’s N-terminus, inducing structural changes that enhance eIF4E’s cap-binding affinity17,18. eIF4A has two domains and alternates between an inactive ‘open’ and an active ‘closed’ conformation, in which both domains form a contiguous RNA-binding surface and the ATP-binding site is at the domain interface19. The low individual helicase activity of eIF4A is strongly enhanced by eIF4G and eIF4B (or eIF4H)20. eIF4G’s HEAT-1 domain stimulates eIF4A’s helicase activity by aligning the DEAD-box motifs in both domains in a productive conformation, whereas HEAT-2, which also binds eIF4A, plays a modulatory role21,22. The topology of eIF4A–eIF4G–eIF4H complexes (Fig.3B) indicates that eIF4H binds the single-stranded mRNA behind eIF4A (relative to the direction of helicase translocation), suggesting that eIF4H (or eIF4B) could stimulate eIF4A’s helicase activity by preventing mRNA re-annealing and promoting processive unidirectional eIF4A movement22. As suggested22, because of the high, but nevertheless limited, processivity of eIF4F–eIF4B (or -eIF4H) complexes, eIF4A eventually dissociates from mRNA but, being anchored to its 5’-end by the eIF4E–cap interaction, these complexes resume another cycle of unwinding, thereby keeping the 5’-proximal region constantly prepared for ribosomal attachment that is likely facilitated by the eIF3–eIF4G interaction23. Thus, recruitment of 43S complexes is ultimately achieved by the cap–eIF4E–eIF4G–eIF3–40S chain of interactions. The ‘open-latch’ conformation of 40S subunits14 I, induced by eIF1 and eIF1A, is likely strongly conducive for attachment.
Figure 3. eIF4G domain structure, interactions, and its position in a scanning 43S complex.
(A) Schematic representation of the longest isoform of eIF4GI (Genbank Acc. NP_937884), of its p100 (C-terminal two-thirds) and p50 (central one-third) fragments, and of p97, showing binding sites for SLBP-interacting protein 1 (SLIP1), PABP, eIF4E, eIF4A, eIF3, and MAP kinase interacting Ser/Thr kinase 1 (Mnk1) or Mnk2 and for RNA (dotted lines below eIF4G1). The interactions of eIF4G with eIF4E and Mnk1 are required for phosphorylation of eIF4E by Mnk1; interactions of eIF4G with PABP and SLIP1 tether eIF4F to the 3’-end of mRNA (see text). The amino acid residues at the N-termini of the PABP-binding domain (PAM-1), 4E–BR (eIF4E–binding domain) and HEAT-1 (also known as MIF4G), HEAT-2 (also known as MA3) and HEAT-3 (also known as W2) domains are indicated, as is the cleavage site in eIF4GI for the picornavirus proteinase 2Apro (Supplementary information S3), which divides eIF4G into an N-terminal domain that binds eIF4E and PABP, and a C-terminal domain that provides all functions of eIF4G required for initiation on Type 1 and Type 2 IRESs (see Box 1). This cleavage event contributes to the switch from host to viral translation during many picornavirus infections (see Supplementary Information S3).
(B) Hypothetical model of the scanning 43S preinitiation complex, viewed from the solvent face, showing associated factors and domains of factors, including eIF4E, eIF4G’s 4E–BR, HEAT-1, HEAT-2 and HEAT-3 domains of eIF4G, the C-terminal and RRM domains of eIF4H and the N-terminal and C-terminal domains of eIF4A. The direction of scanning (5’→3’) is shown by an arrow, and in this model, eIF4A is on the leading (3’) side of the scanning complex. Adapted from Ref. 22 with permission.
Despite these advances, the position of eIF4E in ribosomal complexes and mechanistic aspects of how mRNA enters the mRNA-binding cannel remain unknown. If the cap–eIF4E interaction persists during attachment, it is unlikely that eIF4E-bound mRNA could be threaded through the entire mRNA-binding channel, and loading of 43S complexes would therefore be more compatible with direct positioning of eIF4E–cap at the channel’s E-site side. This would raise the question: from which nucleotide do 43S complexes begin inspecting the 5’-UTR during scanning? The efficiency of initiation on mRNAs with short 5’-UTRs has been investigated in a cell-free system24, However, the data obtained, which showed that about 50% of ribosomes bypass an AUG codon located within 12 nucleotides of the cap, could reflect the susceptibility of initiation complexes potentially forming close to the mRNA’s 5’-end to dissociation by eIF1, as discussed below15, rather than tRNAMeti’s inability to inspect mRNA from certain positions.
Ribosome scanning
After attachment, 43S complexes scan mRNAs downstream of the cap, to the initiation codon. Scanning consists of two linked processes: unwinding of secondary structure in the 5'-UTR and ribosomal movement along it. 43S complexes can scan unstructured 5’-UTRs without factors associated with RNA unwinding and are thus intrinsically capable of movement along mRNA15. Omission of eIF1A substantially reduces this ability and lack of eIF1 almost abrogates this ability15, indicating that movement of 43S complexes requires the scanning-competent conformation induced by eIF1 and eIF1A14. Although eIF3 is indispensable for 48S complex formation, it is difficult to separate its role in scanning from functions such as ribosomal recruitment of eIF2–GTP–Met-tRNAMeti and attachment of 43S complexes. However, eIF3 interacts with mRNA upstream of the E-site (Fig.2A) forming an extension of the mRNA-binding channel that might contribute to scanning25. Even the scanning of 5'-UTRs containing weak secondary structure requires ATP and eIF4A, eIF4G and eIF4B15, and the requirement for ATP and eIF4A is proportional to the degree of secondary structure26,27. Thus, in addition to promoting attachment, eIF4A, eIF4G and eIF4B assist 43S complexes during scanning.
However, the mechanism by which these factors assist scanning remains unknown. Cryo-EM-based modeling placed eIF4G at the 40S subunit’s trailing edge near the E-site8 (Fig.2A), which would be consistent with eIF4A, eIF4G and eIF4B acting by helicase-mediated "ratcheting" of mRNA through the mRNA-binding channel, whereas mRNA secondary structure would be unwound by 40S subunits themselves at their leading edge. However, an alternative model, in which eIF4A, eIF4G and eIF4B unwind mRNA before it enters this channel, has also been suggested22 (Fig.3B).
Although ribosomal attachment is achieved by the cap–eIF4E–eIF4G–eIF3–40S chain of interactions, the fate of each link during the transition from attachment to scanning and during scanning per se is unclear. Their maintenance would cause 5’-UTRs to ‘loop out’, allowing only one 43S complex to scan at a time, whereas breaking of even one link would permit multiple complexes to scan simultaneously on a single 5’UTR.
Another important question concerns the directionality of scanning. The fact that initiation frequency at the 5’-proximal AUG is significantly reduced by the presence of a nearby downstream AUG28 suggests that scanning may consist of forward (5’→3’) thrusts alternating with limited relaxation over distances of a few nucleotides in the reverse direction.
Importantly, recent data obtained using yeast and mammalian systems suggest that initiation involves other DEAD box family members in addition to eIF4A, and that eIF4A can act with p97, a distinct eIF4G-related protein. Mammalian DExH-box protein DHX29 binds 40S subunits directly and is required for efficient scanning through highly structured 5’-UTRs in vitro29. In vivo, silencing DHX29 impairs translation resulting in polysome disassembly and accumulation of mRNA-free 80S monomers30. DHX29 has been suggested to increase scanning processivity by influencing the mRNA-binding channel’s conformation at its entrance29.
Yeast DEAD-box helicase Ded1 has also been implicated in initiation: Ded1 is likely a more potent helicase than eIF4A and their functions are not redundant, suggesting that eIF4A promotes ribosomal attachment, whereas Ded1 assists scanning, particularly on long 5'-UTRs31–33. The involvement in initiation of DDX3, a mammalian Ded1 homologue, is more controversial, although some data suggest that DDX3 depletion specifically affects translation of mRNAs with long structured 5’-UTRs34.
p97, which is ubiquitously expressed in the tissues of mammals, brds and is homologous to the C-terminal two-thirds of eIF4G and binds eIF4A and eIF3, but lacks an eIF4E-binding region35 (Fig.3A). p97 activates translation of uncapped mRNAs in vitro36 and its role in translation of capped mRNAs is not wholly redundant with that of eIF4G: although depletion of eIF4G1 and p97 individually impaired global translation by ~20–30% and co-depletion reduced it by ~60%, depletion of eIF4G1 but not of p97 selectively impaired translation of mRNAs containing upstream Open Reading Frames (uORFs), suggesting that these factors promote initiation on different classes of mRNAs37.
Initiation codon recognition
To ensure the fidelity of initiation, scanning complexes must have a discriminatory mechanism that prevents partial base-pairing of triplets in the 5’-UTR with the Met-tRNAiMet anticodon and promotes recognition of the correct initiation codon, which is usually the first AUG triplet in an optimum context GCC(A/G)CCAUGG, with a purine in −3 and ‘G’ in +4 positions (relative to the A of the AUG codon, which is designated +1)24. eIF1 plays the key role in maintaining the fidelity of initiation. It enables 43S complexes to discriminate against non-AUG triplets and AUG triplets that have poor context or are located within 8 nucleotides of the mRNA 5’-end, and also dissociates ribosomal complexes aberrantly assembled at such triplets in its absence15,38,39. Genetic studies in yeast also identified eIF1 as a determinant of initiation codon recognition40. In a current model, eIF1 in cooperation with eIF1A promotes a scanning-competent ‘open’ conformation of the 43S complex14 (Fig.2C), but to establish stable codon–anticodon base-pairing, ribosomal complexes must undergo conformational changes that are antagonized by eIF1. Establishment of codon–anticodon base-pairing is accompanied by tightening of the eIF1A-40S interaction41 and eIF1’s displacement from near the P-site3,10,42, which switches the complex to a ‘closed’ conformation locked onto the mRNA. Consistently, yeast eIF1 mutants that dissociate more rapidly from 48S subunits enhance initiation at non-AUG codons43. The eIF1A’s N-terminal tail (NTT) and C-terminal tail (CTT), which reach into the P-site13 (Fig.2B) have opposite effects on start-codon selection: the CTT increases its stringency and was proposed to promote the “open” conformation of scanning complexes, whereas the NTT decreases the accuracy of initiation and promotes the “closed” conformation44. The purines at −3 and +4 positions likely affect initiation codon selection by stabilizing conformational changes that occur upon codon–anticodon base-pairing, by interacting with the eIF2α subunit of eIF2 and AA1818–1819 in helix 44 of 18S rRNA, respectively39. In eIF1’s absence, the stability of 48S complexes is not challenged, so that complexes with partial base-pairing can form and participate in subsequent steps in translation. Such complexes cannot maintain their conformation upon binding of eIF1, and mispaired tRNA is likely ejected.
Commitment of ribosomes to a start codon
Initiation codon recognition is followed by a step during which the arrested ribosome becomes committed to initiation at that codon. The commitment step is mediated by eIF5, an eIF2-specific GTPase-activating protein (GAP)1. eIF5 binds eIF2’s β-subunit but induces the GTPase activity of eIF2’s γ-subunit only in eIF2–GTP–Met-tRNAMeti complexes that are bound to 40S subunits. eIF5 has been proposed to act as a classical GAP by providing an arginine finger45. An alternative hypothesis suggests that eIF5 derepresses eIF2γ‘s GTPase activity46. Premature hydrolysis of eIF2-bound GTP in 43S complexes, and particularly subsequent Pi release, are prevented by eIF13,47. Establishment of codon–anticodon base-pairing results in eIF1’s displacement42, which relieves repression of GTP hydrolysis and Pi release3,47. Thus, in addition to its role in initiation codon selection during 48S complex formation, eIF1 also maintains initiation fidelity at a later stage by linking hydrolysis of eIF2-bound GTP with establishment of codon-anticodon base-pairing. Importantly, in addition to eIF1, genetic suppressor studies in yeast also implicated eIF2 and eIF5 in ensuring the fidelity of initiation codon selection40. GTP hydrolysis reduces eIF2’s affinity for Met-tRNAMeti leading to partial dissociation of eIF2–GDP from 40S subunits39,48. eIF2B mediates guanine nucleotide exchange on eIF2, recycling it for the next initiation round1.
Ribosomal subunit joining
Joining of 60S subunits and dissociation of eIF1, eIF1A, eIF3 and residual eIF2–GDP are mediated by eIF5B3,49, a ribosome-dependent GTPase that is homologous to prokaryotic initiation factor IF21. Hydrolysis of eIF5B-bound GTP is not required for subunit-joining, but is essential for eIF5B’s own release from assembled 80S ribosomes1. eIF5B occupies the same region in the intersubunit cleft50 as IF2, which was proposed to promote subunit joining by burying large solvent-accessible surfaces on both subunits12. eIF5B alone can partially displace eIF2–GDP from 40S subunits, whereas complete dissociation occurs only in the presence of 60S subunits during the actual subunit-joining event39. Interaction of eIF5B’s CTD with eIF1A’s CTT51,52, which likely becomes possible only after the latter’s displacement from the P-site (Fig.2B) upon initiation codon recognition13, is required for efficient subunit joining and GTP hydrolysis by eIF5B, indicating that eIF1A remains associated with ribosomal complexes throughout the subunit-joining process and dissociates from assembled ribosomes with eIF5B53,54. Although those eIFs that bind to the 40S subunit’s interface must be released before or at subunit-joining, dissociation of eIF3 and eIF4G, which are largely bound to the solvent side (Fig.2A), may be delayed as discussed below.
Reinitiation after a short upstream ORF
About 45–50% of mammalian genes (but only ~13% of yeast genes) encode mRNAs that have at least one short uORF (typically <~30 codons) upstream of the main protein-coding ORF55–57. In these cases, some (usually <50%) of the ribosomes which have translated the uORF may resume scanning and reinitiate at downstream sites. Post-termination events at uORF stop codons probably proceed conventionally, with release of 60S subunits, followed by deacylated tRNA, but then some 40S subunits remain on the mRNA and resume scanning. At this stage such 40S subunits are incompetent for reinitiation because they lack an eIF2-TC, but this does not prevent scanning during which a new eIF2-TC can be acquired. eIF2-TC availability determines how far 40S subunits migrate before acquiring one.
Rescanning and reinitiation efficiency decreases quite abruptly with increasing length of the uORF58, or if it includes stable RNA secondary structures that cause pausing of elongation59. This suggests that it is the time taken to translate the uORF that is critical, rather than the length per se, which leads to the idea that rescanning might depend on some of the eIF–ribosome interactions that promoted initiation at the uORF AUG, persisting for the time taken to complete uORF translation. The indications are that the critical interactions are those involving eIF4G (and therefore also eIF3, which bridges eIF4G binding to the 40S subunit), because reinitiation is only observed if eIF4F and eIF4B, or at a minimum the eIF4G p50 fragment (Fig.3A) plus eIF4A and eIF4B, actually participated in the primary initiation event at the uORF AUG60. Because eIF3 binds mainly to the 40S subunit’s solvent face8 (Fig.2A), by no means all of the eIF3–40S contacts would need to be broken to allow subunit joining. eIF3 could therefore remain bound transiently to the 40S subunit in a metastable state, and if this and the eIF4G–eIF3 interaction were still in place by the time uORF translation had been completed, it could retain the post-termination 40S subunit on the mRNA and promote its rescanning.
As a general rule the uORF sequence has little influence on reinitiation in mammalian systems, but there are exceptions, and the few well-characterised uORFs in yeast mRNAs are quite strongly sequence-dependent (see Supplementary information S2 for a possible explanation for these differences).
CONTROL OF INITIATION FACTOR ACTIVITY
Mechanisms of regulating initiation fall into two broad categories: (i) mechanisms that impact on the initiation factors (or ribosomes), and therefore affect virtually all scanning-dependent initiation; and (ii) those that impact on the mRNA itself, either via sequence-specific RNA-binding proteins or microRNAs (miRNAs), and are therefore potentially selective for certain mRNAs. The best-established examples of the first type are control of the availability of active eIF2 and eIF4F by reversible protein phosphorylation, but eIF4F’s activity is also regulated by irreversible proteolysis of eIF4G (Supplementary information S3).
There are 4 mammalian protein kinases that phosphorylate eIF2α on Ser-5161: haem-regulated kinase, which is probably significant only in erythroid cells; PKR, which is activated by double-stranded RNAs of >~40 bp, and is important in the anti-viral response; PERK, which is a transmembrane endoplasmic reticulum enzyme with its kinase domain in the cytoplasm, and is activated by ER “stress” (due to misfolded proteins in the ER lumen); and a homologue of yeast GCN2 (the only yeast eIF2 kinase), which is activated by starvation of certain amino acids. Phosphorylated eIF2 is fully capable of forming an initiation-competent eIF2-TC, but following its release, phosphorylated eIF2–GDP tightly binds to and sequesters the guanine nucleotide-exchange factor eIF2B, abrogating its activity. eIF2-TC levels consequently fall and most mRNA translation is reduced, but protein synthesis from certain mRNAs with at least two uORFs of appropriate type and position, can actually be stimulated. The best characterized mammalian examples are the transcription factors ATF4 and ATF5 whose expression is increased ~5-fold by activation of PERK62,63. As shown in Fig.4, this stimulation is explained by the particular uORF configuration shared by both mRNAs, with a very short upstream uORF1, and a longer uORF2 overlapping the ATF4/5 ORF. Yeast GCN4 mRNA translation is regulated in a superficially similar way, but with important differences (Supplementary information S4).
Figure 4. The mechanism of regulation of ATF4 and ATF5 mRNA translation.
(a) Diagram shows the sizes, spacing and disposition of the two upstream open reading frames (uORFs) in human, mouse, rat, cow and chicken activating transciption factor 4 (ATF4) mRNAs and the four mammalian ATF5 mRNAs62, 63. (b) The pattern of translation in control (unstressed) conditions when eIF2-GTP-Met-tRNAi ternary complexes (eIF2-TCs) are abundant. Small (40S) ribosomal subunits with associated eIF2-TCs (blue) scan the mRNA in the direction shown by the short horizontal arrows, and nascent protein chains are shown by the black zig-zag line associated with the large (60S) ribosomal subunit. If eIF2-TCs are abundant, most of the 40S subunits that resume scanning after uORF1 translation will acquire a new eIF2-TC in time to initiate translation of uORF2, and ribosomes that translate this second uORF will be unable to initiate at the ATF4/5 AUG because uORF2 is rather too long to allow rescanning, and also because it would require backwards scanning which doesn’t appear to occur over significant distances59. (c) Pattern of translation in stressed conditions (for example, thapsigargin treatment), when eIF2-TC availability is low due to eIF2 phosphorylation by activated PERK. Consequently, most of the 40S subunits that resume scanning after translating uORF1 acquire a new eIF2-TC only after they have passed the uORF2 initiation codon, but in time to initiate at the next AUG which is at the start of the ATF ORF in both cases.
Phosphorylation also affects the intracellular concentration of the eIF4F complex, but indirectly via eIF4E-binding proteins (4E–BPs)64, of which there are three functionally equivalent homologues in mammals. When hypophosphorylated, a 4E-BP binds eIF4E (in a binary complex) which prevents the eIF4E from associating with eIF4G, but phosphorylation of the 4E-BP on multiple sites, mainly via mTOR, releases eIF4E for assimilation into eIF4F.
eIF4E itself is also subject to phosphorylation (on Ser209) by the Mnk1 and Mnk2 kinases, which bind eIF4G’s C-terminus (Fig.3A) and only phosphorylate eIF4E in cis, that is if the eIF4E is bound to the same eIF4G. Although eIF4E phosphorylation appears to fluctuate in parallel with changes in translation efficiency, the Mnk1 Mnk2 double knock-out mouse shows absolutely no eIF4E phosphorylation, yet exhibits no negative phenotype, showing that phosphorylation-dephosphorylation cycles cannot be essential for translation65. Nevertheless, when haematopoetic stem cells engineered to stably express c-myc plus either an Mnk1 mutant or an eIF4E derivative were injected into irradiated mice, the incidence of lymphomas in the recipient mice was significantly lower with a dominant negative Mnk1 mutant than a constitutively active Mnk1 mutant, and also much lower with the non-phosphorylatable Ser209 to Ala eIF4E mutant than wild type eIF4E66. Thus, it appears that excessive eIF4E phosphorylation can promote malignancy.
Phosphorylation of several other factors (eIF1, eIF2β, eIF2Bε, several eIF3 subunits, eIF4G, eIF4B, eIF4H, eIF5 and eIF5B), and ribosomal protein S6 has also been recorded64, and in many cases increases under conditions where translation is activated, e.g. serum refeeding to quiescent cells. However, there is no solid evidence that any of these phosphorylation events are the cause of such activation. On the contrary, in the case of S6 phosphorylation, although the correlation with increased translation seems particularly striking, cells derived from the embryos of double S6 kinase-1 and −2 knock-out mice, or knock-in of an S6 gene with all 5 phosphorylation sites mutated to alanines, show normal regulation of translation67,68. These cases of eIF4E and S6 phosphorylation should serve as warnings against attaching too much significance to what are merely suggestive correlations.
REGULATION BY RNA-BINDING PROTEINS
Regulation by a given sequence-specific RNA-binding protein is selective for those mRNAs which contain the relevant RNA motif in an appropriate position, and is (almost) invariably inhibitory, except for the interaction of poly(A) binding protein (PABP) with the poly(A) tail. Activation of translation of such mRNAs therefore requires sequestration or degradation of the inhibitory protein, or inactivation of its RNA-binding potential, or disruption of its interactions with essential co-repressor proteins.
Regulation by specific 5’-UTR/protein interactions
Regulation by protein-RNA interactions in the 5’-UTR is surprisingly rare, and there is just one well-studied example, namely ferritin mRNAs69. The general principle to emerge from this paradigm is that strong inhibition of initiation requires the protein-RNA interaction to occur at a cap-proximal location, which prevents loading of the 43S complex on to the mRNA70, but not eIF4F binding to the capped 5’-end. Inhibition is much weaker, or even non-existent, if the critical RNA motif is moved to a more cap-distal position, suggesting that if the 43S complex can be loaded, its subsequent scanning will displace the bound protein71. However, this position effect may depend on the affinity of the protein–RNA interaction, since PABP mRNA translation is autoregulated by excess free PABP binding to clustered oligo(A) motifs ~70–130nt downstream of the cap72. In this case, therefore, bound PABP can apparently block scanning 43S complexes without being displaced by them.
5’-UTR sequences are undoubtedly crucial for the regulation of ribosomal protein and translation elongation factor mRNAs, a large group of abundant and exceedingly important mRNAs73. Their translation is very poor in quiescent cells, but is strongly and rapidly activated on serum refeeding, by insulin, and by amino acid availability. This property can be conferred on a reporter by transplanting any ribosomal protein 5’-UTR, which are unusual in all starting with a C (i.e. m7GpppC…), followed by a run of pyrimidines – hence their name 5’-terminal oligopyrimidine tract (5’-TOP) mRNAs. This 5’-TOP motif is necessary and must be in its native, 5’-terminal position, but may not always be sufficient for proper regulation without the rest of the 5’-UTR. The mechanism of regulation remains rather a mystery, largely because some key parameters differ according to cell type and conditions used. For example, sensitivity of TOP mRNA translation to rapamycin varies from almost no effect to a strong (but never complete) inhibition74. The balance of current evidence suggests that regulation is unlikely to be via a straightforward repressor mechanism similar to the ferritin mRNA paradigm, that S6 kinases and S6 phosphorylation are unlikely to play a direct role, but that the phosphatidylinositol 3-kinase (PI3K) pathway is critical73.
Stimulation by PABP binding to the 3’-poly(A) tail
It is often said that a 3’-poly(A) tail with bound PABP is essential for initiation, and that PABP can therefore be considered a canonical eIF75. As supporting evidence, such statements usually cite the fact that a pab deletion is normally lethal to yeast, but this ignores the important caveat that there are numerous by-pass suppressor mutations, which allow yeast to grow (albeit slowly) in the complete absence of PABP76. Experiments in systems as diverse as yeast poly(A) polymerase mutants and rabbit reticulocyte lysates have shown that the translational advantage of polyadenylated over non-polyadenylated mRNAs is greatest under conditions of strong competition for limiting eIFs and/or ribosomes77,78, suggesting that the PABP–poly(A) effect is stimulatory rather than essential.
This stimulatory effect appears to be mainly due to the potential of PABP’s second RRM domain to interact with the eIF4G component of eIF4F, which would normally be bound to the 5’-end of the mRNA79,80. The resulting circularization of the mRNA, in the so-called “closed loop” configuration, is commonly believed to aid recycling of ribosomes on the mRNA (Supplementary information S5), but there is a simpler explanation. Anchoring of eIF4F to the 3’-poly(A) tail via the PABP bridging interaction ensures that eIF4F will remain tethered to the mRNA even if eIF4F’s contacts with the 5’-end of the mRNA are disrupted, whereas it would be lost in the absence of PABP or a poly(A) tail and would need to be recruited de novo from the free eIF4F pool. This consideration alone is sufficient to explain why poly(A) tails confer a particular advantage under competitive conditions. In effect, the naturally-occurring poly(A)–PABP–eIF4F interactions are equivalent to the tethering experiments discussed in the following two sections. Indeed, translation of a poly(A)− mRNA is greatly enhanced by artificially tethering PABP to its 3’-UTR81.
The importance of the tethering (and the closed loop) is shown by the fact that there are analogous interactions with somatic cell histone mRNAs, which lack a 3’-poly(A) tail yet are very efficiently translated despite the competition from bulk (polyadenylated) mRNA. All replication-dependent histone mRNAs have a conserved stem-loop structure near the 3’-end, which binds stem-loop binding protein (SLBP). SLBP interacts with SLBP-interacting protein 1 (SLIP1), which in turn interacts with the N-terminus of eIF4G close to the PABP-binding site (Fig.3A)82. These interactions, which result in tethering eIF4F to the 3’-end, stimulate histone synthesis.
Regulation by 3’-UTR–specific protein interactions
In contrast to the paucity of examples of regulation of initiation by specific protein–RNA interactions in the 5’-UTR, there are numerous cases, most of them important in development, of control via 3’-UTR–protein interactions. It was once widely believed that such regulation was entirely dependent on changes in poly(A) tail length (which could provide a rationale for why regulatory proteins bind to the 3’-UTR), because the regulated mRNAs usually had a short tail when they were translationally repressed, and activation coincided with lengthening of the tail. However, there are clear exceptions (e.g. mouse protamine-1 mRNA, which maintains a long tail throughout the 7 days when it is repressed during spermatogenesis83,84), and cases where translation can be activated without any lengthening of short poly(A) tails85, which together led to the hypothesis that there must be mechanisms whereby 3’UTR-protein interactions regulate initiation more directly than via changes in polyadenylation status.
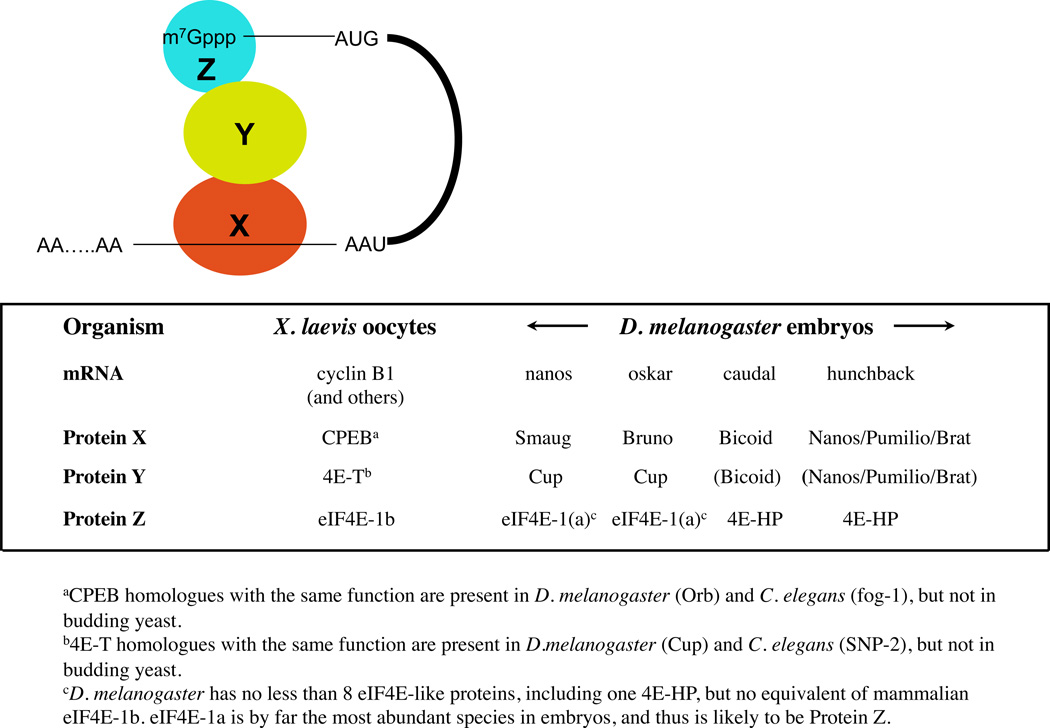
Many of the better understood examples conform to the generic model shown in Box 2 (for specific individual examples, see Ref. 86), in which sequence-specific binding of protein X to the 3’-UTR results in formation of an inhibitory closed loop involving proteins Y and Z. In many cases, protein Y is Cup (in Drosophila melanogaster embryos)86, or its vertebrate homologue, 4E–T86,87, an eIF4E–interacting protein which was first identified as a transporter of eIF4E across the nuclear membrane, but which also has a significant cytoplasmic presence. Protein Z is the canonical eIF4E (eIF4E1a) in some cases, but in the Xenopus laevis CPEB/4E-T system Z is a paralogue, eIF4E1b87, which is restricted to oocytes, eggs and early embryos, and, surprisingly, has rather weak intrinsic affinity for 5’-caps. In D. melanogaster embryos (which lack eIF4E1b) and mouse oocytes, there are examples of repression where Z is another eIF4E paralogue, 4E-HP (eIF4E-homologous protein), which likewise has low intrinsic affinity for caps and cannot bind eIF4G86–88. In addition to the inhibitory closed loop, oligomerization of repressed mRNAs into ill-defined aggregates may provide a further layer of repression89.
Box 2.
Generic model for the regulation of initiation by 3’ UTR-protein interactions
A number of potential co-repressors are often found associated with this protein X–Y–Z complex: a DEAD box helicase (RCK, also known as Xp54 in X. laevis), Pat1p, and two non-specific RNA binding proteins, RAP55 and FRGY287. Homologues of these occur in D. melanogaster embryos (Me31B, PAT1, Trailer hitch and YPS, respectively) and C. elegans (CGH-1, patr-1, CAR-1 and Cey, respectively) and genetic analyses in both organisms have strongly implicated the first three in the mechanism of repression (reviewed in Ref. 87). In the case of X.laevis oocytes, which are not amenable to such genetic analyses, tethering experiments (see Fig.5) have shown that anchoring Xp54, Rap55 or 4E-T to an mRNA causes it to be specifically repressed87,90,91. Interestingly, Pat1p and the DEAD-box helicase appear to act as overlapping regulators of global mRNA translation in yeast92: deletion of both genes prevents the rapid inhibition of initiation that normally occurs on glucose withdrawal.
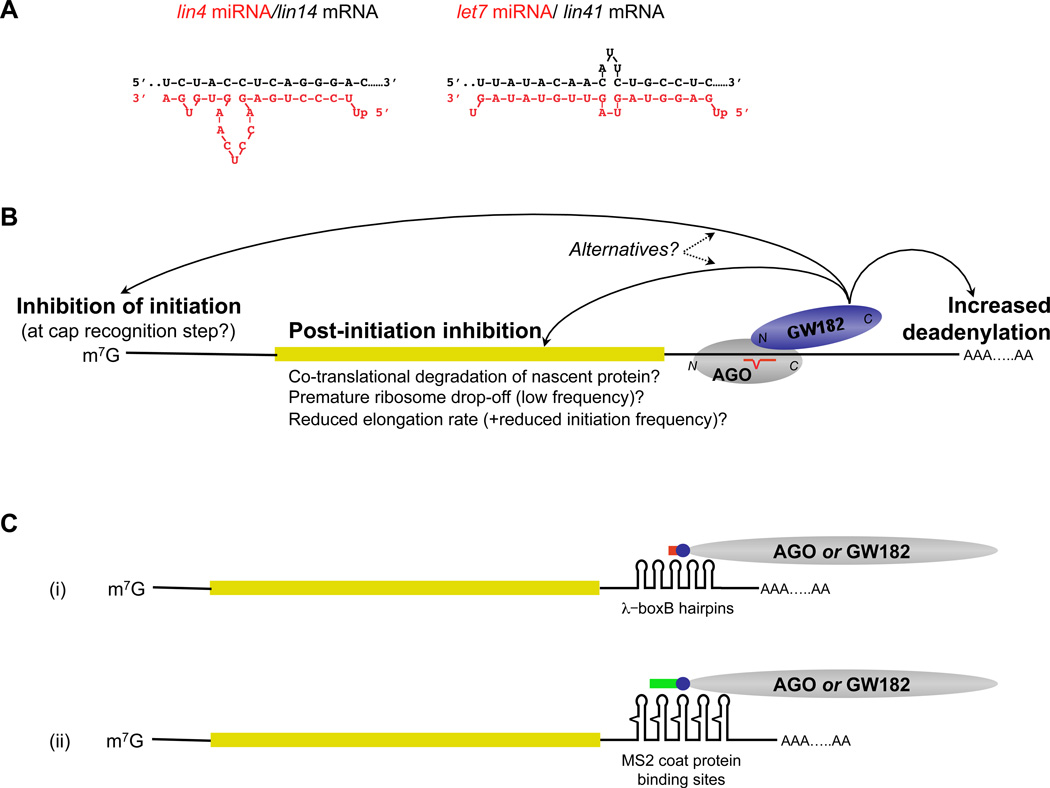
Figure 5. Models of miRNA-mediated repression of translation of target mRNAs.
(a) Examples of the imperfect complementarity between miRNAs (boxed) and their mRNA target sites (upper line) for two validated C. elegans miRNA/mRNA interactions. The interaction typically involves perfect contiguous base-pairing of miRNA residues 2–8 (the seed match), in some cases extending to residues 1–9, followed by mismatch bulges in either the miRNA or mRNA (or both), and then irregular base-pairing of the miRNA 3’-end to the mRNA. (b) Schematic depiction of the different mechanisms by which miRNAs might regulate their target mRNAs. For clarity only a single miRNA target site is shown, and the other proteins in the complex with Ago and GW182 (the most downstream effector of repression identified so far) have been omitted. (c) Tethering experiments showing repression by tethered Ago or GW182. The 3’-UTR of the reporter mRNA has multiple bacteriophage lambda Box B motifs, or bacteriophage MS2 high affinity sites for coat protein; the test protein (Ago or GW182) is expressed as a fusion with an epitope tag (blue), to allow monitoring of expression levels, and either lambda N-peptide (red) or MS2 coat protein (green). Controls have the epitope tag but lack N-peptide or MS2 coat protein sequences. Tethering a translational activator to the 3’-UTR by the same method results in stimulation of translation, e.g. tethering PABP to a poly(A)- mRNA81.
On progesterone-induced maturation of frog oocytes, many of these players become phosphorylated (CPEB, 4E-T, Pat1p and Maskin) followed by extensive degradation87 (at least for CPEB and Pat1p), and these changes are likely to be the key to activation of translation, which must involve disruption of the inhibitory closed loop.
It is striking that the same proteins are implicated in all organisms from worms to vertebrates (Box 2 Table), suggesting a universal mechanism subject to relatively minor variations. Moreover, although these models are based on regulation in development, they are unlikely to be confined to such situations. For example, CPEB paralogues have been found in somatic cells, particularly neuronal tissue, where they are thought to play an important role in synaptic plasticity93.
There are two cases of regulation via the 3’-UTR in somatic cells which do not conform to the above model, but point to a defect in a late stage in initiation, at or soon after the first commitment step. First, binding of the KH-domain proteins hnRNP-K and hnRNP-E1 to tandem 19nt CU-rich repeats in the 3’-UTR of erythroid 15-lipoxygenase mRNA represses its translation until the late reticulocyte stage94. Second, binding of ZBP (another KH-domain protein) to the ~54nt “zip-code” motif located downstream of the stop codon of β-actin mRNA represses translation until the mRNA is properly localized in the lamellipodia of fibroblasts95. In both cases, repression can be recapitulated in an in vitro system in which formation of 80S initiation complexes in the presence of GTP is strongly inhibited, whereas 48S complex formation (with the 40S subunit at the initiation codon) in the presence of GMPPNP is not. Taken at face value, this suggests that the eIF5B-catalysed reaction and/or subunit joining is abortive, leading to unproductive release of 40S subunits from the mRNA. Before this model is taken as gospel, however, we need to be sure that 48S complex formation is also unaffected when such complexes are formed in the presence of GTP (but with eIF5B and 60S subunits absent), to eliminate the possible artefact of GMPPNP stabilising an intermediate that doesn’t actually exist when GTP is used. Nevertheless, the possibility that the eIF5B reaction might be regulated is raised by the provocative finding that the D.melanogaster embryo DEAD-box helicase, Vasa, binds eIF5B, and that this interaction is required for activation of gurken mRNA translation96.
TRANSLATION REGULATION BY miRNAs
miRNAs are another means of repression via the 3’-UTR, and can even act in conjunction with sequence-specific RNA-binding proteins, as has been found for CAT-1 mRNA regulation in liver cells97. The interaction of the ~21nt miRNA with its target sites takes the form shown in Fig.5A. The degree of repression increases with increasing number of miRNAs associated with the 3’-UTR, irrespective of whether or not they are identical98. Repression efficiency may also be influenced by the distance and sequence between miRNA target sites, and by their position in the 3’-UTR.
An Argonaute protein (Ago), of which there are 4 mammalian isoforms, is intimately associated with the paired miRNA–mRNA interaction, and many other proteins are present more peripherally, including the p54 helicase discussed in the previous section and GW182, of which there are three mammalian paralogues, commonly designated TNRC6A, TNRC6B and TNRC6C98. miRNAs therefore act as adaptors, which in effect confer sequence-specific mRNA binding on Ago. In fact, repression can be recapitulated, even in the absence of any miRNA target site, by tethering Ago to the 3’-UTR (Fig.5C)99. Moreover, tethering any of the three human GW182 paralogues can by-pass the requirement for both Ago and miRNA100,101. These assays show that repression is mediated by the C-terminal ~33% of GW182 (the silencing domain)100, whereas the GW-repeat-containing N-terminal domain binds Ago101. Thus, miRNAs recruit Ago, which in turn recruits GW182 — the most downstream effector identified so far.
The mechanism of repression seems to have two components98: first, a true repression of mRNA translation, and second, an accelerated rate of mRNA degradation via the normal deadenylation-dependent pathways102. The relative importance of these two components seems to vary between different miRNA–mRNA pairs for unknown reasons, but in tethering assays the same GW182 silencing-domain was necessary and sufficient for both outcomes101. Two recent reports have shown that this GW182 domain binds PABP103,104 (although they disagree over which domain of PABP is involved), and this in turn can recruit the complex of deadenylating enzymes.
The actual mechanism of true repression of translation remains controversial. Some authors find the repressed mRNA displaced from large polysomes into small polysomes or sub-polysomal particles, indicative of inhibited initiation. Others find the repressed mRNA in polysomes of similar size as when not repressed, implying inhibition at a post-initiation stage. A recent provocative report, yet to be independently confirmed, suggests that the initiation or post-initiation outcome is determined by the identity (but not the efficiency) of the promoter used to drive reporter mRNA synthesis105, for reasons that remain unknown.
The mechanism underlying the post-initiation lesion remains a mystery. One suggestion is specific co-translational degradation of the nascent protein, because polysome-associated nascent protein N-terminal sequences could not be detected by immunoprecipitation106. If confirmed, this eliminates other suggestions of premature ribosome drop-off (which would have to be infrequent to maintain polysome size), or a reduced rate of elongation (which would cause polysomes to increase unless it were coupled with a quantitatively similar reduction in initiation frequency).
As for inhibition of initiation, previous suggestions that Ago itself may interact with the 5’-cap, or that the repression mechanism might impact on eIF6, seem to have both been soundly refuted107. Those who observe inhibition of initiation are generally agreed that strong repression is only seen if the mRNA has the normal m7Gppp.. cap, and not if this is replaced by Appp…, nor if the mRNA has a viral IRES, suggesting that it may be the cap-eIF4F interaction that is the proximal target of the repression mechanism108–110, not unlike the model depicted in Box 2 for regulation by other 3’-UTR–protein interactions. Because the GW182-PABP interaction mentioned above seems to compete against the eIF4G–PABP interaction that maintains the closed loop103,104, the consequent disruption of the closed loop may contribute to translational repression, but this cannot be the complete answer, as translation of poly(A)- mRNAs can also be repressed by miRNAs104,111.
CONCLUSIONS and PERSPECTIVES
The picture which emerges from this review is one of steady progress on most fronts. On mechanism(s) of initiation, further advances can be expected from the approaches that have previously proved most informative: yeast genetics and kinetic analysis in yeast cell-free systems, and mammalian in vitro systems which recapitulate all the steps of translation with purified protein factors. Further insights into initiation complex structure can be expected from cryo-electronmicroscopy and biochemical mapping of initiation factor binding sites on 40S subunits, relying on modeling based on the eubacterial ribosome crystal structure (because crystal structures of eukaryotic ribosomes are unlikely to be available for some time).
A rather urgent problem is to resolve the controversies over the mechanism(s) of miRNA-mediated repression, so that a solid molecular interpretation can be placed on the current flood of bioinformatic, micro-array and proteomic data aimed at elucidating the regulatory networks dependent on the several hundred miRNAs present in vertebrates. Apart from D.melanogaster and C.elegans genetic analyses, further pursuit of what proteins interact with Ago, and more especially with GW182, would seem to be the most promising way forward, coupled with tethered function assays (Fig.5C). The same approaches also appear the best for gaining further insights into mechanisms of regulation by protein/3’-UTR interactions. RNA interference and antisense approaches to knock down specific proteins will undoubtedly also play an increasingly important role.
The biggest gap in knowledge remains the mechanism of regulation of vertebrate TOP mRNAs, where lower eukaryote genetics cannot offer any insights. One possible reason why this topic is proving so difficult may be that we don’t yet fully understand how the PI3K signaling-pathway impacts on translation, and it may also be that regulation of this group of mRNAs is so extremely important that there are multiple overlapping, partially redundant pathways, as a type of fail-safe mechanism.
Supplementary Material
TEXTBOX 1.
IRES-MEDIATED TRANSLATION INITIATION
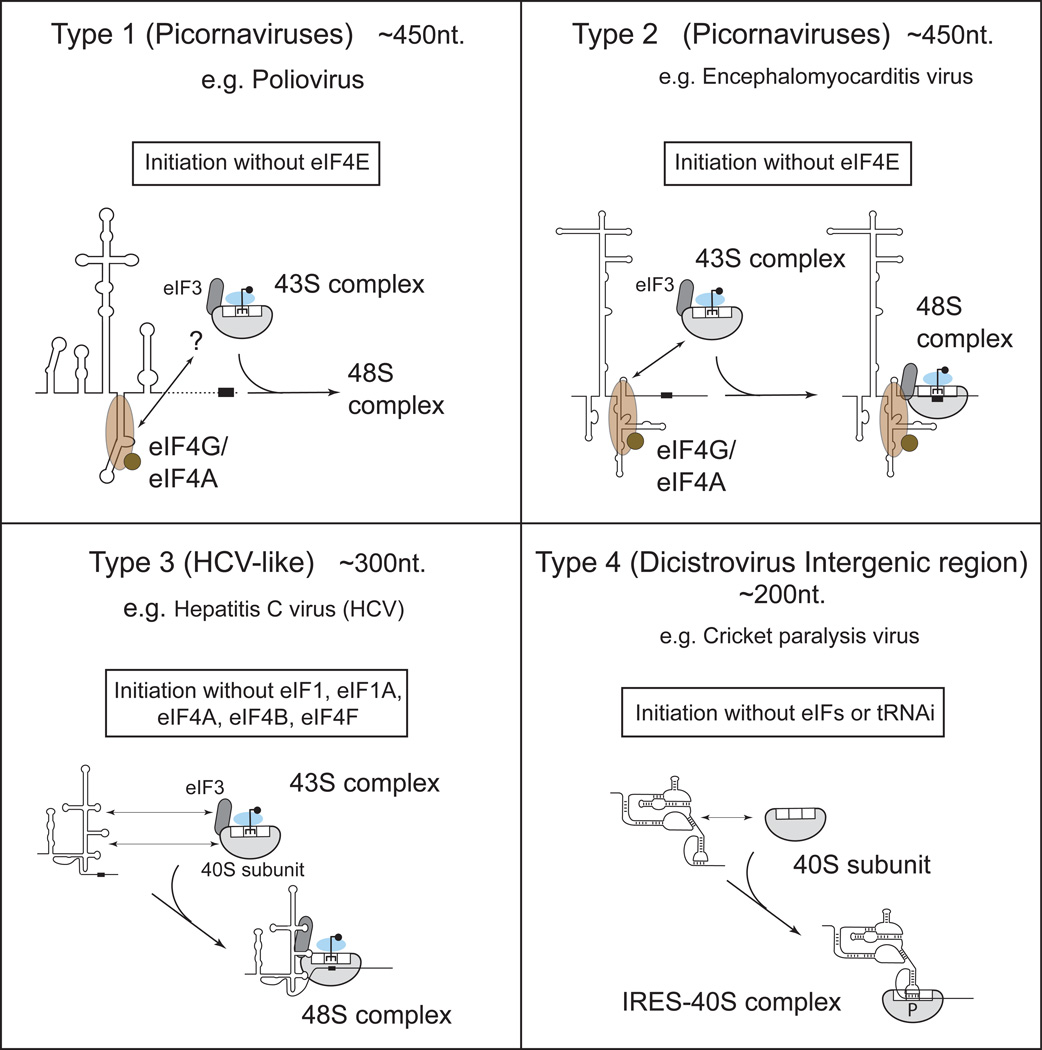
Internal ribosomal entry sites (IRESs) are RNA elements that mediate end-independent ribosomal recruitment to internal locations in mRNA. Structurally-related viral IRESs use distinct mechanisms, based on non-canonical interactions with eIFs and/or 40S subunits (see the Figure). Initiation on Type 1 and Type 2 IRESs involves their specific binding to eIF4G’s central p50 domain (Fig.3A), which is enhanced by eIF4A112–114, on Type 3 IRESs involves their interaction with eIF3 and 40S subunit components of 43S complexes8, 115, and on Type 4 IRESs involves their binding to 40S subunits7, 116. The eIF4G–eIF4A complex recruits 43S complexes to Type 1 and Type 2 IRESs without the involvement of eIF4E, Type 3 IRESs directly attach 43S complexes to the initiation codon independently of eIF4F, eIF4B, eIF1 and eIF1A, whereas Type 4 IRESs initiate without eIFs or tRNAiMet (the 40S subunit’s P-site is occupied by an IRES domain that mimics codon-anticodon base-pairing). Hence, IRES-mediated initiation may be resistant to cellular regulatory mechanisms, such as eIF2 phosphorylation (Type 4 IRESs) and/or eIF4E sequestration (all Type of IRESs)116. Initiation on some IRESs also requires IRES trans-acting factors (ITAFs), RNA-binding proteins that are thought to stabilize the optimal three-dimensional IRES conformation117.
The list of cellular mRNAs believed to contain IRESs grows almost daily, and although a recent stringent test has questioned some of these claims118, it would be prudent to presume that many are still valid. Cellular IRESs show little structural relationship to each other and their underlying mechanism remains largely unknown, but probably follows the picornavirus paradigm of binding the [eIF4G/eIF4A] complex. Importantly, cellular IRES-containing mRNAs can also be translated by the scanning mechanism, which raises the critical question of what regulates the switch between these modes of initiation. One key parameter might be the intracellular concentration of eIF4G. The concentration of eIF4G (but not eIF4E) is highly elevated in many advanced breast cancers, and in inflammatory breast cancer this results in efficient IRES-dependent translation of p120-catenin and vascular endothelial growth factor (VEGF) mRNAs119. In other breast cancer cell lines with high eIF4G levels, overexpression of 4E–BP1 (to sequester eIF4E), coupled with hypoxia, activates VEGF and hypoxia-inducible factor 1α (HIF1α) IRESs120. Another parameter that may determine which mechanism predominates is the intracellular concentration of ITAFs.
TEXTBOX 2.
Generic model for the regulation of translation initiation by sequence-specific 3’-UTR binding proteins. Protein X binds in a sequence-specific manner to critical 3’-UTR motif(s), and interacts with an intermediate bridging protein (Y), which in turn interacts with a cap-binding protein (Z), leading to the formation of an inhibitory closed loop that precludes access of eIF4F to the 5’-end. As protein X is the only sequence-specific RNA-binding protein amongst the three, the identity of protein X in the complex differs more widely between different mRNAs or groups of mRNAs, than the identities of protein Y and protein Z (see the Table). The functions of proteins X and Y are embodied in a single protein (for example, Bicoid), or in a group of proteins (Nanos, Pumilio and Brat)86. It should be noted that although Maskin has been claimed to be protein Y in Xenopus laevis oocytes121, its interactions with cytoplasmic polyadenylation element-binding protein (CPEB) have not been seen in some laboratories87, the motif by which it is supposed to interact with eIF4E is not conserved in Maskins from other species, and it is only expressed in the late stages of oogenesis87.
ACKNOWLEDGEMENTS
We thank Jennifer Doudna, Assen Marintchev and Lori Passmore for figures. Research in the authors’ laboratories is supported by grants from the BBSRC and The Wellcome Trust (RJJ), and the NIH-NIAID (CUTH) and the NIH-NIGMS (TVP).
GLOSSARY
- Met-tRNAMeti
the unique initiator tRNA that, aminoacylated with methionine, is used to initiate protein synthesis. Its anticodon is complementary to the AUG initiation codon, it forms a specific ternary complex with eIF2 and GTP and it binds to the ribosomal P-site
- P-site
the site on the ribosome that holds tRNA that is linked to the growing peptide chain (peptidyl-tRNA)
- IRES (Internal ribosome entry site)
A structure that is located in the 5' UTR or open reading frame of some mRNAs of cellular or viral origin. It mediates translation initiation independently of the mRNA’s 5’-end by recruiting the ribosome directly to an internal position on the mRNA.
- ATP-binding cassette (ABC) proteins
typically contain two nucleotide-binding domains (NBDs) that form two composite nucleotide-binding sites. The transition between the closed ATP-bound and open ADP-bound states induce a tweezer-like powerstroke between NBDs that cause conformational changes in associated domains and/or macromolecules
- 18S rRNA
Ribosomal RNA of the 40S ribosomal subunit. It determines the overall shape of the 40S subunit and is the main component of its decoding center. It is also involved in formation of the main contacts between 40S and 60S ribosomal subunits
- A-site
the site on the ribosome that holds the new incoming aminoacyl-tRNA
- E-site
the site on the ribosome that accommodates deacylated tRNA before it is released from the ribosome
- DEAD-box RNA helicase
An RNA helicase that contains the DEAD (Asp-Glu-Ala-Asp) or DEXD/H (Asp-Glu-X-Asp/His, where X represents any amino acid) motif. These proteins use the energy of ATP hydrolysis to unwind RNA
- RRM domain
The RRM (RNA-recognition motif) contains two short consensus sequences embedded in a structurally conserved region of approximately 80 amino acids
- HEAT domain
A protein domain of 37-47 amino acids consisting of tandemly-repeated pairs of antiparallel alpha helices. HEAT domains have a superhelical structure and often function as protein-protein interaction surfaces
- GTPase-activating protein
A protein that stimulates the intrinsic ability of a GTPase to hydrolyse GTP to GDP
- Arginine finger
A catalytic residue that was first defined for Ras-GTPase-activating proteins (RasGAPs), and that supplies a catalytic arginine residue into the active site of Ras to increase the reaction rate
- MicroRNA
A small RNA of ~ 21 nucleotides that regulates the expression of mRNAs to which it is partially complementary in sequence
- KH domain
(K-homology domain). Originally identified in the human hnRNP K protein, this domain is important for RNA binding and probably binds RNA directly
- Argonaute
A family of proteins that are characterized by the presence of two homology domains, PAZ and PIWI. These proteins are essential for diverse RNA silencing pathways
Footnotes
DATABASES
The following terms in this article are linked online to: UniProtKB: http://www.uniprot.org
eIF1 | eIF1A | eIF2α | eIF2β | eIF2γ | eIF2Bα | eIF2Bβ | eIF2Bγ | eIF2Bδ | eIF2Bε | eIF3a | eIF3b | eIF3c | eIF3d | eIF3e | eIF3f | eIF3g | eIF3h | eIF3i | eIF3j | eIF3k | eIF3l | eIF3m | eIF4A | eIF4B | eIF4E | eIF4G | eIF4H | eIF5 | eIF5B | eIF6 | p97 | DHX29 | DDX3 | PABP | ABCE1 | eRF1 | eRF3 | 4E-BP1 | 4E-BP2 | 4E-BP3 | 4EHP | eIF4E-T | hnRNP-E | hnRNP-K | HCR | PERK | PKR | hGCN2 | TNRC6A | TNRC6B | TNRC6C | hAGO1 | RCK/p54 | xRAP55 | CUP |
REFERENCES
- 1.Pestova TV, Lorsch JR, Hellen CUT. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2007. pp. 87–128. [Google Scholar]
- 2.Pisarev AV, Hellen CUT, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon- anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. 2004. This biochemical study shows that eIF1 is a negative regulator of hydrolysis of eIF2-bound GTP which inhibits the commitment step in initiation until codon–anticodon base-pairing is established: eIF1 thus ensures the fidelity of initiation both after and during scanning
- 4.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 5.Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat. Rev. Mol. Cell. Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 7. Schüler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. 2006) This cryoEM reconstruction of a dicistrovirus IRES bound to the 80S ribosome showed details at subnanometer resolution of the IRES-ribosome interaction and reveals the potential for conformational changes in the IRES that enable it to promote successive steps in an exceptional mechanism of internal initiation
- 8.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 9.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl. Acad. Sci. USA. 2007;104:18445–18450. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. The first mapping of an eIF-binding site on the ribosome by directed hydroxyl radical probing, which showed that eIF1 binds the 40S subunit platform near the P-site, in a position that enables it to maintain the fidelity of initiation codon selection indirectly, by influencing the conformation of the platform and the positions of mRNA and initiator tRNA in initiation complexes
- 11.Simonetti A, et al. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 12.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli . Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Research. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. CryoEM reconstructions of yeast 40S subunits bound to eIF1 and eIF1A, showing induced conformational changes that open the mRNA-binding channel in a scanning-competent conformation which reverses upon initiation codon recognition and consequent eIF1 release to clamp down on the mRNA
- 15.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 17.Gross JD, et al. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 18.Volpon L, Osborne MJ, Topisirovic I, Siddiqui N, Borden KL. Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. EMBO J. 2006;25:5138–5149. doi: 10.1038/sj.emboj.7601380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 20.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 21.Schütz P, et al. Crystal structure of the yeast eIF4A–eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marintchev A, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. 2009) In this study, modeling based on known structures of factor domains, NMR, quantitative binding assays and site-directed mutagenesis were used to derive a model of the eIF4A–eIF4G–eIF4H (or eIF4A–eIF4G–eIF4B) helicase complex and to propose hypotheses for its dynamic organization, location and modus operandi on the scanning ribosomal complex
- 23.LeFebvre AK, et al. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 25.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CUT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson RJ. The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur. J. Biochem. 1991;200:285–294. doi: 10.1111/j.1432-1033.1991.tb16184.x. [DOI] [PubMed] [Google Scholar]
- 27.Svitkin YV, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5’ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda D, Dreher TW. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12:1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pisareva VP, et al. Translation initiation on mammalian mRNAs with structured 5’UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. Identifies the DExH-box protein DHX29 as a novel initiation factor that promotes ribosomal scanning, particularly on highly structured 5’-UTRs, in a translation system reconstituted from highly purified factors
- 30.Parsyan A, et al. The helicase protein, DHX29 promotes translation initiation, cell proliferation and tumorigenesis. Proc. Natl. Acad. Sci. USA. 2009 Dec 11; doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. Reports the importance of DHX29 for initiation in vivo
- 32.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E–dependent translation in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsden S, Nardelli M, Linder P, McCarthy JE2006. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J. Mol. Biol. 2006;361:327–335. doi: 10.1016/j.jmb.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Tarn WY, Chang TH. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 35.Imataka H, Olsen HS, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell. Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 39.Pisarev AV, et al. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donahue TF. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2000. pp. 487–502. [Google Scholar]
- 41.Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 42. Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. Start codon recognition induces conformational change in yeast ribosomal initiation complexes and the displacement of eIF1, likely resulting in closure of the mRNA-binding channel (Ref. 14) and in triggering of hydrolysis of eIF2-bound GTP (Ref. 3), respectively, thereby committing the arrested 48S complex to the initiation codon
- 43.Cheung Y-N, et al. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fekete CA, et al. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulin FE, Campbell LE, O’Brien K, Loughlin J, Proud CG. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr. Biol. 2001;11:55–59. doi: 10.1016/s0960-9822(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 46.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 47.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 49.Pestova TV, et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 50.Unbehaun A, et al. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen DS, et al. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc. Natl. Acad. Sci. USA. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acker MG, Shin BS, Dever TE, Lorsch JR. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 2006;281:8469–8475. doi: 10.1074/jbc.M600210200. [DOI] [PubMed] [Google Scholar]
- 54.Acker MG, et al. Kinetic analysis of late steps of eukaryotic translation initiation. J. Mol. Biol. 2009;385:491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resch AM, Ogurtsov AY, Rogozin IB, Shabalina SA, Koonin EV. Evolution of alternative and constitutive regions of mammalian 5’UTRs. BMC Genomics. 2009;10:162. doi: 10.1186/1471-2164-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawless C, et al. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009;10:7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luukkonen BGM, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pöyry TAA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. Data obtained using a panel of mRNAs that have different eIF requirements, suggesting that eIF3 and eIF4G remain weakly associated with ribosomes during translation of short uORFs and that these factors then promote the resumption of scanning, leading to reinitiation
- 61.Dever TE, Dar AC, Sicheri F. In: Translational control in biology and medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. New York, USA: Cold Spring Harbor Laboratory Press; 2007. pp. 319–344. [Google Scholar]
- 62. Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. Shows how the configuration of the two uORFs in ATF4 mRNA results in stimulated ATF synthesis following PERK activation
- 63.Zhou D, et al. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 64.Raught B, Gringras A-C. In: Translational control in biology and medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. New York, USA: Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- 65.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eIF4E but not for cell growth or development. Mol. Cell. Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wendel HG, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pende M, et al. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5’- terminal oligopyrmidine tract mRNA translation and reveal a mitogen-activated protein kinase- dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ruvinsky I, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. Shows that transgenic knock-in mice homozygous for non-phosphorylatable rpS6 are viable and normal, and show proper regulation of ribosomal protein mRNA translation
- 69.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 71.Paraskeva E, Gray NK, Schläger B, Wehr K, Hentze MW. Ribosomal pausing and scanning arrest as mechanisms of translational regulation from cap-distal iron-responsive elements. Mol. Cell. Biol. 1999;19:807–816. doi: 10.1128/mcb.19.1.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Melo Neto OP, Standart N, Martins de Sa C. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 1995;23:2198–2205. doi: 10.1093/nar/23.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPS and their regulation. Biochem. Soc. Trans. 2006;34:12–16. doi: 10.1042/BST20060012. [DOI] [PubMed] [Google Scholar]
- 74.Patursky-Polischuk I, et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol. Cell. Biol. 2009;29:640–649. doi: 10.1128/MCB.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kahvejian A, Svitkin YV, Sukareieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 77.Proweller A, Butler JS. Ribosome concentration contributes to discrimination against poly(A)- mRNA during translation initiation in Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:6004–6010. doi: 10.1074/jbc.272.9.6004. [DOI] [PubMed] [Google Scholar]
- 78.Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G–PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5’-end. Nucleic Acids Res. 2000;28:4068–4075. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol. Cell. Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. SLIP1 is shown to interact with both SLBP (bound to the 3’ stem-loop of histone mRNAs) and the eIF4G subunit of eIF4F, forming a ‘closed loop’ that stimulates histone mRNA translation
- 83.Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989;106:367–373. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- 84.Braun RE, Peschon JJ, Behringer R. Brinster, R. L. & Palmiter, R. D. Protamine 3’untranslated sequences regulate temporal translational control and subcellular localization of growth hormone in spermatids of transgenic mice. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 85.Standart N, Dale M, Stewart E, Hunt T. Maternal mRNA from clam oocytes can be specifically unmasked in vitro by antisense RNA complementary to the 3’-untranslated region. Genes Dev. 1990;4:2157–2168. doi: 10.1101/gad.4.12a.2157. 1990. [DOI] [PubMed] [Google Scholar]
- 86.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E–T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. Repression of maternal mRNAs in vertebrate (X. laevis) oocyctes is shown to involve the binding of a large CPEB-containing protein complex to the 3’-UTR, and a CPEB – 4E–T – eIF4E-1b tripartite interaction relay, which blocks eIF4F access to the 5’-cap
- 88.Villaescusa JC, et al. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One. 2009;4:e5213. doi: 10.1371/journal.pone.0005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 90.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanaka KJ, et al. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 2006;281:40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- 92.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klann E, Richter JD. In: Translational control in biology and medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. New York, USA: Cold Spring Harbor Laboratory Press; 2007. pp. 485–506. [Google Scholar]
- 94.Ostarek DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: The 3’UTR regulatory complex controls 60S subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 95.Hüttelmaier S, et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 96.Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 97. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. One of the few studies of miRNA-mediated repression of an endogenous vertebrate mRNA, showing an inhibition of initiation that is relieved on amino acid starvation
- 98.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007 doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 99.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. Tethering the C-terminal domain of GW182 to the 3’-UTR is shown to recapitulate miRNA-mediated repression of translation
- 101.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR:NOT deadenylase and DCP1:DCP2 decapping enzymes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. Tethering GW182 to the 3’-UTR in the absence of AGO or miRNAs is show to result in both translational repression and accelerated mRNA degradation via the normal deadenylation- dependent route
- 103.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zekri L, Huntzinge E, Heimstädt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of miRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kong YW, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl. Acad. Sci. USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 107.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 108. Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap- binding complex elF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. Recapitulation of miRNA-mediated repression in a mouse cell-free extract suggests that miRNAs directly or indirectly inhibit eIF4F function
- 109.Thermann R. & Hentze, M.W., Drosphila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 110.Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 111.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pestova TV, Shatsky IN, Hellen CUT. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wilson JE, Pestova TV, Hellen CUT, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 117.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 118.Baranick BT, et al. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl. Acad. Sci. USA. 2009;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Silvera D, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell. Biol. 2009;11:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 120.Braunstein S, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 121.Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB- associated factor that transiently interacts with eIF4E. Mol. Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.