Abstract
A general model has been proposed for the fusion mechanisms of class I viral fusion proteins. According to this model a metastable trimer, anchored in the viral membrane through its transmembrane domain, transits to a trimeric prehairpin intermediate, anchored at its opposite end in the target membrane through its fusion peptide. A subsequent refolding event creates a trimer of hairpins (often termed a six-helix bundle) in which the previously well-separated transmembrane domain and fusion peptide (and their attached membranes) are brought together, thereby driving membrane fusion. While there is ample biochemical and structural information on the trimer-of-hairpins conformation of class I viral fusion proteins, less is known about intermediate states between native metastable trimers and the final trimer of hairpins. In this study we analyzed conformational states of the transmembrane subunit (TM), the fusion subunit, of the Env glycoprotein of the subtype A avian sarcoma and leukosis virus (ASLV-A). By analyzing forms of EnvA TM on mildly denaturing sodium dodecyl sulfate gels we identified five conformational states of EnvA TM. Following interaction of virions with a soluble form of the ASLV-A receptor at 37°C, the metastable form of EnvA TM (which migrates at 37 kDa) transits to a 70-kDa and then to a 150-kDa species. Following subsequent exposure to a low pH (or an elevated temperature or the fusion promoting agent chlorpromazine), an additional set of bands at >150 kDa, and then a final band at 100 kDa, forms. Both an EnvA C-helix peptide (which inhibits virus fusion and infectivity) and the fusion-inhibitory agent lysophosphatidylcholine inhibit the formation of the >150- and 100-kDa bands. Our data are consistent with the 70- and 150-kDa bands representing precursor and fully formed prehairpin conformations of EnvA TM. Our data are also consistent with the >150-kDa bands representing higher-order oligomers of EnvA TM and with the 100-kDa band representing the fully formed six-helix bundle. In addition to resolving fusion-relevant conformational intermediates of EnvA TM, our data are compatible with a model in which the EnvA protein is activated by its receptor (at neutral pH and a temperature greater than or equal to room temperature) to form prehairpin conformations of EnvA TM, and in which subsequent exposure to a low pH is required to stabilize the final six-helix bundle, which drives a later stage of fusion.
The fusion-promoting glycoproteins of orthomyxo-, paramyxo-, filo-, corona-, and retroviruses are grouped as class I fusion proteins because their fusion subunits possess a common postfusion form (12, 19, 20, 57). The structure they have in common consists of a fusion peptide at or near the N-terminal end, a central trimeric coiled coil, a chain reversal segment, a C-terminal region (in many cases a helix) that packs in the grooves of the central coiled coil, a transmembrane domain, and a cytoplasmic tail. In the compact postfusion form, the two hydrophobic elements, the fusion peptide and the transmembrane domain, lie in close proximity. All class I fusion proteins start out as metastable trimers on the virus surface. In only one case, that of the influenza virus hemagglutinin (HA), do we know the atomic structure of the complete ectodomain of the metastable trimer (63). Nonetheless, a common model has emerged for how class I fusion proteins mediate fusion. In essence, the metastable trimer is anchored in the viral membrane through its transmembrane domain. Following a triggering event (interaction with receptors or exposure to a low endosomal pH), the metastable trimer converts to a prehairpin intermediate in which the newly exposed fusion peptide binds to the target bilayer. The prehairpin is envisioned as a straight trimeric coiled-coil rod with the fusion peptide at one end (in the target membrane) and the transmembrane domain at the opposite end (in the viral membrane). Through conformational changes that essentially fold the prehairpin in half, the two hydrophobic elements (and their attached bilayers) are brought together to initiate membrane merger. Support for the common prehairpin intermediate has come from the use of C-helix peptide inhibitors. C-helix peptides from class I fusion proteins that function at neutral pH (e.g., those of paramyxoviruses, coronaviruses, and retroviruses) inhibit fusion and infectivity (6, 18-20, and references therein). Experiments with C-helix peptide inhibitors at fusion-arrested states have supplied evidence for prehairpin conformations of human immunodeficiency virus (HIV) Env (44) and a paramyxovirus F protein (52). For both of these proteins a prehairpin intermediate has been trapped biochemically by coimmunoprecipitating the fusion proteins with a cognate epitope-tagged C-helix peptide during fusion activation (22, 52).
Our laboratory has been studying the fusion mechanism of a model retrovirus, the subtype A avian sarcoma/leukosis virus (ASLV-A). The Env glycoprotein of ASLV-A (EnvA) is predicted to be a class I fusion protein with structural similarity between its fusion subunit and those of human T-cell leukemia virus Env and Ebola virus GP (24, 36). The fusion peptide is sequestered in the native EnvA trimer but is exposed and binds to target membranes following interaction with the host cell receptor Tva (2) at a temperature greater than or equal to room temperature (T ≥ RT) (26, 28, 29). Our work indicates that EnvA (on virus particles) can mediate both the target bilayer binding and lipid-mixing stages of fusion at neutral pH and T ≥ RT (18, 25, 28). Other work indicates that reverse transcripts of the virus are not produced if infected cells are maintained in the continued presence of agents (e.g., bafilomycin) that raise the pH of endosomes (45; S. Matsuyama, L. Earp, and J. M. White, unpublished data). The latter findings suggest that a low pH is needed for either a later stage of fusion or viral uncoating.
As part of our ongoing effort to delineate the precise stage of ASLV entry that requires a low pH, we have modified and extended an analysis (45) of conformational intermediates in the fusion subunit (the transmembrane subunit [TM]) of EnvA. Our modified approach entailed assessing sequential effects of a minimal soluble receptor (62) and a low pH on the EnvA TM found on virus particles. For this analysis we conducted Western blot assays of viral samples prepared under mildly denaturing conditions. This analysis, in conjunction with specific fusion-inhibitory (e.g., a potent C-helix peptide inhibitor [18]) and fusion-promoting agents, revealed sequential intermediates in the pathway of fusion that represent prehairpin and hairpin forms of EnvA. Our findings are consistent with a model in which interaction with receptor at neutral pH activates EnvA to form prehairpin intermediates and in which a low pH is needed to stabilize a final six-helix bundle (hairpin) form of EnvA at a later stage of fusion.
MATERIALS AND METHODS
Virus, soluble receptor, and peptide R99.
ASLV-A was purified from DF-1 cells chronically infected with RCASBP(A)AP as described previously (18). A soluble form of the receptor (sTva) was prepared as described previously (62). Peptide R99 was described previously (18).
Lipids and liposomes.
l-α-Phosphatidylcholine (PC; egg; Avanti Polar Lipids), l-α-phosphatidylethanolamine (PE; egg; Avanti Polar Lipids), sphingomyelin (Sph; brain; Avanti Polar Lipids), and cholesterol (Chol; Sigma Chemical Co.) were stored as described previously (28). Lipids (1:1:1:1.5 molar PC-PE-Sph-Chol ratio) were mixed, dried under N2 gas in a glass tube, and lyophilized overnight. After addition of buffer (20 mM morpholineethanesulfonic acid, 20 mM HEPES, pH 7.4), the lipid suspension was vortexed, sonicated in a water bath sonicator, and then extruded 25 times through a 0.1-μm-pore-size Nuclepore filter in an Avanti Mini-Extruder. Liposomes (5.6 mM lipid on the basis of the input lipid) were stored at 4°C and used within 1 week.
Membrane-perturbing reagents.
LPC (lysophosphatidylcholine) and CPZ (chlorpromazine) were purchased from Sigma Chemical Co. Fresh stocks of LPC and CPZ (25 mM in 20 mM morpholineethanesulfonic acid-20 mM HEPES, pH 7.4) were prepared just before use.
TM oligomer assay.
The TM oligomer assay developed by P. Bates and coworkers was performed as described in reference 45, with minor modifications. In brief, 2.5 μl of freshly harvested and freshly concentrated ASLV-A and 0.5 μl (300 ng) of sTva were mixed on ice. After 30 min, 2.5 μl of liposomes was added on ice. For one-step incubations, samples were warmed to 37°C for 30 min (unless stated otherwise). For two-step incubations, samples were warmed to 37°C for 30 min (at neutral pH) and then adjusted to pH 5 (unless stated otherwise) with a predetermined amount of 0.1 M HCl. After 5 min at 37°C, 0.5 μl of 1 M Tris (pH 8.0) was added to neutralize low-pH-treated samples. Samples were then treated with buffer to yield final concentrations of 0.1% sodium dodecyl sulfate (SDS), 2% β-mercaptoethanol, and 6% glycerol, incubated for 5 min at 37°C, and then loaded onto SDS-7% polyacrylamide gel electrophoresis gels. Following electrophoresis, proteins were transferred to nitrocellulose and probed with a rabbit polyclonal antibody against a peptide corresponding to the cytoplasmic tail of EnvA (18). Primary antibody was detected with horseradish peroxidase-conjugated anti-rabbit antibody (Amersham Biosciences).
RESULTS
Receptor and a low pH sequentially induce different conformational states of EnvA TM.
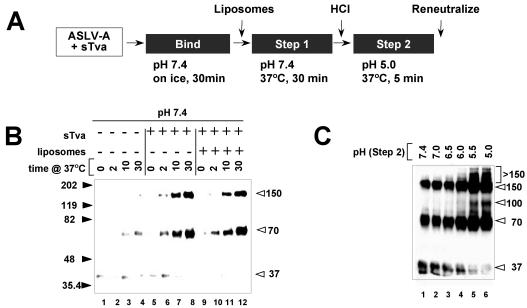
We set out to compare conformational states of the EnvA TM subunit (the fusion subunit) formed during each step of a two-step protocol (Fig. 1A). In all cases, freshly prepared ASLV particles were premixed with sTva, a soluble form of the EnvA receptor (62), on ice for 30 min at neutral pH. After the addition of PC-PE-Sph-Chol liposomes, samples were incubated at 37°C for 30 min at neutral pH (step 1). The pH was then adjusted to 5.0, and the mixture was incubated for an additional 5 min at 37°C (step 2). Samples were then neutralized, mixed with sample buffer to a final concentration of 0.1% SDS, incubated at 37°C for 5 min, and analyzed by Western blot assays for forms of the TM (fusion) subunit. The EnvA TM subunit from starting virions was present as a 37-kDa band. This band is identical in size to the monomeric band obtained when virions are boiled with sample buffer containing 1% SDS. When virions with prebound receptor were incubated at 37°C, bands of 70 and 150 kDa appeared sequentially in a time-dependent manner (Fig. 1B, lanes 5 to 8). A small amount of the 70-kDa band formed upon warming in the absence of receptor (lanes 3 and 4). Receptor-induced formation of the 70- and 150-kDa bands occurred equally well in the absence (lanes 5 to 8) or presence (lanes 9 to 12) of liposomes and equally well with 1/400 of the amount of receptor (S. Matsuyama, Z. Chen, and J. M. White, unpublished data). Moreover, incubation of receptor-bound virions for longer times (60 to 90 min) at 37°C at neutral pH did not change the gel pattern seen in lanes 8 and 12 (Matsuyama, Chen, and White, unpublished).
FIG. 1.
Sequential formation of distinct EnvA TM conformational states. (A) Schematic of the procedure used to generate various conformational states of EnvA TM. Unless stated otherwise, virions (ASLV-A) were incubated on ice for 30 min in the presence of receptor (sTva). Liposomes were then added, and the samples were warmed to 37°C at neutral pH for 30 min (step 1). As indicated (step 2), the pH was adjusted to 5.0 for 5 min. Low-pH-treated samples were then reneutralized and processed to visualize EnvA TM as described in Materials and Methods. (B) Virions were subjected to step 1 conditions in the absence of either receptor or liposomes (lanes 1 to 4), with receptor only (lanes 5 to 8), or with receptor and liposomes (lanes 9 to 12) for the indicated times at 37°C and then processed as described above. (C) Receptor-bound virion-liposome complexes from step 1 (panel B, lane 12) were exposed to the indicated pH during step 2 and then processed as described above. The values on the left and right of panels B and C are molecular sizes in kilodaltons.
We note that although equal amounts of virus were loaded per lane, the 70- and 150-kDa bands appeared proportionately more prominent than the 37-kDa band. Although our ability to detect the 37-kDa band is variable from experiment to experiment, this observation suggests that the cytoplasmic tail epitope of EnvA TM is hyperreactive in the 70- and 150-kDa bands in this gel system.
We next exposed virus-receptor-liposome complexes from step 1 to pH values ranging from 7.4 to 5.0 for 5 min at 37°C (step 2). After treatment at pHs 5.5 and 5.0 a new EnvA TM band at 100 kDa and a mixture of bands at >150 kDa were clearly observed (Fig. 1C, lanes 5 and 6). Smaller amounts of the 100- and >150-kDa bands were seen in samples treated at pH 6.5 (lane 3) or 6.0 (lane 4). Bands at 70 and 150 kDa were still present after the two-step protocol. This may be due, in part (see Discussion), to the previously noted apparent hypersensitivity of these forms of EnvA TM to the anti-EnvA cytoplasmic tail antibody on immunoblots. On the basis of the pH dependence observed in Fig. 1C, we used pH 5.0 for the low-pH treatment in subsequent experiments. Liposomes were present in all subsequent experiments.
To rule out the possibility that any of the higher-molecular-weight bands of EnvA TM (70, 150, 100, or >150 kDa) resulted from stable association with internal viral components, immunoblots of samples treated by either step 1 or the sequential step 1-step 2 protocol were probed with antibodies against the matrix and capsid proteins. None of the EnvA species reacted with anti-matrix or anti-capsid antibodies (data not shown).
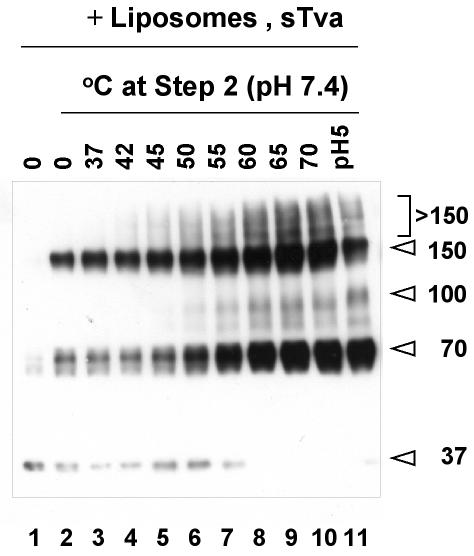
An elevated temperature can mimic the effects of a low pH at step 2.
In the case of influenza virus, an elevated temperature can substitute for a low pH to trigger fusion-relevant conformational changes (7, 51). Similarly, an elevated temperature can substitute for the need for the HN protein for SV5-mediated fusion (47). We therefore asked whether an increased temperature could mimic the effects of a low pH in step 2. Accordingly, complexes were subjected to the step 1 treatment, followed by incubation for 5 min at increasing temperatures (at pH 7.4). As shown in Fig. 2, the >150-kDa bands began to form at 42°C and the 100-kDa band began to form at 50°C, suggesting that the >150-kDa bands form prior to the 100-kDa band. The >150- and 100-kDa bands increased in intensity with increasing temperature.
FIG. 2.
Effects of an increased temperature, in lieu of a low pH, at step 2 on conformational states of EnvA TM. Virions (lane 1) were treated to step 1 conditions (lane 2) and then to the indicated temperature (instead of a low pH) at step 2 (lanes 3 to 10). Lane 11 is a sample of virions treated to the two-step protocol. Samples were processed to visualize EnvA TM as described in the legend to Fig. 1. The values on the right are molecular sizes in kilodaltons.
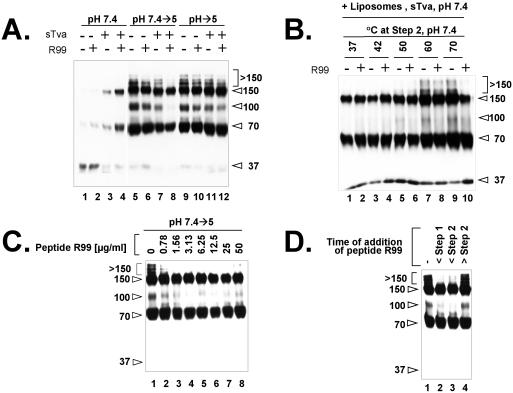
Effects of peptide R99 on the formation of different conformational states of EnvA TM.
Peptide R99 is a synthetic analogue of the EnvA C helix, which inhibits viral infectivity, virus-cell fusion, and EnvA-mediated cell-cell fusion (18). We therefore asked whether peptide R99 affects the formation of any of the higher-molecular-weight forms of EnvA TM. To do this we performed step 1 and 2 incubations (Fig. 1A) in the presence or absence of peptide R99. As shown in Fig. 3A (lane 4), peptide R99 did not inhibit the formation of either the 70- or the 150-kDa band at neutral pH (step 1). However, when virions were subjected to step 2 of the procedure in the presence of peptide R99, the >150- and 100-kDa bands did not form (Fig. 3A, lane 8). Peptide R99 did not inhibit the formation of the >150- and 100-kDa bands if virions were exposed to a low pH in step 2 in the absence of receptor (lane 6) or if they were directly warmed to pH 5.0 in the presence or absence of receptor (lanes 10 and 12). Hence, receptor activation at neutral pH (T ≥ RT) is a prerequisite for forming the >150- and 100-kDa bands along a pathway that is inhibitable by peptide R99. The ability of an increased temperature (in lieu of a low pH) at step 2 to induce formation of the >150 and 100-kDa bands was also inhibited by peptide R99 (Fig. 3B), albeit apparently less effectively than when a low pH was used in step 2 (Fig. 3A).
FIG. 3.
Effects of peptide R99 on conformational states of EnvA TM. (A) Virions were incubated under step 1 conditions (lanes 1 to 4) or under step 1 and then step 2 conditions (lanes 5 to 8) or directly warmed at pH 5 (lanes 9 to 12) in the absence (lanes 1, 2, 5, 6, 9, and 10) or presence (lanes 3, 4, 7, 8, 11, and 12) of receptor and in the absence (odd-numbered lanes) or presence (even-numbered lanes) of peptide R99 (50 μg/ml). Liposomes were present in all samples. (B) Receptor-bound virion-liposome complexes that had been subjected to step 1 conditions were treated to increasing temperatures (in lieu of a low pH) at step 2 in the absence (odd-numbered lanes) or presence (even-numbered lanes) of peptide R99 (50 μg/ml). (C) Receptor-bound virion-liposome complexes were subjected to the two-step protocol in the presence of the indicated amount of peptide R99. (D) Peptide R99 (50 μg/ml) was added to samples before step 1 (lane 2), after step 1 but before step 2 (lane 3), or after step 2 (lane 4). Lane 1 contains a sample subjected to the two-step protocol in the absence of peptide R99. Samples were processed to visualize EnvA TM as described in the legend to Fig. 1. The values on the left or right are molecular sizes in kilodaltons.
The experiments described in Fig. 3A and B were performed with 50 μg of peptide R99 per ml, a concentration that completely inhibits the lipid-mixing activity of ASLV-A particles (18). However, as shown in Fig. 3C, concentrations of peptide R99 as low as 0.78 μg/ml inhibited the formation of the >150- and 100-kDa bands. We next asked if we could define a point in the protocol at which the >150- and 100-kDa bands become refractory to peptide R99. As shown in Fig. 3D, peptide R99 inhibited the formation of the >150- and 100-kDa bands if it was added either before step 1 (warming of virus-receptor complexes at neutral pH) (lane 2) or before step 2 (subsequent incubation at a low pH) (lane 3). If peptide R99 was added after the low-pH incubation step (lane 4), formation of the >150- and 100-kDa bands was not inhibited.
Effects of fusion modulators on different conformational states of EnvA TM.
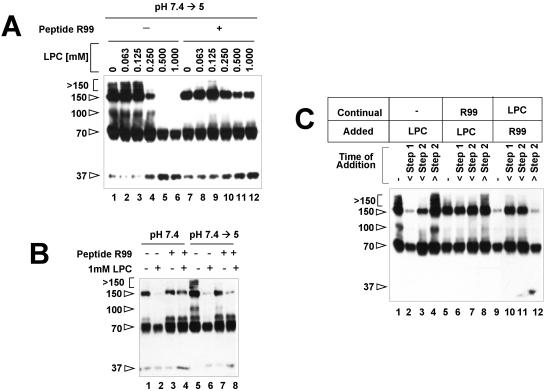
Agents that affect membrane curvature can inhibit or promote membrane fusion (9). We therefore tested the effects of a fusion-inhibiting and a fusion-promoting agent on the formation of the various EnvA TM conformational states. LPC is a lipid analogue that inhibits membrane merger in many systems (9, 10, 33). Virions were treated by the two-step method (Fig. 1A) in the presence of increasing concentrations of LPC. As shown in Fig. 4A, with concentrations of LPC of ≥0.25 mM, formation of the 150-, >150-, and 100-kDa bands was increasingly inhibited and the amount of the 37-kDa band increased. When the same experiment was performed in the presence of peptide R99 (lanes 7 to 12), the 150-kDa band, but not the >150- and 100-kDa bands, was protected (compare lanes 11 and 12 with lanes 5 and 6). Hence, LPC inhibits the formation of the 150-, >150-, and 100-kDa bands produced during the two-step protocol. Of these, the 150-kDa species is protected from LPC inhibition by peptide R99.
FIG. 4.
Effects of LPC on conformational states of EnvA TM. (A) Receptor-bound virions were mixed with liposomes, the indicated amount of LPC was added in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of peptide R99, and mixtures were then subjected to the two-step protocol. (B) Receptor-bound virions were subjected to step 1 only (lanes 1 to 4) or the two-step protocol (lanes 5 to 8); reaction mixtures contained no inhibitors (lanes 1 and 5), LPC (lanes 2, 4, 6, and 8), or peptide R99 (lanes 3, 4, 7, and 8). (C) LPC was added to the virion-receptor-liposome mixtures at the stages indicated in the absence (lanes 1 to 4) or continual presence of peptide R99 (lanes 5 to 8). Alternatively, R99 was added to the mixture at the indicated stage in the continual presence of LPC (lanes 9 to 12). Samples were then processed as described in the legend to Fig. 1. The values on the left are molecular sizes in kilodaltons.
We next tested the effects of LPC during step 1 of the two-step protocol (Fig. 1A). As shown above (for example, Fig. 1B), incubation of receptor-bound virions at neutral pH and 37°C leads to the formation of 70- and 150-kDa bands (Fig. 4B, lane 1). Formation of the 150-kDa band at neutral pH (i.e., during step 1) was inhibited by LPC (Fig. 4B, lane 2). Also, peptide R99 protected the 150-kDa band from inhibition by LPC during step 1 (Fig. 4B, lane 4 versus lane 2). Collectively, the results in Fig. 4A and B suggest that LPC has two effects in this system (see below and Discussion).
We next sought to determine when LPC had to be added to have its inhibitory effects. As shown in Fig. 4C, if LPC was added before step 1, the >150- and 100-kDa bands did not form and less of the 150-kDa band formed (Fig. 4C, compare lane 2 with lane 1). If LPC was added after step 1 but before step 2, more of the 150-kDa band formed but formation of the >150- and 100-kDa bands was still inhibited (Fig. 4C, lane 3). Adding LPC after step 2 had no effect on any of the EnvA TM bands (Fig. 4C, lane 4). If peptide R99 was present continually (lanes 5 to 8), LPC, added before step 1 or before step 2, inhibited the formation of the >150- and 100-kDa bands but did not inhibit the formation of the 150-kDa band (Fig. 4C, lanes 6 and 7). If peptide R99 was added either before step 1 or before step 2, in the continual presence of LPC (lanes 9 to 12), the 150-kDa band was protected (Fig. 4C, lanes 10 and 11). If added after step 2, peptide R99 was no longer able to protect the 150-kDa band (Fig. 4C, lane 12).
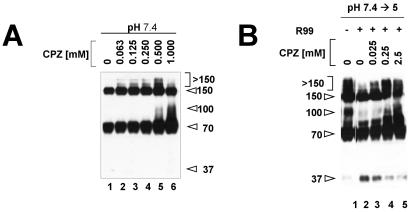
We next assessed the effects of CPZ, a reagent that promotes membrane fusion (38, 42), on the formation of the higher-molecular-weight forms of EnvA TM. In particular we asked if CPZ could promote the formation of the >150- and 100-kDa bands at neutral pH. Indeed, as shown in Fig. 5A, CPZ induced the formation of the >150- and 100-kDa bands at neutral pH. As with increasing temperature in lieu of a low pH at step 2 (Fig. 2), the >150-kDa bands appeared to form first (Fig. 5A, lanes 2 to 5) and then to chase into the 100-kDa band as the CPZ concentration was increased (Fig. 5A, lane 6 compared to lane 5). Interestingly, the presence of CPZ could overcome the inhibitory effect of peptide R99 on the formation of the >150- and 100-kDa bands (Fig. 5B, lanes 3 to 5; see Discussion). As expected, CPZ did not affect the formation of the 70- or 150-kDa bands at neutral pH (Fig. 5A), nor did it affect the formation of any of the higher-molecular-weight bands formed during the two-step protocol (not shown).
FIG. 5.
Effects of CPZ on conformational states of EnvA TM. (A) Receptor-bound virions were mixed with liposomes, the indicated amount of CPZ was added, and the mixtures were then incubated under step 1 conditions. (B) Samples were subjected to the two-step protocol in the presence (lanes 2 to 5) or absence (lane 1) of peptide R99 and in the presence (lanes 3 to 5) or absence (lanes 1 and 2) of the indicated concentration of CPZ. Samples were then processed as described in the legend to Fig. 1. Lanes 1 and 2 were not adjacent in the original gel. The values on the left or right are molecular sizes in kilodaltons.
Stability of EnvA TM conformational states.
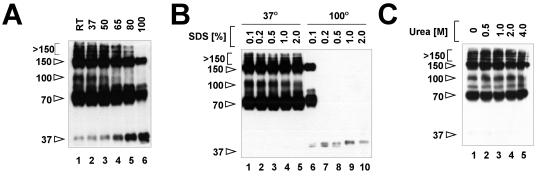
We tested the stability of the conformational states of EnvA TM when subjected to an elevated temperature, SDS, and urea. As shown in Fig. 6A, the 70- and 150-kDa bands were stable at 80°C in 0.1% SDS (lanes 1 to 5) and only modestly diminished after boiling (lane 6). The >150- and 100-kDa bands were less stable at an elevated temperature in 0.1% SDS, with loss beginning at 65°C. All of the higher-molecular-weight forms of EnvA TM were resistant to SDS at concentrations at least as high as 2% when samples were incubated at 37°C (Fig. 6B, lanes 1 to 5). When samples were boiled in 0.1% SDS, the >150- and 100-kDa bands were not observed (Fig. 6B, lane 6). The 70- and 150-kDa bands were observed in samples boiled in 0.1% SDS (Fig. 6B, lane 6) but not in samples boiled in higher concentrations of SDS (Fig. 6B, lanes 7 to 10). As with increasing temperature (Fig. 6A), loss of the higher-molecular-weight forms of EnvA TM was accompanied by appearance of the 37-kDa monomer band. All of the higher-molecular-weight forms of EnvA TM were stable in up to at least 4 M urea at 37°C (Fig. 6C).
FIG. 6.
Stability of EnvA TM oligomers. Virus samples treated to the two-step protocol were either warmed at the indicated temperature in sample buffer containing 0.1% SDS for 5 min (A), incubated in sample buffer containing the indicated amount of SDS at either 37°C (lanes 1 to 5) or 100°C (lanes 6 to 10) for 5 min (B), or incubated in the indicated amount of urea at 37°C for 5 min (C). Samples were then electrophoresed and subjected to Western blotting as described in Materials and Methods. The values on the left are molecular sizes in kilodaltons. RT, room temperature.
DISCUSSION
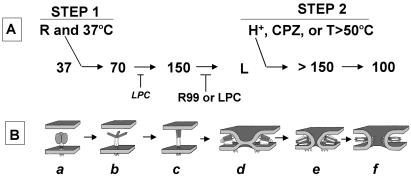
In this study we identified a series of conformational states of the EnvA TM (fusion) subunit that arise sequentially following interaction with receptor at neutral pH (step 1) and then exposure to a low pH (step 2). To do this we analyzed virus samples prepared for SDS gels with sample buffer containing 0.1% SDS and warmed to 37°C. In native virions the EnvA TM subunit appeared as a 37-kDa band. Following interaction with soluble receptor at neutral pH and 37°C, a 70-kDa band and then a 150-kDa form appeared (Fig. 1B). Following subsequent exposure to a pH of <6.0, a group of bands at >150 kDa and then a 100-kDa band formed (Fig. 1C). A C-helix peptide that inhibits virus-cell fusion, EnvA-mediated cell-cell fusion, and virus infection (18) inhibits the formation of the >150- and 100-kDa bands. The >150- and 100-kDa bands can form at neutral pH if, following receptor activation (at neutral pH), samples are treated at an elevated temperature (e.g., 50°C) or with the fusion-promoting agent CPZ. LPC, a fusion-inhibitory lipid analogue, inhibits the formation of the >150- and 100-kDa bands. The opposing effects of CPZ and LPC suggest that virus-liposome fusion occurs in this system. The promoting effect of CPZ and the inhibitory effects of LPC and peptide R99 also indicate that the >150- and 100-kDa bands are relevant to virus fusion. The conditions that generate the higher-molecular-weight forms of EnvA and their temporal relationships are depicted in Fig. 7A.
FIG. 7.
Working model of the roles of distinct conformational states of EnvA TM in distinct steps of fusion. (A) Conformational states (apparent molecular masses in kilodaltons) of Env A TM detected on mildly denaturing SDS gels and the conditions that promote or inhibit their formation. (B) Model of what the distinct conformations may look like and how they may mediate distinct stages of fusion. The TM subunit of starting virions migrates as a 37-kDa band (drawing a). (In drawing a, the SU subunits, which hold the TM subunit in its metastable state, are shown as gray ovals. For clarity, the SU subunits, which have presumably moved out of the way, are not shown in subsequent drawings.) Incubation of virions with soluble receptor at neutral pH and 37°C (step 1) results in the sequential formation of two new bands of 70 and 150 kDa (drawings b and c). We propose that the 150-kDa band represents the fully formed prehairpin conformation of EnvA TM. The 70-kDa band may be a prehairpin precursor. Upon exposure to a low pH (or an increased temperature or CPZ; step 2), a set of >150-kDa bands and then a 100-kDa band form (drawings e and f). Peptide R99 inhibits the formation of the >150- and 100-kDa bands. LPC inhibits the formation of the 150-, >150-, and 100-kDa bands. We propose that LPC inhibits the formation of the >150- and 100-kDa bands because it inhibits fusion. We propose that LPC inhibits the formation of the 150-kDa band owing to interaction with the exposed fusion peptide (see text). A distinct conformational state corresponding to the peptide R99-inhibitable lipid-mixing stage of fusion (drawing d) has not been detected in our gel system. Hence we designate it L for lipid mixing (see text). (The arrows are not meant to imply irreversibility.) R, receptor.
Model for steps of fusion mediated by distinct conformational states of EnvA TM.
We suggest that the 70- and 150-kDa bands that form following incubation with receptor at 37°C represent, respectively, intermediate and then fully formed EnvA TM prehairpins (drawings b and c in Fig. 7B). Several observations support this hypothesis. For one, the C-helix analogue peptide R99 does not inhibit the formation of either band. In fact, peptide R99 stabilizes the 150-kDa band in the presence of LPC (discussed below). These observations are consistent with our prior finding that peptide R99 does not inhibit the fusion peptide-mediated association of EnvA with target membranes (18). Further support that the 150-kDa band represents the target membrane binding prehairpin is that we have recently observed (Z. Chen and J. M. White, unpublished data) that the receptor concentration needed to form the 150-kDa band (in step 1) is the same as that needed to induce ASLV particles to bind to target membranes (18).
As stated above, LPC (added either before step 1 or before step 2) inhibits the formation of the >150- and 100-kDa bands, consistent with an inhibition of fusion. An initially surprising finding was that when LPC was added to virion-receptor-liposome mixtures before step 1, formation of the 150-kDa band was also inhibited (Fig. 4). Our hypothesis to explain this secondary effect of LPC is as follows. Evidence has been presented that LPC can bind to exposed fusion peptides and inhibit their association with target membranes (27, 58). If LPC binds to the exposed EnvA fusion peptide (14, 26, 28), it may sterically inhibit coiled-coil formation in the N-terminal end of the prehairpin (gray part of protein in drawing b of Fig. 7B). As noted above, peptide R99 can overcome the LPC block to forming the 150-kDa band (Fig. 4). This result suggests that the affinity of peptide R99 for the N-helix coiled coil is greater than the affinity of LPC for the fusion peptide. For these reasons, we propose that the C-helix analogue peptide R99 can not only bind to the N-terminal end of the fully formed prehairpin (drawing c in Fig. 7B) but also stabilize the prehairpin conformation.
When ASLV particles activated by receptor at neutral pH are subsequently exposed to a low pH, a temperature of >50°C, or CPZ, a 100-kDa band forms. We propose that the 100-kDa band represents the final fully formed EnvA TM hairpin, its six-helix bundle (Fig. 7B, drawing f). This proposal is supported by the observations that the 100-kDa band is the last species formed under any of the post receptor activation conditions that we have explored (a low pH, an elevated temperature, or treatment with CPZ) and that formation of the 100-kDa band is inhibited by the C-helix analogue peptide R99. Interestingly, CPZ is able to drive the formation of the 100-kDa band in the presence of R99 (at the concentration of R99 tested; Fig. 5B). Importantly, peptide R99 does not inhibit the formation of the 100-kDa band if receptor-bound virions (produced and maintained at 4°C) are directly warmed to 37°C at pH 5 or if virions are subjected to the two-step protocol in the absence of receptor (Fig. 3A). These findings, coupled with the need for sequential activation by receptor at neutral pH, followed by a low-pH shock to overcome the block to reverse transcript production imposed by bafilomycin (45) (Matsuyama and White, unpublished), suggest that receptor activation at neutral pH (and T ≥ RT) is needed to keep EnvA on a productive fusion pathway. We further propose that conversion to the 100-kDa form mediates the final stage of fusion, fusion pore enlargement (drawing f in Fig. 7B; see below).
A set of bands at >150 kDa appear to be intermediates on the pathway between the 150-kDa band (proposed prehairpin) and the 100-kDa band (proposed final six-helix bundle). This is evident when either CPZ (Fig. 5A) or an elevated temperature (Fig. 2) is used to drive conformational changes that occur post receptor activation. We suggest that the >150-kDa bands mediate the opening of small fusion pores (Fig. 7B, drawing e). The >150-kDa species may be analogous to higher-order oligomers that form transiently during baculovirus gp64-mediated fusion (40). Formation of higher-order oligomers has been proposed for many viral fusion systems (4, 13, 16, 39, 40). Upon fusion pore expansion, the higher-order transient oligomers may disperse and transform into unitary six-helix bundles. The transition of the >150-kDa bands to the 100-kDa band may drive conversion from small, labile fusion pores to large, robust fusion pores, as has recently been described for HIV Env (35, 37).
All of the higher-molecular-weight forms of EnvA TM are stable in at least 2% SDS at 37°C (Fig. 6B, lanes 1 to 5) and in at least 4 M urea at 37°C (Fig. 6C). We were surprised, however, to find that the 70- and 150-kDa bands appear to be more stable than the >150- and 100-kDa bands if gel samples are prepared at ≥80°C in 0.1% SDS. Six-helix bundle cores (lacking transmembrane domains and fusion peptides) are highly stable at elevated temperatures in aqueous solutions. However, to our knowledge, there have not been studies comparing the stability of prehairpin and hairpin conformations of intact class I viral fusion proteins in SDS. The apparent persistence of the prehairpin forms of EnvA TM (70- and 150-kDa bands) on SDS gels of low-pH-treated samples (which may be exaggerated by the apparent hyperreactivity of the 70- and 150-kDa bands on our immunoblots; see Results) may indicate that not all EnvA TM prehairpins convert to hairpin conformations. Remaining prehairpins may help buttress fusion pores, as has recently been modeled for HIV Env (37).
We have observed that ASLV particles can reach the lipid-mixing stage of fusion with target cells at neutral pH (drawing d in Fig. 7B) and that lipid mixing, but not target bilayer association, can be inhibited by peptide R99 (18). However, we have not visualized an EnvA TM species (on an SDS gel) that is inhibited by peptide R99 and that forms following interaction with receptor at neutral pH and T ≥ RT. We therefore propose that there is a species, designated L in Fig. 7A, that mediates the lipid-mixing stage of virus-cell fusion. Species L (for lipid mixing) could be one or more of the >150-kDa species that are unstable in 0.1% SDS without prior treatment with a low pH, CPZ, or an elevated temperature. Alternatively, lipid-mixing species L could be a distinct conformation of EnvA TM, such as that depicted in drawing d of Fig. 7B (which is not stable in 0.1% SDS). As drawn, the C helices in species L make trimeric contacts with each other (perhaps as in the metastable trimer). Such trimeric contacts would have to be broken so that the three C helices could pack individually into the grooves of the N-terminal end of the coiled coil (as in drawings e and f). Breaking of such trimeric contacts, and transiting to a symmetric conformation such as that shown in drawing e, could be an energy-requiring step that may be facilitated by a low pH, an elevated temperature, or CPZ. Multiple sequences within the C helix (23, 41), within the disulfide-bonded chain reversal region (36, 54), within the transmembrane (35, 59) and juxtamembrane (53) regions, and within the cytoplasmic tail (see below), could influence the d-to-e transition.
Physiological relevance of the two-step fusion process.
We have previously shown that receptor activation can cause ASLV to reach the lipid-mixing stage of virus-cell fusion (drawing d in Fig. 7B) at neutral pH (18). The data presented here are consistent with a need for subsequent exposure to a low pH, presumably after endocytosis, to complete the fusion process (15, 45, 46). Several important questions remain regarding this novel two-step fusion process. First, although it is likely, it is still unclear if a low pH is the true cellular trigger for the second step of fusion. The strongest evidence of a need for a low pH is that the continued presence of 10 nM bafilomycin, an inhibitor of the endosomal H+-ATPase, blocks the production of reverse transcripts from incoming ASLV-A particles (Matsuyama, Earp, and White, unpublished). However, in addition to its effect on endosomal acidification, bafilomycin can interfere with membrane trafficking events, such as movement from early to late endosomes (3, 11).
A second important question is why a virus would have evolved to interrupt its fusion process midstream (i.e., at the lipid-mixing stage). Perhaps ASLV evolved the two-step mechanism to deliver viral cores to a specific location in the target cell, for example, below the cortical cytoskeleton and close to the nucleus (32), as a means to increase the efficiency of reverse transcription or subsequent steps of replication. Other receptor-activated viruses (e.g., HIV) appear to have evolved specific proteins (e.g., Nef) to accomplish this topological task (8, 61). A need for a low pH for virus-cell fusion may also have evolved as a mechanism to prevent extensive cell-cell fusion of infected EnvA expressing cells at neutral pH. In the case of murine retroviruses, syncytium formation is inhibited by the presence of an extended cytoplasmic tail sequence that is proteolytically processed by the viral protease, during viral budding, to activate virus-cell fusion activity (1, 5, 34, 43, 49, 60). Interestingly, the fusion activity of a paramyxovirus F protein that has a long cytoplasmic tail was recently shown to be activated either by experimental truncation of the cytoplasmic tail or by exposure to a low pH (55). In both cases, specific sequence motifs within the cytoplasmic tail appear to influence the fusion phenotype (34, 56, 60). The cytoplasmic tails of other class I viral fusion proteins also seem to modulate late stages of fusion (17, 19, 21, 30, 31).
A third important question is whether this two-step fusion process is unique to avian retroviruses or whether it is used by other viruses. It has recently been shown that two other retroviruses, foamy virus (48) and mouse mammary tumor virus (MMTV) (50), show reduced infectivity in the presence of lysosomotropic agents. The receptor for foamy virus is not known, but the receptor for MMTV is the transferrin receptor (50), whose trafficking pathway through low-pH endosomes has been extensively characterized. It will be interesting to determine if interaction between transferrin receptor and MMTV Env, at the cell surface at neutral pH, induces conformational changes in MMTV Env that are prerequisites for subsequent low-pH-induced conformational changes that mediate later stages of fusion.
In conclusion, the results of our study have led to a model (Fig. 7) in which receptor and a low pH act sequentially to form distinct conformations of EnvA that mediate specific stages of fusion. To our knowledge, this represents the first biochemical identification of a series of distinct prehairpin and hairpin conformations for a class I viral fusion protein. The major significances of this study are (i) that it lends support to the notion that avian retroviruses use a novel two-step fusion process (45) and (ii) that it provides a framework for future investigations. With this framework in hand, one can evaluate the precise mechanistic role of a low pH in ASLV-A fusion and entry. Furthermore, our studies suggest that ASLV-A will be an ideal model virus with which to independently study the molecular mechanisms of early and late stages of fusion. Lastly, one should be able to use similar biochemical approaches to test whether other viruses use this novel two-step fusion process, i.e., receptor activation at neutral pH followed by a low-pH completion step.
Acknowledgments
This work was funded by an NIH grant (AI22470 to J.M.W.).
We thank J. Gruenke, L. Earp, and especially H. Park for help with Fig. 7B and H. Park for many helpful comments on the manuscript.
REFERENCES
- 1.Aguilar, H. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J. Virol. 77:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, P., J. A. T. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, N., D. Schober, E. Prchla, R. F. Murphy, D. Blaas, and R. Fuchs. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645-9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobkova, M., J. Stitz, M. Engelstadter, K. Cichutek, and C. J. Buchholz. 2002. Identification of R-peptides in envelope proteins of C-type retroviruses. J. Gen. Virol. 83:2241-2246. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chazal, N., G. Singer, C. Aiken, M.-L. Hammarskjold, and D. Rekosh. 2001. Human immunodeficiency virus type 1 particles pseudotyped with envelope proteins that fuse at low pH no longer require Nef for optimal infectivity. J. Virol. 75:4014-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernomordik, L. V., and M. M. Kozlov. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175-207. [DOI] [PubMed] [Google Scholar]
- 10.Chernomordik, L. V., S. S. Vogel, A. Sokoloff, H. O. Onaran, E. A. Leikina, and J. Zimmerberg. 1993. Lysolipids reversibly inhibit Ca2+-, GTP-, and pH-dependent fusion of biological membranes. FEBS Lett. 318:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Clague, M. J., S. Urbe, F. Aniento, and J. Gruenberg. 1994. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J. Biol. Chem. 269:21-24. [PubMed] [Google Scholar]
- 12.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell. Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 13.Danieli, T., S. L. Pelletier, Y. I. Henis, and J. M. White. 1996. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 133:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delos, S. E., and J. M. White. 2000. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J. Virol. 74:9738-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Griffero, F., S. A. Hoschander, and J. Brojatsch. 2002. Endocytosis is a critical step in entry of subgroup B avian leukosis viruses. J. Virol. 76:12866-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutch, R. E., and R. A. Lamb. 2001. Deletion of the cytoplasmic tail of the fusion protein of the paramyxovirus simian virus 5 affects fusion pore enlargement. J. Virol. 75:5363-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earp, L. E., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. The many mechanisms of viral membrane fusion proteins. In M. Marsh (ed.), Current topics in microbiology and immunology. Membrane trafficking in viral replication, in press. Springer Verlag, New York, N.Y. [DOI] [PMC free article] [PubMed]
- 20.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 21.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 23.Gallagher, T. M., C. Escarmis, and M. J. Buchmeier. 1991. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J. Virol. 65:1916-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallaher, W. R. 1996. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 85:477-478. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert, J., D. Mason, and J. White. 1990. Fusion of Rous sarcoma virus with host cells does not require low pH. J. Virol. 64:5106-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert, J. M., L. D. Hernandez, J. W. Balliet, P. Bates, and J. M. White. 1995. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J. Virol. 69:7410-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunther-Ausborn, S., A. Praetor, and T. Stegmann. 1995. Inhibition of influenza-induced membrane fusion by lysophosphatidylcholine. J. Biol. Chem. 270:29279-29285. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez, L. D., R. R. Peters, S. E. Delos, J. A. T. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez, L. D., and J. M. White. 1998. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J. Virol. 72:3259-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, F. J., N. Manel, Y. Boublik, J. L. Battini, and M. Sitbon. 2003. Human T-cell leukemia virus type 1 envelope-mediated syncytium formation can be activated in resistant mammalian cell lines by a carboxy-terminal truncation of the envelope cytoplasmic domain. J. Virol. 77:963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozerski, C., E. Ponimaskin, B. Schroth-Diez, M. F. Schmidt, and A. Herrmann. 2000. Modification of the cytoplasmic domain of influenza virus hemagglutinin affects enlargement of the fusion pore. J. Virol. 74:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100:9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leikina, E., and L. V. Chernomordik. 2000. Reversible merger of membranes at the early stage of influenza hemagglutinin-mediated fusion. Mol. Biol. Cell 11:2359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, M., C. Yang, and R. W. Compans. 2001. Mutations in the cytoplasmic tail of murine leukemia virus envelope protein suppress fusion inhibition by R peptide. J. Virol. 75:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, X., C. A. Derdeyn, R. Blumenthal, J. West, and E. Hunter. 2003. Progressive truncations C terminal to the membrane-spanning domain of simian immunodeficiency virus Env reduce fusogenicity and increase concentration dependence of Env for fusion. J. Virol. 77:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maerz, A. L., R. J. Center, B. E. Kemp, B. Kobe, and P. Poumbourios. 2000. Functional implications of the human T-lymphotropic virus type 1 transmembrane glycoprotein helical hairpin structure. J. Virol. 74:6614-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2003. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell 14:926-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2000. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell 11:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markovic, I., E. Leikina, M. Zhukovsky, J. Zimmerberg, and L. V. Chernomordik. 2001. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J. Cell Biol. 155:833-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markovic, I., H. Pulyaeva, A. Sokoloff, and L. V. Chernomordik. 1998. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J. Cell Biol. 143:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGinnes, L. W., T. Sergel, H. Chen, L. Hamo, S. Schwertz, D. Li, and T. G. Morrison. 2001. Mutational analysis of the membrane proximal heptad repeat of the Newcastle disease virus fusion protein. Virology 289:343-352. [DOI] [PubMed] [Google Scholar]
- 42.Melikyan, G. B., S. Lin, M. G. Roth, and F. S. Cohen. 1999. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 10:1821-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and F. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 46.Narayan, S., R. J. Barnard, and J. A. Young. 2003. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J. Virol. 77:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 48.Picard-Maureau, M., G. Jarmy, A. Berg, A. Rethwilm, and D. Lindemann. 2003. Foamy virus envelope glycoprotein-mediated entry involves a pH-dependent fusion process. J. Virol. 77:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross, S. R., J. J. Schofield, C. J. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 99:12386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruigrok, R. W., S. R. Martin, S. A. Wharton, J. J. Skehel, P. M. Bayley, and D. C. Wiley. 1986. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology 155:484-497. [DOI] [PubMed] [Google Scholar]
- 52.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salzwedel, K., J. T. West, and H. E. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders, D. A. 2003. Ancient viruses in the fight against HIV. Drug Discov. Today 8:287-291. [DOI] [PubMed] [Google Scholar]
- 55.Seth, S., A. Vincent, and R. W. Compans. 2003. Activation of fusion by the SER virus F protein: a low-pH-dependent paramyxovirus entry process. J. Virol. 77:6520-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seth, S., A. Vincent, and R. W. Compans. 2003. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J. Virol. 77:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 58.Stegmann, T. 1993. Influenza hemagglutinin-mediated membrane fusion does not involve inverted phase lipid intermediates. J. Biol. Chem. 268:1716-1722. [PubMed] [Google Scholar]
- 59.Taylor, G. M., and D. A. Sanders. 1999. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol. Biol. Cell 10:2803-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor, G. M., and D. A. Sanders. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312:295-305. [DOI] [PubMed] [Google Scholar]
- 61.Tobiume, M., J. E. Lineberger, C. A. Lundquist, M. D. Miller, and C. Aiken. 2003. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J. Virol. 77:10645-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tonelli, M., R. J. Peters, T. L. James, and D. A. Agard. 2001. The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus. FEBS Lett. 509:161-168. [DOI] [PubMed] [Google Scholar]
- 63.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-372. [DOI] [PubMed] [Google Scholar]