Abstract
Parapoxviruses can be morphologically distinguished from other poxviruses in conventional negative staining electron microscopy (EM) by their ovoid appearance and the spiral tubule surrounding the virion's surface. However, this technique may introduce artifacts. We have examined Orf virus (ORFV; the prototype species of the Parapoxvirus genus) by cryoelectron microscopy (cryo-EM) and cryo-negative staining EM. From these studies we suggest that the shape and unique spiral tubule are authentic features of the parapoxviruses. We also constructed an ORFV mutant deleted of a gene encoding a 10-kDa protein, which is an orthologue of the vaccinia virus (VACV) 14-kDa fusion protein, and investigated its ultrastructure. This mutant virus multiplied slowly in permissive cells and produced infectious but morphologically aberrant particles. Mutant virions lacked the spiral tubule but displayed short disorganized tubules similar to those observed on the surface of VACV. In addition, thin extensions or loop-like structures were appended to the ORFV mutant particles. We suggest that these appended structures arise from a failure of the mutant virus particles to properly seal and that the sealing activity is dependent on the 10-kDa protein.
Members of the Parapoxvirus genus within the poxvirus family can infect a variety of wildlife and domestic mammals. These viruses are best known for their ability to infect ruminants, cattle, and occasionally humans, usually leading to localized lesions of the skin and of the mucosal membranes (4). Orf virus (ORFV), a virus of sheep and goats, also known as contagious pustular dermatitis virus or contagious ecthyma virus, is the prototype member of the Parapoxvirus genus. The ORFV genome displays a high G+C content and a relatively small size for poxviruses (140 to 160 kbp). Its basic organization is very similar to that of other poxviruses (19), but it encodes a unique set of immunomodulatory functions (13). ORFV morphogenesis appears to resemble that of other poxviruses; however, the mature viral particles are clearly distinguishable. Conventional electron microscopy (EM) displays ORFV virions with an ovoid shape (∼260 nm long by 160 nm wide) and a slightly smaller size than the brick-shaped orthopoxviruses (∼350 nm long by 270 nm wide) (1, 21). They are often surrounded by a tightly apposed envelope, presumably derived from the trans-Golgi network (TGN) as described in detail for vaccinia virus (VACV), the prototype for orthopoxviruses (29). The most striking feature, which readily enables identification of ORFV as well as other parapoxviruses, is a tubule-like structure that surrounds the particle in a spiral fashion (21). This tubule appears uninterrupted on parapoxvirus particles and is thicker than the short tubules noticed on the surface of orthopoxviruses by conventional EM (22). Whether the tubule-like structures visualized on the surface of parapoxviruses or orthopoxviruses are actually tubules or simply ridges in the virus membrane has not yet been determined. In fact, when VACV was embedded in vitreous water and observed by EM under low-dose conditions, without prior negative staining, no tubule structures were observed on its surface, suggesting that they may be artifacts due to the desiccation of the samples (9). Keeping that in mind, we have reexamined the surface structure of ORFV strain D1701-V particularly to determine whether the characteristic spiral tubule is genuine or not. For this study we have employed an ORFV mutant (D1701-Vr10) deleted of a viral gene encoding a 10-kDa protein predicted to be important for the surface structure of ORFV. Prior sequence analysis of the ORFV genome identified this gene as a homologue of the VACV A27L gene encoding the 14-kDa protein (20). The 14-kDa VACV protein is expressed on the surface of the intracellular mature virus (IMV) particle (7) and is a target of virus-neutralizing antibodies (5, 24). It has also been shown to mediate virus-cell fusion (25), to act as a ligand for the binding of the IMV particle to the cell receptor heparin sulfate (15), and is essential for transport of IMV particles on microtubules (28) and their wrapping by TGN membranes (26).
Using cryoelectron microscopy (cryo-EM) methods, we demonstrate that the ORFV spiral tubule is a genuine structure whose integrity depends on the presence of the gene encoding the 10-kDa protein. We also show that the 10-kDa protein, unlike its VACV counterpart, is not required for wrapping by TGN-derived virus envelopes but is essential for the formation of spiral tubules, correct virus assembly, and release of infectious virus. These studies have led us to postulate a role for the 10-kDa protein in ORFV assembly.
MATERIALS AND METHODS
Cells and virus.
The attenuated ORFV strain D1701-V and the mutant virus D1701-Vr10 were propagated and titrated in Vero cells as described recently (11) or in fetal bovine esophagus cells (KOP) kindly provided by R. Riebe (Federal Research Center for Virus Diseases of Animals, Insel Riems, Germany). The use of KOP cells, which were maintained in Dulbecco's modified Eagle medium supplemented with 10% pestivirus-free fetal bovine serum (PAA, Pasching, Austria) allowed longer cultivation times than Vero cells, and, therefore, a more accurate discrimination between the cytopathic effect of the wild-type (WT) D1701-V and D1701-Vr10 strains was possible. Virus was purified from the combined supernatants and cell lysates by sucrose gradient centrifugation (17).
Generation of ORFV mutant D1701-Vr10.
The EcoRI fragment E of D1701-V containing the ORFV of the gene encoding the 10-kDa protein (referred to here as the 10-kDa gene) was cloned into plasmid pSPT-18dH, which was derived from plasmid pSPT-18 (Roche Biochemicals, Mannheim, Germany) after HindIII cleavage, filling in the generated cohesive ends, and religation. The resulting plasmid, pRZ-E1, contains a single HindIII site approximately in the middle of the 10-kDa gene, which could be used to open it and subsequently remove portions of the 10-kDa gene by bidirectional Bal31 digestion. Blunt-end ligation of the EcoRV linker introduced a new, unique EcoRV restriction site to generate pd10K-1. A lacZ-Xgpt gene cassette driven by the synthetic early-late VACV promoter elP4 was inserted into the EcoRV site of plasmid pd10K-1 to generate p10dLX-4 (supplemental data for the entire cloning strategy is available at http://www.efs-alsace.fr/pub/orfplasmid.tif). DNA sequencing of plasmid p10dLX-4 confirmed the deletion of amino acids 10 to 68 of the 10-kDa gene and insertion of the marker gene cassette in the 10-kDa gene orientation (data not shown).
Transfection of D1701-V-infected Vero cells with p10dLX-4 and subsequent screening for lacZ-positive virus-infected cells were performed as described recently (11). Briefly, infected Vero cells were incubated in selective modified Eagle medium (10) containing 15 μg of hypoxanthine per ml, 0.2 μg of aminopterine per ml, and 4 μg of thymidine per ml, supplemented with 250 μg of xanthine per ml and 40 μg of mycophenolic acid per ml. The use of selective medium resulted in approximately a 4 log titer reduction of D1701-V (data not shown). Thereafter, the blue foci of D1701-Vrd10-infected KOP cells were picked, and after five rounds of isolating blue cell foci, homogeneous D1701-Vr10 virus was obtained as shown by Southern blot hybridization and 10-kDa gene-specific PCR (data not shown). Further propagation of D1701-Vr10 was then performed in either KOP or Vero cells under normal growth conditions.
Growth curve and mixed infection experiments.
Since infection with D1701-Vr10 did not lead to the typical plaque formation observed with the parental virus, infected cell foci of the mutant deleted of the 10-kDa gene were visualized by Bluo-Gal staining (300 μg per ml; Invitrogen Life Technologies). Virus growth kinetics were assayed with separated cell lysates and cell-free supernatants from infected cells (11). For direct comparison of the growth characteristics of D1701-Vr10 and D1701-V, both viruses were mixed in ratios of 1,000:1, 100:1, 10:1, and 1:1, respectively, and used for the infection of KOP cells. At different times postinfection (p.i.), cell lysates were harvested and titrated on KOP cells. After 4 to 5 days, titration was stopped by fixation with 0.25% (vol/vol) glutaraldehyde in phosphate-buffered saline containing 0.02% (vol/vol) Nonidet-P40 and 0.01% sodium-deoxycholate. After Bluo-Gal staining and washing with phosphate-buffered saline containing 0.1% (vol/vol) Triton X-100, the cells were immunostained with monoclonal antibody 4D9 (diluted 1:200) directed against the 37-kDa major envelope protein of ORFV (6). This double staining allowed a clear distinction between D1701-V plaques and the blue cell foci of D1701-Vr10 (data not shown).
Conventional EM.
Vero cells were infected with 1 to 5 PFU for 24 to 72 h and fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2). The samples were then post-fixed in 1% osmium tetroxide, dehydrated in acetone, and embedded in Araldite resin. Ultrathin sections were stained with 0.5% uranyl acetate and lead citrate and observed with a LEO EM 910 electron microscope operated at 80 kV. Negative staining of purified virions was performed with 1% aqueous uranyl acetate.
Cryo-EM.
Samples were prepared for cryo-EM as previously described (3). Homemade holey carbon films were used. These films were first mounted on 200-mesh copper grids, and a thin layer of Au-Pd was sputtered on one side of the grid. After sputtering, the plastic holey film was dissolved with ethyl acetate. For vitrification of the native sample, a drop of solution was applied to the metal-coated face of the grid without previous glow discharge. The grid was mounted on a plunger, blotted, and vitrified in liquid ethane. For cryo-negative staining EM, the procedure was the same, except for the fact that the grid with a drop of the sample was put on the staining solution for 30 seconds before being mounted on the plunger and then immediately blotted and vitrified (2). The stain was prepared by using a saturated ammonium molybdate solution (0.9 ml), with the addition of 0.1 ml NaOH 10 M to obtain a pH of 7.2. The samples were then observed in an FEI CM100 electron microscope equipped with a Gatan cold stage and observed at liquid nitrogen temperature. Pictures were taken under low-dose conditions after the detection of interesting areas by using the low-dose mode.
RESULTS
Isolation and characterization of an ORFV mutant deleted of the 10-kDa gene.
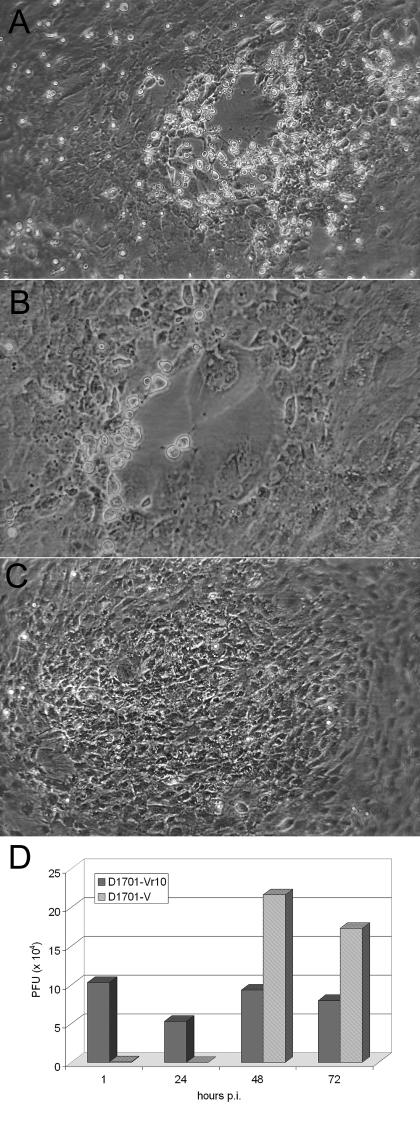
To examine whether the ORFV 10-kDa protein plays a role in morphogenesis similar to its VACV counterpart, we generated a virus mutant by replacing amino acids 10 to 68 of the 10-kDa open reading frame by a lacZ/gpt cassette. The resulting virus mutant D1701-Vr10 clearly exhibited different growth characteristics compared to the parental strain D1701-V. The typical ORFV plaque observed 2 and 4 days p.i. (Fig. 1A and B) was not visible in the case of the mutant, but instead the cytopathic effect became visible at 2 days p.i. as a focal rounding of cells, increasingly affecting nearly the whole cell monolayer by 6 to 7 days p.i. (Fig. 1C). Growth curve experiments conducted with a multiplicity of infection of 1 or 10 demonstrated at least a 100-fold reduced production of intracellular infectious D1701-Vr10 virus compared to D1701-V over a 3-day period although the mutant virus yield nearly reached the level obtained with the parental strain 6 days p.i. (data not shown). Titration of cell-free supernatants taken from infected cells 6 days p.i. revealed virtually no release of infectious D1701-Vr10, whereas WT virus-infected cells, which displayed 100% cytopathic effect, produced approximately 107 PFU per ml of extracellular virus (not shown). The growth disadvantage of the ORFV mutant deleted of the 10-kDa gene was further proven by mixed infection experiments as described in Material and Methods. Even after the infection of cells with a mixture representing a ratio of 1,000:1 of D1701-Vr10 and D1701-V, at 48 h p.i. (hpi) the production of WT virus clearly exceeded that of mutant virus. As shown in Fig. 1D, the titration of lysates derived from the mixed-infected cells revealed at least a 24-h eclipse phase for the WT virus, followed by a rapid rise in the infection titer at 48 hpi and no increase thereafter. The titer of the mutant virus decreased slightly at 24 hpi and then reached the level of the input titer after 48 h. Thus, despite the 1,000-fold higher input dose of D1701-Vr10, no significant increase in the titer of the mutant was detected over this time period. The mixed infection experiment could not be carried out for more than 3 days because the cells were completely destroyed by WT virus after this period of time. Nevertheless, it should be recalled that infection with the mutant alone for a longer period of time (6 days) led to a yield nearly equal to that of the WT virus in cell lysates. Taken together, these results indicate a delayed production of intracellular infectious progeny and a defect in the release and spread of infectious virus due to the deletion of the ORFV 10-kDa gene.
FIG. 1.
Multiplication of the ORFV D1701-V and D1701-Vr10 strains in KOP cells. Infection of KOP cells (multiplicity of infection, 5) with WT D1701-V resulted in increasing plaque formation from 2 (A) to 4 days p.i. (B), whereas 6 days p.i., D1701-Vr10 led only to foci of infected, β-galactosidase-positive cells (C). Cell morphology was recorded under phase-contrast microscopy. The result of mixed infection experiments is shown in panel D. After simultaneous infection of cells with strains D1701-Vr10 and D1701-V in a ratio of 1,000:1, cell lysates were harvested at the indicated times p.i. and titrated. The number of PFU of D1701-Vr10 virus was determined by counting β-galactosidase-positive foci and the number of WT (D1701-V) PFU was directly visualized by counting WT virus plaques.
Examination of infected cells after embedding and conventional EM.
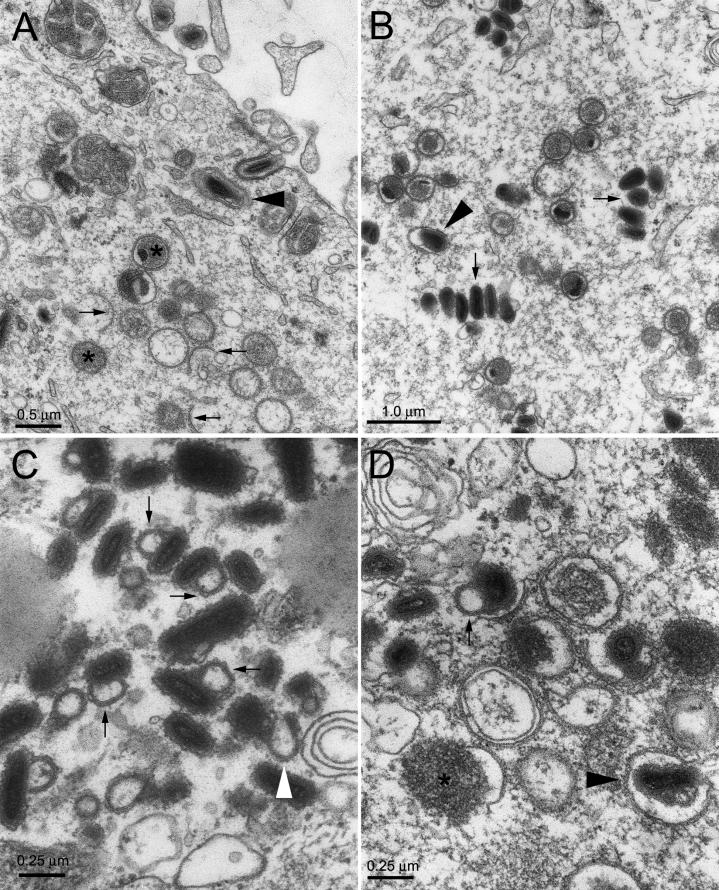
EM analysis of Vero cells infected with D1701-V demonstrated the entire spectrum of ORFV assembly intermediates, namely, membrane crescents surrounding an electron-dense viroplasm, spherical immature virus (IV) enclosing light viroplasm, or dense nucleoprotein surrounded by lipid bilayers, as well as numerous mature particles viewed under their long or short axis and sometimes surrounded by additional envelopes presumably derived from the TGN (Fig. 2A and B). We could also visualize double-membrane crescents or double-membrane IV, consistent with the view that the IMV particles are surrounded by two apposed membranes (23, 30) rather than a single membrane (14). The same spectrum of virus intermediate and mature particles was observed in cells infected with the mutant virus D1701-Vr10 (data not shown). However, most if not all of the mature D1701-Vr10 particles had an appended loop-like structure enclosing an area of light electron density (Fig. 2C and D). In some instances, the particles displayed a thin extension which did not enclose a light area (Fig. 2C, white arrow), suggesting that the loop-like structures could be derived from such extensions that had folded back onto another end of the virus particles. Since the tips of such extensions were occasionally at a distance nearly half the particle's diameter, they could not correspond to incompletely viewed loop-like structures. However, it was usually not possible to determine if the loop-like structures had originated at one or the other end of the virus particle or if they were in continuity with the virus particle at both ends. In those instances where membranes were visualized around the loop-like structures or extensions, we could distinguish a series of overlapping membranes (Fig. 2C, arrows).
FIG. 2.
EM of Vero cells infected with D1701-V and D1701-Vr10. Vero cells were infected with strain D1701-V or D1701-Vr10 and thin sections were examined by electron microscopy. Panels A and B illustrate two different regions of cells infected with D1701-V. In panel A, arrows indicate membrane crescents, stars are positioned over IV particles, and an arrowhead indicates enveloped virus within the cell. In panel B, arrows indicate groups of IMVs, and the arrowhead points to an enveloped mature virus. Panels C and D illustrate cell regions infected with D1701-Vr10. Thin arrows point to particles with appended loop-like structures, and the white arrow points to a particle displaying an extension with a visible end. In panel D a star is positioned over a viroplasm partially surrounded by a viral crescent membrane.
EM of purified virus.
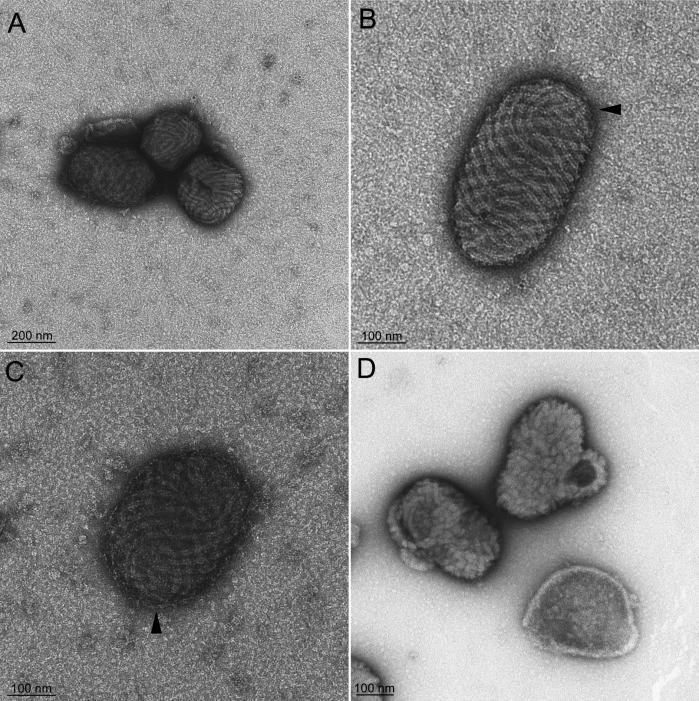
Purified ORFV (D1701-V) observed by EM after staining with uranyl acetate displayed the classical “ball-of-wool” structure with its regularly spaced spiral tubule (Fig. 3A to C). According to analytical geometry, a vector field covering a spherical or elliptical body presents at least two discontinuities. In fact, we observed regions of disruption of the regular spiral geometry (Fig. 3B and C, arrowheads). ORFV particles were surrounded by a tightly apposed envelope characteristic of extracellular enveloped viruses. Mutant particles observed by the same EM method lacked a spiral tubule but instead displayed short disorganized tubules reminiscent of the surface structure of VACV when observed by conventional EM (Fig. 3D). Furthermore, the loop-like structure visible in thin sections was appended to most of the virions, but no extensions with visible tips were noticed. The envelope of the mutant virions was more difficult to distinguish than that of the parental virus but could be clearly visualized by cryo-EM (see below).
FIG. 3.
EM of negatively stained purified ORFV. Panels A to C display ORFV. An envelope surrounds some of the particles and is distinctly visible in panels B and C. Areas of apparent discontinuity in the ball-of-wool structure are indicated (arrowheads). Panel D shows the mutant deleted of the 10-kDa gene with the loop-like structure viewed at different spatial positions; short disorganized tubules are visualized.
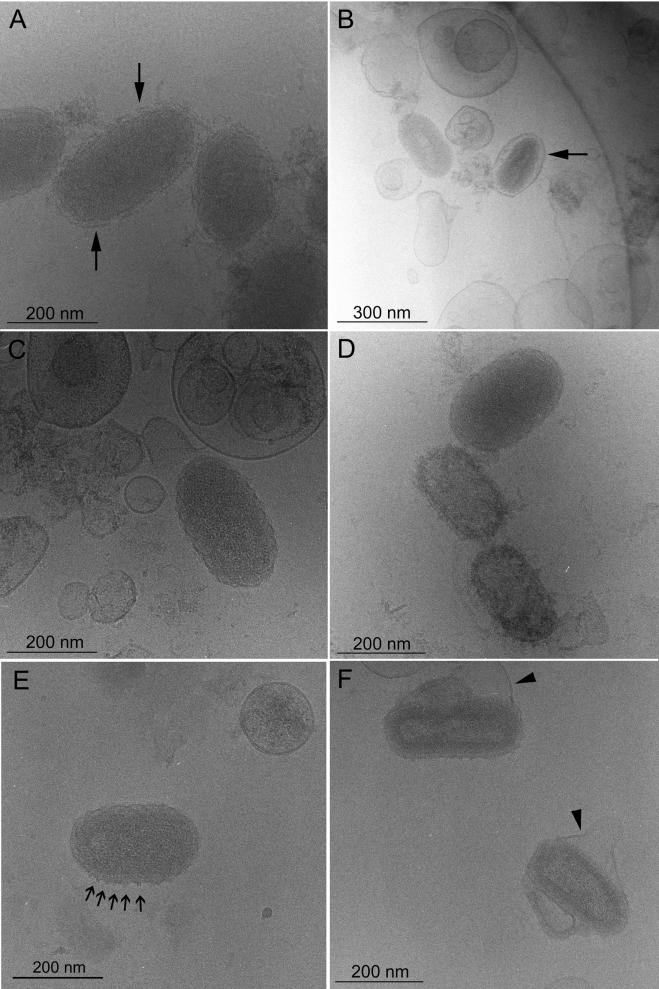
To assess the structure of ORFV in a more native state, unstained samples were embedded in a thin layer of vitreous water and observed by cryo-EM (3). The virus suspensions (Fig. 4A to E) contained liposome-like particles which may correspond to envelopes that were damaged prior to embedding. Observation of such structures confirmed the excellent preservation of the material by the cryo-EM method since the double-leaflet membranes of the liposomes could be readily visualized. As expected, unstained virus particles appeared more poorly contrasted under cryo-EM conditions than after conventional negative staining EM. The majority of the virions were surrounded by an envelope. The most striking features compared to previously published views of VACV examined by cryo-EM (9) were the crenellated edges and a faintly visible spiral tubule over the entire surface of the particles (Fig. 4A). However, the surface domain, palisade layer, and core domain described for VACV were not observed. When the virus was submitted to mild sonication, envelopes were removed, and the crenellated edges as well as the continuity of these edges with an overlaying tubule were more clearly visible (Fig. 4C to E). Occasionally, less-dense virus particles were noticed (Fig. 4D), and they may represent empty particles. Double filaments (18 ± 2 nm [mean ± standard deviation]) were visualized on the edges of a few particles (Fig. 4E) and appeared to be repeated at distances similar to the spacing between the surface crenellations, indicating that they correspond to the spiral tubule viewed along the side.
FIG. 4.
Cryo-EM of ORFV. Panels A and B show views of untreated WT ORFV. Most particles are enveloped by TGN-derived membranes (arrows). Panels C and E show views of ORFV after prior sonication. The virus particles display a crenellated surface, a faint spiral tubule, and in panel E a few short double filaments on their periphery (arrows). Due to sonication, the envelopes have been removed and most likely recircularized into liposome-like particles. Panel D shows less-dense particles which correspond to empty viral particles. Panel F shows two particles of the virus mutant with their loop-like structures, loosely surrounded by TGN-derived envelopes (arrowheads).
In contrast to the parental WT virus, the mutant D1701-Vr10 virus displayed internal structures resembling a surface domain, palisade domain, and amorphous core domain (Fig. 4F) as previously described for VACV. Moreover, crenellations were seen on the edges of the D1701-Vr10 virus, and a few double filament-like structures were observed which could correspond to the disorganized tubules seen by conventional negative staining. Importantly, the mutant particles exhibited the characteristic loop-like structures and were frequently enveloped (Fig. 4F, arrowheads).
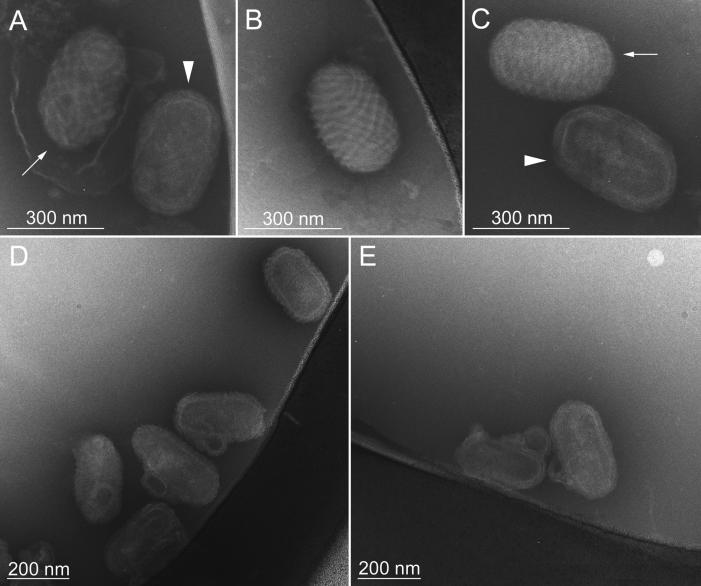
As a compromise between conventional negative staining EM and cryo-EM, we employed cryo-negative staining EM, in which the negative stain, ammonium molybdate, is applied to the sample without air drying and immediately before vitrification. This results in an improvement of the signal-to-noise ratio as well as in improved resistance to beam damage (2, 8). Envelopes were removed by the staining procedure or only weakly attached, as shown in Fig. 5A. Under these conditions, we observed two types of particles in the WT virus preparations which closely resembled ORFV particles viewed by standard negative staining EM. Some particles displayed a very distinct spiral tubule and crenellated edges on the particle surface, due to outlining of the particle by the negative stain (Fig. 5A to C, arrows). Other particles revealed a core domain, probably as a result of the penetration of the stain (Fig. 5A and C, arrowheads). Examination of the virus deleted of the 10-kDa gene revealed the appended loop-like structures and loss of the regularly spaced spiral tubule (Fig. 5D and E).
FIG. 5.
Cryo-negative staining EM of ORFV. Panels A to C are views of the WT ORFV. Panels D and E are views of the mutant D1701-Vr10.
DISCUSSION
The surface appearance of parapoxvirus particles is a distinctive diagnostic property of this genus. The purpose of this study was to reassess this unique structural feature and to investigate the role of the 10-kDa protein in virus morphogenesis. Cryo-EM observations of ORFV enabled us to observe surface tubules and crenellated edges in continuity with the surface tubule, features that collectively provide evidence that the spiral tubule is a bona fide structure. Cryo-negative staining EM considerably enhanced the contrast of the particles and confirmed the presence of the spiral tubule. These results should be discussed in light of the report by Dubochet et al., who suggested that the VACV tubules are artifacts of the negative staining method since they could not be observed by cryo-EM (9). If VACV tubules are bona fide structures, they may be more difficult to visualize by cryo-EM than the ORFV tubule since they are more disorganized; furthermore, as discussed by Malkin et al. (18), 20-nm tubules may be invisible on a background of a cryo-EM image which is a projection of the entire thickness of a particle about 240 to 290 nm. In fact, Malkin et al. (18) were recently able to detect VACV tubules by using in situ atomic force microscopy, a technique that allows the visualization of the surface of unstained samples. In the case of ORFV, the finding that the spiral tubule is absent from the virus mutant with the 10-kDa gene deleted provides additional evidence for the authenticity of such structures. Whether the 10-kDa protein is a constituent of the spiral tubule or is simply required for its appearance needs further investigation. It is important to stress that although our studies support the notion that the spiral tubule is a genuine feature of ORFV, they do not provide any indication as to its origin. In fact, ORFV may be surrounded by a true tubule, or the tubular appearance may be due to a thickening of membrane domains in a very regular fashion.
In contrast to the VACV 14-kDa protein, the ORFV 10-kDa protein is not necessary for the formation of intracellular enveloped particles since we readily observed TGN-enveloped particles. However, the 10-kDa protein is clearly important for the proper morphogenesis of ORFV, as documented by the appearance of thin extensions and loop-like structures on virus particles lacking the 10-kDa protein. This is particularly surprising since no unusual features were reported for IMV particles produced by a VACV mutant expressing highly reduced levels of the 14-kDa protein (16, 26). Interestingly, the loop-like structures often appeared to comprise a series of overlapping membranes even when the virus particles displayed no evidence of being enveloped by TGN-derived membranes. This observation, together with the fact that we could observe double membranes surrounding IVs, may be pertinent to the controversy about the presence of one or two membranes around poxvirus particles (31) and argues that the latter theory is the correct one. Removal of the 10-kDa gene did not only affect proper ORFV maturation but also prevented the formation of virus plaques. Moreover, infectious virus progeny was not detectable in supernatants of D1701-Vrd10-infected cells nor was extracellular enveloped virus observed in thin sections of these cells, indicating that the 10-kDa protein may play a role in virus release.
One of the critical events in poxvirus assembly may be the final sealing of the membranes that enclose viroplasmic material. This could occur upon completion of spherical immature virus or simultaneously with the maturation of the virus into the more condensed form. We hypothesize that this sealing event, which probably requires membrane fusion and is concomitant with tubule formation, is inefficient in the absence of the ORFV 10-kDa protein but nevertheless occurs beyond its normal site. This would result in an extension of the particle, which could then fold back onto another end of the same particle to create a loop-like structure. According to this hypothesis, the 10-kDa protein not only participates in tubule organization but also aids sealing. The fact that the 10-kDa protein is an orthologue of a VACV fusion protein makes this hypothesis attractive. A less conventional model of poxvirus assembly has pictured the formation of mature virus particles without any final membrane fusion event but, instead, the sealing of opposing membranes by a protease and dithiothreitol-sensitive plug (27). In the framework of this model, one may speculate that the 10-kDa protein is important for the plugging event. A variant of the latter model proposes that a series of overlapping membranes ensures virus integrity without the requirement for actual closing of the structure (12). According to the latter model, one may view the ORFV mutant described here as defective in the folding events required for mature virus formation but not for virus infectivity. Obviously, additional studies are required to investigate the precise role of the ORFV 10-kDa protein. The recent development of cryoelectron tomography may be valuable to distinguish between the different alternatives that have been proposed for the organization of poxvirus membranes.
Acknowledgments
We greatly appreciated the generous supply of purified viruses and support in the generation of mutant virus as well as the critical comments on the manuscript given by Mathias Büttner. We are also grateful to Jacques Dubochet (CME, Lausanne Switzerland), Marc Schmutz (ICS, Strasbourg, France), and Patrick Schultz (IGBMC, Strasbourg, France) for helpful discussions and advice. The excellent technical assistance of Berthilde Bauer and Angelika Braun is gratefully acknowledged as well as the help of Eduard Hettich in transfection experiments. The lacZ-Xgpt gene cassette was generously provided by Falko Falkner (Baxter, Vienna, Austria).
Part of this work was funded by BAYER AG, Monheim-Leverkusen, Germany.
REFERENCES
- 1.Abdussalam, M., and V. E. Cosslett. 1957. Contagious pustular dermatitis virus I. Studies on morphology. J. Comp. Path. 67. [DOI] [PubMed]
- 2.Adrian, M., J. Dubochet, S. D. Fuller, and J. R. Harris. 1998. Cryo-negative staining. Micron 29:145-160. [DOI] [PubMed] [Google Scholar]
- 3.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. [DOI] [PubMed] [Google Scholar]
- 4.Buttner, M., and H. J. Rziha. 2002. Parapoxviruses: from the lesion to the viral genome. J. Vet. Med. B 49:7-16. [DOI] [PubMed] [Google Scholar]
- 5.Czerny, C. P., S. Johann, L. Holzle, and H. Meyer. 1994. Epitope detection in the envelope of intracellular naked orthopox viruses and identification of encoding genes. Virology 200:764-777. [DOI] [PubMed] [Google Scholar]
- 6.Czerny, C. P., R. Waldmann, and T. Scheubeck. 1997. Identification of three distinct antigenic sites in parapoxviruses. Arch. Virol. 142:807-821. [DOI] [PubMed] [Google Scholar]
- 7.Dallo, S., J. F. Rodriguez, and M. Esteban. 1987. A 14K envelope protein of vaccinia virus with an important role in virus-host cell interactions is altered during virus persistence and determines the plaque size phenotype of the virus. Virology 159:423-432. [DOI] [PubMed] [Google Scholar]
- 8.De Carlo, S., C. El-Bez, C. Alvarez-Rua, J. Borge, and J. Dubochet. 2002. Cryo-negative staining reduces electron-beam sensitivity of vitrified biological particles. J. Struct. Biol. 138:216-226. [DOI] [PubMed] [Google Scholar]
- 9.Dubochet, J., M. Adrian, K. Richter, J. Garces, and R. Wittek. 1994. Structure of intracellular mature vaccinia virus observed by cryoelectron microscopy. J. Virol. 68:1935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkner, F. G., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, T., O. Planz, L. Stitz, and H. J. Rziha. 2003. Novel recombinant parapoxvirus vectors induce protective humoral and cellular immunity against lethal herpesvirus challenge infection in mice. J. Virol. 77:9312-9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths, G., R. Wepf, T. Wendt, J. K. Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated-particle analysis. J. Virol. 75:11034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haig, D. M., J. Thomson, C. McInnes, C. McCaughan, W. Imlach, A. Mercer, and S. Fleming. 2002. Orf virus immuno-modulation and the host immune response. Vet. Immunol. Immunopathol. 87:395-399. [DOI] [PubMed] [Google Scholar]
- 14.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao, J. C., C. S. Chung, and W. Chang. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 72:8374-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiao, J. C., C. S. Chung, and W. Chang. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73:8750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 18:9-18. [DOI] [PubMed] [Google Scholar]
- 18.Malkin, A. J., A. McPherson, and P. D. Gershon. 2003. Structure of intracellular mature vaccinia virus visualized by in situ atomic force microscopy. J. Virol. 77:6332-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer, A. A., K. Fraser, G. Barns, and A. J. Robinson. 1987. The structure and cloning of orf virus DNA. Virology 157:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Naase, M., B. H. Nicholson, K. M. Fraser, A. A. Mercer, and A. J. Robinson. 1991. An orf virus sequence showing homology to the 14K ′fusion' protein of vaccinia virus. J. Gen. Virol. 72:1177-1181. [DOI] [PubMed] [Google Scholar]
- 21.Nagington, J., and R. W. Horne. 1962. Morphological studies of Orf and Vaccinia viruses. Virology 16:248-260. [DOI] [PubMed] [Google Scholar]
- 22.Nagington, J., A. A. Newton, and R. W. Horne. 1964. The structure of Orf virus. Virology 23:461-472. [DOI] [PubMed] [Google Scholar]
- 23.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez, J. F., R. Janeczko, and M. Esteban. 1985. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J. Virol. 56:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez, J. F., E. Paez, and M. Esteban. 1987. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J. Virol. 61:395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez, J. F., and G. L. Smith. 1990. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 18:5347-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roos, N., M. Cyrklaff, S. Cudmore, R. Blasco, J. Krijnse-Locker, and G. Griffiths. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 15:2343-2355. [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson, C. M., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 81:47-58. [DOI] [PubMed] [Google Scholar]
- 29.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 30.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van 't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]