Abstract
Background
Common variable immunodeficiency (CVID) is characterized clinically by inadequate quantity and quality of serum immunoglobulins with increased susceptibility to infections resulting in significant morbidity and mortality. Only a few genes have been uncovered and the genetic background of CVID remains elusive to date for the majority of patients.
Objective
To seek novel associations of genes and genetic variants with CVID.
Methods
We performed association analyses in a discovery cohort of 164 CVID cases and 19,542 healthy control subjects genotyped on the Immuno BeadChip from Illumina (iCHIP); replication of findings were examined in an independent cohort of 135 CVID cases and 2,066 healthy control subjects, followed by meta-analysis.
Results
We identified 11 SNPs at the 16p11.2 locus associated with CVID at genome-wide significant level in the discovery cohort. The most significant SNP, rs929867 (p = 6.21×10−9) is in the gene FUS (fused-in-sarcoma) with four other SNPs mapping to ITGAM (integrin CD11b). Results were confirmed in our replication cohort. Conditional association analysis suggests a single association signal at the 16p11.2 locus. A strong trend of association was also seen by 38 SNPs (p < 5×10−5) in the MHC region, supporting that this is a genuine CVID locus. Interestingly we found that 80% of patients with the rare ITGAM variants have reduced counts of switched-memory B-cells.
Conclusion
We report a novel association of CVID with rare variants at the FUS/ITGAM (CD11b) locus on 16p11.2. The association signal is enriched for promoter/enhancer markers in the ITGAM gene. ITGAM encodes the integrin CD11b, a part of complement receptor 3 (CR3/Mac-1), a novel candidate gene implicated here for the first time in the pathogenesis of CVID.
Keywords: immunodeficiency, immunogenetics, genome-wide association study, ITGAM, rare variants
Introduction
Common variable immunodeficiency (CVID) is characterized by susceptibility to bacterial infections, caused by inadequate quantity and quality of immunoglobulins.1 CVID is a primary immunodeficiency disease that is highly heterogeneous in nature, albeit believed to result from intrinsic immunological deficits. Significant phenotype heterogeneity in CVID is demonstrated by variability in the age at presentation as well as related comorbidities, including autoimmune disease, malignancy, chronic lung disease and gastrointestinal disease.2, 3
CVID, although relatively rare (prevalence of 1:25,000 – 1:50,000), is clinically an important cause of immunodeficiency, because of the significant morbidity and mortality conferred by its comorbidities. Given its clinical heterogeneity, it is not surprising that the etiology of CVID remains poorly understood and complex. A number of different genes have been implicated as candidates for CVID susceptibility in specific patient subsets. These include genes that act in B-cell-receptor (BCR) signaling (CD19, CD20, CD21, CD81, PLCγ2, TWEAK and PKCδ), or pathways of isotype switching and somatic recombination (BAFFR, ICOS, MSH5, TACI and NFκB2).4–18 Unbiased approaches including genome-wide-association studies (GWAS) and whole-genome copy number variation (CNV) studies of CVID to-date have had limited success, but among the associations identified is the MHC (major histocompatibility complex) region and ORC4L.8, 19, 20 However, the above associations only account for a minority of CVID cases. Gaining a better understanding of the genetic variability underlying CVID may be a first and critical step to developing personalized approaches to treatment, comorbidity monitoring, and overall care of patients with CVID. The search for novel genetic susceptibility loci in CVID is therefore a burning object of interest in the world of immunology.
Methods
Patients
As CVID is a rare disease, many of the previous studies were underpowered and with limited coverage of immunology specific genes. Our study included 360 CVID cases, the diagnosis of which was confirmed with standard diagnostic criteria. The study was approved by institutional review boards of each participating site and informed consent for DNA collection and genotyping was obtained from all participants. The CVID patients were from four participating sites and reported in a previous study.19 The 21,610 healthy controls were from the IBD Consortium and previously described in a study by Jostins L, et al. in 2012.21
SNP genotyping and Quality control
All CVID cases and all controls were genotyped on the iCHIP,22, 23 an Infinium Immuno BeadChip from Illumina, with 196,524 SNPs and small indels targeting genes that are implicated in autoimmune and inflammatory diseases based on previous GWAS.21 The design of this chip has been reported previously.21 This novel array is designed to provide dense coverage of rare polymorphisms and strong candidate genes for major autoimmune and inflammatory diseases from prior GWAS studies. We sought to associate these rare immune gene SNPs with CVID, as well as with the clinical phenotypes to address the heterogeneity of CVID, in our ongoing study aimed at identifying susceptibility loci for CVID.
We obtained the cluster egt file from IBD consortium and applied it to our data. In the quality control (QC) filtering step, 18,401 SNPs were excluded because of low genotyping rate (<95%) and 30 samples (29 cases and 1 control) were removed from further analysis because of low sample call rate (<95%). We further filtered out SNPs that deviate from Hardy-Weinberg equilibrium (HWE test p-value < 10−6). To adjust for population stratification, we evaluated ancestry informative markers for all samples using principal component analysis (PCA) implemented in EIGENSTRAT24 software, based on the results of which we removed an addition of 4 outliers (3 cases and 1 control). We further performed genome-wide identity-by-descent analysis with LD-pruned SNPs. By a cutoff of Pi-hat 0.25, 29 cryptic related pairs of cases were detected and one sample from each pair was removed from the analysis. An additional 6,813 SNPs fall into the IBD consortium’s SNP exclusion list or failed our manual review of genome studio cluster plots and were also excluded from further analysis.
According to principal components plot (Figure E1), we divided our dataset into a discovery cohort and a replication cohort. The discovery cohort was composed of 164 cases and 19,542 controls. SNP QC was conducted again within the discovery cohort, which left with 170,212 SNPs including both common and rare variants. The replication cohort consists of 135 cases and 2,066 controls. Similar SNP QC resulted in a final collection of 169,914 SNPs in the replication cohort. We also further conducted principal component analysis within discovery cohort and replication cohort separately.
Association analysis
After sample and SNP based QC, association testing was carried out in the discovery cohort. Logistic regression under the additive model was employed for association analysis, including the first 3 principal components as covariates. The quantile-quantile plot of null SNP p-values (Figure E2) and an estimated genomic inflation factor lambda of 1.03 do not suggest any substantial population stratification. Conditional regression analysis, LD estimation and pairwise epistasis tests were conducted via PLINK25. Association analysis was similarly conducted in the replication cohort. Meta-analysis was performed with software PLINK25 and METAL.26
Imputation
Haplotype phasing was first performed via SHAPEIT v227 and local imputation at the locus of 16p11.2 was carried out with the reference of 1000 Genome (http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html) using IMPUTE2.28, 29 Afterwards, missing data likelihood score test implemented in software SNPTEST V229 was conducted for investigating SNP association with CVID phenotype, including the first 3 principal components as covariates.
Results
Novel locus associated with CVID
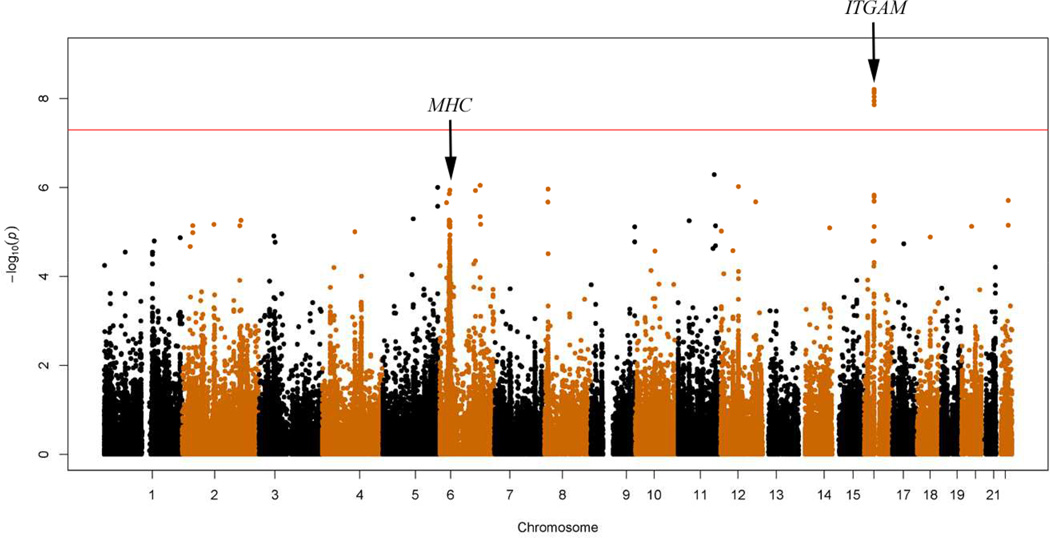
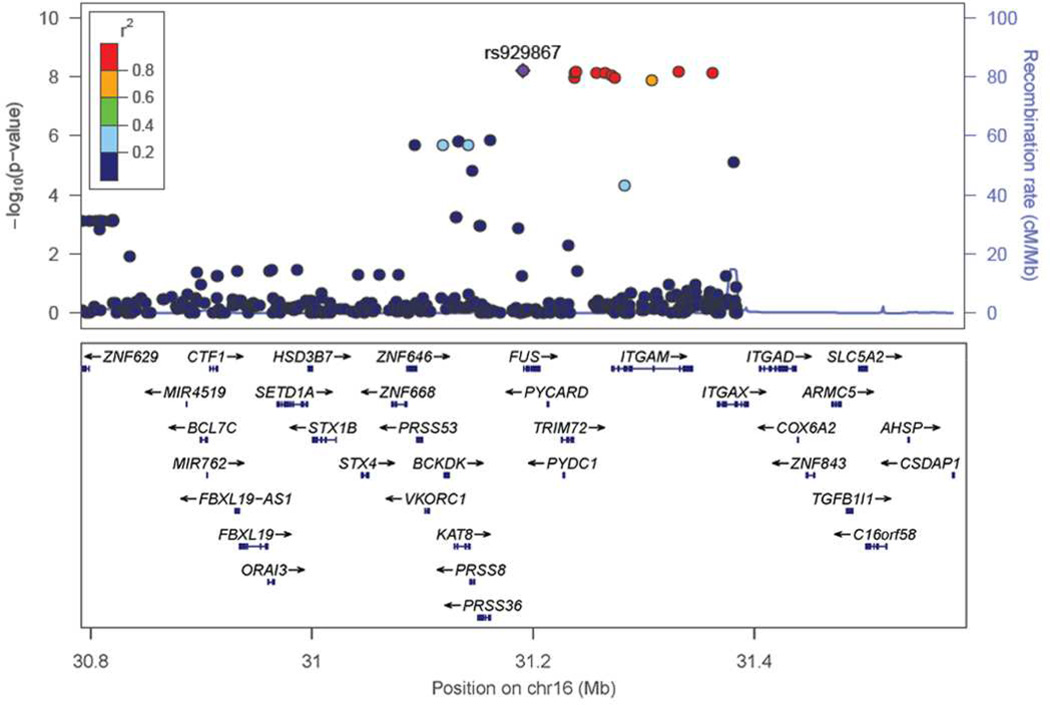
We identified a locus on chromosome 16 (Figure 1), with 11 SNPs surpassing genome-wide significance, following logistic regression (Table 1) to be associated with CVID. The most significant SNP, rs929867 (p-value = 6.21×10−9, OR = 89.8), is in the gene FUS (fused-in-sarcoma). This maps to a linkage disequilibrium block on 16p11.2, which is a gene rich region (Figure 2) containing 10 other SNPs also associated with CVID in our analysis (p-value range of 1.39×10−8 to 6.79×10−9; OR range = 58.55–87.84)(Table 1). Four of the identified SNPs reaching genome-wide significance are mapped in the gene ITGAM (encoding integrin CD11b); with two additional SNPs being close to this gene as well. Three of the variants in ITGAM are intronic, whereas a fourth (rs8056264) is a synonymous variant in the coding region of ITGAM, see Table E1 for details. Detailed inspection of the genome studio cluster plots of each SNP confirmed distinct clusters for each SNP (Figure E3). Consistent with the minor allele frequency (MAF) of controls in our dataset, in the European population of the 1000-genome-project, each of these SNPs had a MAF < 0.0001 (Table E2). By applying conditional logistic regression on the top significant SNP, rs929867, all other associations were ablated (Table E3), suggesting a single association signal at the 16p11.2 locus. In concordance with this, no epistatic interaction was noted among the genome wide significant SNPs at this locus (all epistasis test p-values > 0.05).
Figure 1.
Manhattan plot of genome-wide association results for CVID in the Caucasian discovery cohort. Red line indicates the genome-wide significance threshold.
Table 1.
Genome-wide significant SNPs at 16p11.2.
| SNP | BP (hg19) |
Closest Gene |
Minor /Major Allele |
Dis MAF cases |
Dis MAF controls |
Dis OR | Dis P | Rep P | Meta OR |
Meta P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs750953 | 31094089 | ZNF646 | C/T | 0.0061 | 0.000102 | 66.7 | 2.05×10−6 | 1.84×10−3 | 24.91 | 4.25×10−8 |
| rs929867 | 31191482 | FUS | A/G | 0.00926 | 0.000102 | 89.8 | 6.21×10−9 | 5.50×10−3 | 37.33 | 5.49×10−10 |
| rs7192718 | 31237920 | TRIM72* | T/C | 0.00915 | 0.000128 | 67.94 | 1.15×10−8 | 2.91×10−4 | 33.37 | 3.64×10−11 |
| rs10871455 | 31238690 | TRIM72* | T/G | 0.00915 | 0.000102 | 87.84 | 7.39×10−9 | 2.89×10−4 | 36.61 | 3.19×10−11 |
| rs12934814 | 31239855 | TRIM72* | T/C | 0.00943 | 0.000128 | 72.13 | 7.11×10−9 | 2.42×10−4 | 37.9 | 1.65×10−11 |
| rs4889637 | 31258238 | ITGAM* | C/T | 0.00915 | 0.000128 | 72.23 | 7.54×10−9 | 2.77×10−4 | 35.19 | 2.35×10−11 |
| rs4889641 | 31265541 | ITGAM* | A/G | 0.00915 | 0.000103 | 87.6 | 7.55×10−9 | 2.89×10−4 | 37.29 | 3.11×10−11 |
| rs8063978 | 31271700 | ITGAM | C/T | 0.00938 | 0.000128 | 70.39 | 9.16×10−9 | 5.64×10−3 | 32.76 | 6.67×10−10 |
| rs4889541 | 31274551 | ITGAM | T/C | 0.00915 | 0.000128 | 68.29 | 1.12×10−8 | 7.98×10−3 | 30.51 | 1.29×10−9 |
| rs9929801 | 31283472 | ITGAM | C/T | 0.00915 | 0.000744 | 12.12 | 4.88×10−5 | 1.34×10−4 | 9.22 | 2.96×10−8 |
| rs8045402 | 31308281 | ITGAM | T/C | 0.00915 | 0.000154 | 58.55 | 1.39×10−8 | 3.38×10−5 | 30.34 | 4.73×10−12 |
| rs8056264 | 31332655 | ITGAM | T/C | 0.00915 | 0.000128 | 73.24 | 6.79×10−9 | 2.96×10−4 | 35.38 | 2.31×10−11 |
| rs11574629 | 31362892 | ITGAX* | G/A | 0.00915 | 0.000103 | 87.75 | 7.44×10−9 | 3.28×10−4 | 37.26 | 3.50×10−11 |
Not directly on gene;
BP=base pair; Dis MAF cases=minor allele frequency in cases of the discovery cohort; Dis MAF controls=minor allele frequency in controls of the discovery cohort; Dis OR=odds ratio in the discovery cohort; Dis P=P-value in the discovery cohort; Rep P=P-value in the replication cohort; Meta OR= odds ratio in the meta-analysis; Meta P= P-value in the meta-analysis.
Figure 2.
Regional association plot at the 16p11.2 (ITGAM) locus. SNPs are plotted by chromosomal position in a 400-kb window against their −log10 transformed P values for association with CVID phenotype in the discovery cohort. The color of each SNP reflects their LD with the most significant rs929867. The r2 values were calculated from the discovery cohort in PLINK.
Next, we sought to replicate these findings in the independent replication cohort. All 11 SNPs identified in the discovery cohort replicated with nominal p-values < 0.05 (Table 1). When the two cohorts were combined by meta-analysis, all 11 SNPs were found to be genome-wide significant, and two additional genome-wide significant SNPs were uncovered (p-values: 2.96×10−8 and 4.25×10−8; OR values: 9.22 and 24.91, respectively) (Table 1). The genotype status of each CVID subject carrying the rare variant allele was further validated by Sanger sequencing together with randomly selected CVID subjects without the rare variant allele who were confirmed to be negative (Figure E4).
Using the 1000-genome as reference, we conducted imputation around the 16p11.2 locus and uncovered 23 additional SNPs of genome-wide significance in our meta-analysis (Table E4), further supporting the importance of this locus.
Association of the MHC region with CVID
The MHC region has previously been reported to be significantly associated with CVID.19 We found two SNPs in the MHC region that reached genome-wide significant p-values in the discovery cohort and a third SNP reached a marginally significant p-value < 5×10−7. However, these SNPs (rs2517625, rs6933596, rs7773051) were excluded from the final analysis, due to abnormal genome-studio cluster plots. Despite this, we did appreciate a strong trend of association by 38 SNPs (p-value < 5×10−5) in this region (chr6:28Mb–34Mb) in the discovery cohort (Figure 1), with the strongest association at rs879882 (p-value = 1.156×10−6). Meta-analysis yielded 20 SNPs in this region with p-value < 5×10−5 (Table 2). These results support the MHC region as a genuine CVID locus as previously reported19. Additionally, SNPs in the genes CR2, ICOS, MSH5 and TACI were found to be of nominal significance (p-values <0.05) in our meta-analysis (Table 3). Although known TACI carriers were included in our patient population these SNPs are not covered by the iCHIP and thus did not reach higher significance in our analysis. We detected a nominally significant SNP rs61061086 in gene FLJ16124 (Table 3) which was reported to be one of the top genes associated with CVID in a previous GWAS study.19 Furthermore, the aforementioned conditional analysis on rs929867 did not ablate the association seen with SNPs across the MHC region, and there were no epistatic interactions observed between SNPs at the 16p11.2 and the MHC loci. This suggests that the two loci independently affect the risk of CVID.
Table 2.
SNPs in the MHC region with meta-analysis p-value < 5×10−5.
| SNP | CHR | BP (hg19) | Minor Allele | Meta P | Meta OR |
|---|---|---|---|---|---|
| rs3130724 | 6 | 29118530 | A | 6.39×10−6 | 1.55 |
| rs3129151 | 6 | 29138315 | T | 7.31×10−6 | 1.55 |
| rs3129152 | 6 | 29138777 | C | 1.39×10−5 | 1.52 |
| rs760804 | 6 | 29171039 | T | 3.86×10−6 | 1.57 |
| rs3117426 | 6 | 29272012 | T | 3.15×10−6 | 1.58 |
| rs3131036 | 6 | 30728290 | A | 1.05×10−5 | 1.49 |
| rs3094117 | 6 | 30737486 | C | 1.56×10−5 | 1.47 |
| rs3129981 | 6 | 30758857 | T | 3.05×10−6 | 1.67 |
| rs3095089 | 6 | 30933794 | T | 2.66×10−5 | 1.54 |
| rs3131932 | 6 | 30940328 | A | 1.88×10−5 | 1.44 |
| rs3132579 | 6 | 30940989 | C | 3.03×10−5 | 1.57 |
| rs2073721 | 6 | 31129616 | A | 3.73×10−5 | 0.61 |
| rs2073723 | 6 | 31130078 | T | 3.38×10−5 | 0.61 |
| rs1065461 | 6 | 31130502 | T | 1.00×10−5 | 0.58 |
| rs3132528 | 6 | 31131569 | C | 4.81×10−5 | 0.61 |
| rs9263805 | 6 | 31135735 | A | 3.15×10−5 | 0.61 |
| rs3130501 | 6 | 31136453 | A | 3.48×10−5 | 0.61 |
| rs3130502 | 6 | 31136666 | A | 4.87×10−5 | 0.62 |
| rs6923313 | 6 | 31241370 | C | 3.63×10−5 | 0.68 |
| rs2507997 | 6 | 31314781 | G | 3.69×10−5 | 1.47 |
CHR= Chromosome; BP=base pair; Meta P=Meta-analysis P-value; Meta OR=Meta-analysis odds ratio.
Table 3.
Association results for candidate genes.
| SNP | CHR | BP (hg19) | Gene | Distance | Minor /Major Allele |
Meta P | Meta OR |
|---|---|---|---|---|---|---|---|
| rs4618970 | 1 | 207661640 | CR2 (CD21) | 0 | C/T | 0.0429 | 1.27 |
| rs10932030 | 2 | 204803483 | ICOS | 0 | C/T | 0.0134 | 1.94 |
| rs9968356 | 4 | 151471789 | LRBA | 0 | A/G | 0.0944 | 1.42 |
| rs1144708 | 6 | 31710020 | MSH5 | 0 | T/C | 0.0236 | 1.22 |
| rs7897947 | 10 | 104157711 | NFKB2 | 0 | G/T | 0.192 | 1.14 |
| rs35979293 | 16 | 28944700 | CD19 | 0 | T/G | 0.235 | 1.11 |
| rs4889393 | 16 | 81844013 | PLCG2 | 0 | A/G | 0.128 | 0.738 |
| rs7214221 | 17 | 16859829 | TNFRSF13B (TACI) | 0 | A/G | 0.0107 | 2.39 |
| rs61061086 | 2 | 65710170 | FLJ16124 | 0 | G/A | 2.85×10−3 | 1.33 |
CHR=Chromosome; BP=base pair; Distance=Distance between the SNP and the gene; Meta P=Meta-analysis P-value; Meta OR= Meta-analysis odds ratio.
Functional annotation of the significant SNPs in FUS/ITGAM locus
A number of SNP polymorphisms identified to be associated with complex diseases have been shown to affect the disease through regulation of gene expression via the DNA macromolecular complex. To determine if any of the identified SNPs from this study may have a regulatory role, we searched for prior identified or predicted functional annotations30, 31 of these SNPs using the software Haploreg32. Our results show that the identified genome-wide significant SNPs associated with CVID susceptibility are enriched for enhancer markers (Table E5) and DNase sites (Table E6) in certain immune cell types, such as B-lymphocytes. Furthermore, genomic annotation (Table E7) using Haploreg32 shows each of the genome-wide significant SNPs to overlap with promoter/enhancer histone markers or transcription regulatory motifs, especially in CVID relevant immune cell types, suggesting these SNPs may impact transcriptional regulation in immune cells.
Network and pathway analysis of ITGAM gene
Since multiple significant SNPs were located in the ITGAM gene, we conducted a network and pathway analysis around this gene to further understand its biological function. By FunCoup33, 34 search, we found multiple known and predicted ITGAM functional association partners (ALOX5AP, SELL, C3, FCER1G, ICAM1, ITGAX, ITGB2, LYN, NCF1, NCF2, NCF4, PLAUR, TYROBP, VAV1, SPI1, CYBA). Five of these contain SNPs with nominal significant association with CVID in our study (Table E8). To further explore the biological implication of ITGAM association with CVID, we input ITGAM, the nominally significant CVID susceptibility genes (CR2, ICOS, MSH5, TNFRSF13B, FLJ16124) and the nominally significant ITGAM functional association partners (ITGAX, C3, LYN, VAV1 and NCF2) into FunCoup33, 34 for association network analysis, the result of which are shown in Figure E5. We also input these genes into STRING,35, 36 another protein-protein interaction network analysis tool, with similar results. The pathway analysis tool implemented in FunCoup,33, 34 demonstrated enrichment of multiple B-cell and T-cell signaling pathways, which are highly relevant to CVID pathophysiology (Table E9).
Clinical phenotypes of the CVID patients carrying rare ITGAM variants
To determine if these susceptibility SNPs are associated with certain clinical characteristics or a subgroup of CVID patients, we examined the available phenotype information for individuals with the rare ITGAM variant. Results showed that 50% were females and 80% (n=4/5, data not available for n=1) have low switched-memory B-cells (<2% of B-cells3), which is a higher percentage than that seen among the rest of the CVID cases (60%, n=105/175 with data available), or that in the Euroclass study (58%)3, see Table 4. Reviewing the clinical phenotypes of these individuals we found only one subject to have autoimmune cytopenia (ITP), this patient also developed nodular-regenerative-hyperplasia of the liver. Two patients developed lung disease, one had bronchiectasis, and another was diagnosed with lung cancer. A second patient developed cancer, Burkitt’s lymphoma, and this patient also had SLE (systemic lupus erythematosus) with lupus nephritis, requiring renal transplant.
Table 4.
Phenotypic information for CVID patients with the ITGAM rare variants.
| Patient | Age at onset of symptoms |
Sex | FHx of PIDD* |
IgG** (mg/dL) |
IgM** (mg/dL) |
IgA** (mg/dL) |
% CD19 B-cells |
% Switched memory B-cells (CD19+/CD27+/IgM−/IgD−) |
|---|---|---|---|---|---|---|---|---|
| 1 | 3 years | M | No | 80 | 0 | 14 | 14.0 | 0.00 |
| 2 | N/A | M | No | 150 | 16 | <7 | 2.1 | 7.32 |
| 3 | N/A | F | No | 197 | 6 | <4 | 17.8 | 0.35 |
| 4 | N/A | F | No | 186 | 16 | 6 | <1.0 | 0.00 |
| 5 | N/A | M | Yes | N/A | N/A | N/A | N/A | N/A |
| 6 | 11 years | F | No | 120 | N/A | N/A | N/A | 1.90 |
Family history of Primary Immunodeficiency Disease
Laboratory values were obtained time of diagnosis.
Sanger sequencing of FUS, ITGAM and ITGAX
To elucidate the genetic association of 16p11.2 to CVID, we performed Sanger sequencing of the plausible candidate genes at this locus. We evaluated two CVID cases carrying the rare variant at all the genome-wide significant SNPs at the 16p11.2 locus and one CVID case with none of these rare variants. All exons of the three plausible candidate genes (FUS, ITGAM, and ITGAX) in strong linkage disequilibrium with the most significant SNPs were interrogated by Sanger sequencing. We confirmed again the C>T synonymous variant of rs8056264 in exon 15 of gene ITGAM. However, no exonic nucleotide bases were uncovered that demonstrated corresponding rare variant non-synonymous changes which would lead to an amino acid change in the translated protein product and directly impact the gene function in CVID patients, suggesting that the association signal at the locus is capturing some other regulatory effects on gene function yet to be established.
Discussion
We describe a novel association of CVID, with a rare variant, using the iCHIP platform that densely covers immune-disease related genes. The genome-wide-significant locus discovered at 16p11.2 presents the first SNP association in CVID in a genome wide study in a non-MHC region. This locus is downstream of and in separate LD blocks from the 16p11.2 locus (29.5–30.1Mb) previously associated with autism, schizophrenia, and obesity37– 39 (Figure E6). The single association signal at this locus and genomic annotation suggests these rare SNPs may impact transcriptional regulation in lymphocytes. The most significant SNP, rs929867, maps to the gene FUS, encoding a multifunctional protein component of the heterogenous-nuclear-ribonuceoprotein complex (hnRNP). FUS, belonging to the FET family of RNA binding proteins, has an important role in pre-mRNA splicing, nuclear export of processed mRNA, and may consequently help to maintain genomic integrity.40, 41 Mutations in FUS have been reported in amyotrophic lateral sclerosis type 6.42, 43
Two SNPs that are significantly associated with CVID in our study are of particular interest. The SNP rs929867 with the best p-value in our discovery cohort is located in the 5'-UTR of the FUS gene. This region is crucial for translational regulation in many species44 by regulating translation efficiency and mRNA stability through secondary structures and RNA binding proteins.44, 45 It could function as the binding site for several transcription factors (Table E7), such as CTCF and NFκB which both play essential roles in normal B cell functions46, 47. NFκB has also been associated with the development of CVID18. Another SNP rs8056264 causes a synonymous substitution in the ITGAM gene. Although synonymous mutations do not alter the amino acid sequence they may impact mRNA transport, splicing and even translation due to codon usage bias.48, 49 Through software Haploreg32, we found both SNPs are conserved sites by SiPhy-omega (Site-specific PHYogenetic analysis) algorithm.50 Whether these rare variants truly affect the mRNA or protein level of FUS or ITGAM needs to be further explored.
Among the genes in the 16p11.2 locus, the ITGAM is the most attractive candidate with four genome-wide-significant SNPs mapping to this gene and 2 additional SNPs being close to it. ITGAM encodes the integrin αM (CD11b). Integrins are heterodimeric integral membrane proteins with key roles in adhesion and cell contact. CD11b, together with β2 integrin (ITGB2/CD18) forms the leukocyte-specific heterodimer; macrophage-receptor-1/complement receptor 3 (Mac-1/CR3). CR3 is expressed on mononuclear phagocytes, neutrophils, NK cells, a subset of B cells and some T-cells.51– 54 Its main ligands are ICAM-1, ICAM-2, inactivated-C3b (iC3b) and fibrinogen. CR3 ligation results in pro-inflammatory responses such as leukocyte adhesion, migration, recruitment and superoxide release.55 Importantly, CD11b is involved in phagocyte adhesion and migration as well as in phagocytosis of opsonized pathogens/particles. Thus the ITGAM variant may impair these critical functions of the immune system and altered trafficking and cell-cell interactions may predispose to defective immune responses in CVID. The immunophenotype of ITGAM knockout mice supports the importance of its function for normal immune responses. These mice have reduced leukocyte and neutrophil adhesion, impaired neutrophil degranulation, phagocytosis and oxidative burst. Also the numbers of CD4+ T-cells are reduced as well as T-cell proliferation. Importantly susceptibility and mortality from pneumococcal infection is significantly increased in these mice (Table E10) (http://www.informatics.jax.org/diseasePortal/).
Interestingly, the nominally significant ITGAM association partners, direct or indirect, identified by our network analyses include a tyrosine kinase involved in BCR signaling (LYN) and a guanine nucleotide exchange factor important for actin cytoskeletal rearrangements involved in both B- and T-cell development and activation (VAV1). Thereby, our network analyses demonstrates enrichment of B- and T-cell signaling pathways, including regulation of activation of both cell types. Finally, FcγR signaling, that has roles in phagocytosis of opsonized pathogens also is implicated.
Previous studies have demonstrated reduced numbers of dendritic cells and defective TLR signaling in CVID, especially through TLR 7 and 9.56, 57 TLR7 and 9 are expressed on both naïve and memory B-cells and can mediate powerful stimulation and potentiate B-cell activation, possibly allowing for autoreactivity to self RNA/DNA. Interestingly, CD11b can both positively and negatively regulate TLR signaling, depending on the experimental conditions,58– 64 furthermore, CR3 can interact with Fc receptors on neutrophils/macrophages and modulate the effects of immune complex binding.65 CR3 ligation can also suppress TLR4 and TLR7/8 signaling through degradation of downstream components (MyD88 and TRIF),61, 66 suggesting negative cross-talk between the TLR and CR3 pathways. This cross-talk and effects of TLR stimulation on B-cells might influence responses to self antigens and thus autoimmunity. Finally, these interactions may affect B-cell activation through other pathways.
Previous GWAS studies have revealed associations between different SNPs in the ITGAM locus and SLE susceptibility,65, 67–71 where studies found phagocytosis by monocytes/macrophages and firm adhesion of neutrophils to be impaired.65–67, 72– 74 In SLE, ITGAM variants influence risk of renal disease, autoantibody production and immunological manifestations,59 possibly by impacting clearance of apoptic cells and leukocyte trafficking. Mac-1 can also induce tolerance by its effects on dendritic cells (DCs), including maturation restriction, antigen presenting cell (APC) function and T-cell activation.75, 76 Hence, decreased phagocytosis could through decreased binding of Mac-1 to iC3b contribute to both immune dysfunction and autoimmunity through reduced clearance of apoptic cells, activation of DCs, production of inflammatory cytokines and uptake and presentation of self-antigens to T-cells. Also, DC-SIGN, on DCs, can bind CR3 on neutrophils during cross-talk between these cells.77, 78 Another role for CD11b in tolerance is by restriction of Th17 differentiation, a cell type implicated in autoimmunity.79–81
Our limited phenotypic information does not definitively indicate a specific immunophenotype for patients carrying the ITGAM variant, however, our sample size may be too small to identify such an association. Importantly, 4/6 patients had CVID comorbidities, including cancer and lung disease. Also of note, the majority (80%) had low switched-memory B-cells, compared to 60% of CVID patients without the ITGAM variant. This is interesting in the context of the possible association of ITGAM dysfunction and autoimmunity, since CVID patients with low switched-memory B-cells are at higher risk of autoimmune disease.
To determine if rare non-synonymous exonic variants might be tagged by the SNP microarray association signal at the 16p11.2 locus, we conducted Sanger sequencing of the three plausible candidate genes in this region. However, no corresponding rare variant non-synonymous mutation was found. Therefore, we hypothesize that gene regulation at the expression level of these proteins is the more likely genetic mechanism of perturbation in CVID, rather than direct amino acid change in a protein product. In addition to the two aforementioned SNPs, rs929867 and rs8056264, the genotyped SNP, rs4889637 and the imputed SNPs rs114798394, rs7202897, are predicted to alter binding motifs for STAT30, a component of the JAK-STAT signaling pathway that is critical for many key functions of the immune system82. Furthermore, amongst the imputed SNPs, binding factor YY1 at rs88955230 is required for early B cell development in mice83; and rs2098730 is located at the binding motif for NFκB30. Therefore the rare variants we found could be functional variants themselves contributing to the pathogenesis of CVID. A parallel example in this regard would be the SNP rs7903146 in intron 3 of TCF7L2 that is considered a plausible causal variant for type 2 diabetes84, 85. Furthermore, intronic variants in FUS, ITGAM, and ITGAX could impact alternative splicing, which would not be captured by our Sanger sequencing analysis. In view of the above, coupled with the finding that CVID individuals with the associated variant show a lower percentage of switched memory B-cells, our leading hypothesis is that the associated variants are impacting gene regulation and subsequent protein dosage. Alternatively, it is possible that more distal genes in linkage disequilibrium with the gene rich region of 16p11.2, or even more distal genes based on chromosome conformation in three dimensional space, are acting as enhancers through chromosome looping.
As in our previous GWAS,19 the MHC locus stands out at or close to the genome wide significance level and SNPs in the gene FLJ16124 were nominally significant in both studies. The unique SNP coverage in the iCHIP platform allowed identification of the ITGAM locus in this study, whereas the previously applied platform (HumanHap 610) had few SNPs in this region (53 in the region of chr16:31,100,000–31,500,000). Additionally, previously the focus was exclusively on common variants (MAF > 5%). Similarly, the nominally significant signals of other loci in our previous study only have sparse SNP coverage on the iCHIP (1–2 common variants at each locus) and therefore associations at those loci were not detected by this study.
In summary, our results suggests that the variants uncovered at the 16p11.2 locus involve CD11b and thus could impact many important pathways, such as TLR signaling, with modulating effects on immune responses, B-cell and T-cell activation and autoimmunity in CVID. This is supported by our pathway and network analyses, phenotype data and known knockout mouse phenotypes making ITGAM an exciting candidate gene for functional evaluation. Further understanding of the autoimmune phenomena in CVID would greatly impact care of these patients, since to date prediction models and treatments for these are scarce. Hopefully, further understanding of this pathway and its implications in CVID can improve the future care of these patients in the future.
Supplementary Material
Key messages.
We report results of an association analysis of CVID through dense genotyping of immune-related loci, the first of its kind in CVID.
We identified an association of CVID with rare variants at the FUS/ITGAM (CD11b) locus on 16p11.2 in a cohort of 360 CVID patients and 21,610 healthy controls.
The locus is enriched for promoter/enhancer histone markers, supporting that it may contribute to the pathogenesis of CVID.
ITGAM encodes the integrin CD11b, an important molecule for cellular contact and adhesion. Knockout mice models in the literature support the importance of ITGAM function for normal immune responses.
Acknowledgements
We gratefully thank all the participants enrolled in this study. We also thank the IBD consortium for access to the genotype information of the controls used in this study.
Funding: This work was supported by an Institute Development Fund from CHOP, U01HG006830 and a donation from the Kubert Estate Foundation
Abbreviations used
- CVID
Common Variable Immunodeficiency
- SNPs
Single Nucleotide Polymorphisms
- GWAS
Genome Wide Association Study
- iCHIP
Immuno bead-chip from Illumina
- PCA
Principal Component Analysis
- QC
Quality Control
- MAF
Minor Allele Frequency
- MHC
Major Histocompatability Cluster
- SLE
Systemic Lupus Erythematosus
- DC
Dendritic Cell
- APC
Antigen Presenting Cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None
References
- 1.Chapel H, Cunningham-Rundles C. Update in understanding common variable immunodeficiency disorders (CVIDs) and the management of patients with these conditions. Br J Haematol. 2009;145:709–727. doi: 10.1111/j.1365-2141.2009.07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92:34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 3.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111:77–85. doi: 10.1182/blood-2007-06-091744. [DOI] [PubMed] [Google Scholar]
- 4.Bacchelli C, Buckridge S, Thrasher AJ, Gaspar HB. Translational mini-review series on immunodeficiency: molecular defects in common variable immunodeficiency. Clin Exp Immunol. 2007;149:401–409. doi: 10.1111/j.1365-2249.2007.03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzer U, Chapel HM, Webster AD, Pan-Hammarstrom Q, Schmitt-Graeff A, Schlesier M, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37:820–828. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 6.Castigli E, Wilson SA, Garibyan L, Rachid R, Bonilla F, Schneider L, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 7.Pan-Hammarstrom Q, Salzer U, Du L, Bjorkander J, Cunningham-Rundles C, Nelson DL, et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat Genet. 2007;39:429–430. doi: 10.1038/ng0407-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olerup O, Smith CI, Bjorkander J, Hammarstrom L. Shared HLA class II-associated genetic susceptibility and resistance, related to the HLA-DQB1 gene, in IgA deficiency and common variable immunodeficiency. Proc Natl Acad Sci U S A. 1992;89:10653–10657. doi: 10.1073/pnas.89.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimbacher B, Hutloff A, Schlesier M, Glocker E, Warnatz K, Drager R, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4:261–268. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 10.Kanegane H, Agematsu K, Futatani T, Sira MM, Suga K, Sekiguchi T, et al. Novel mutations in a Japanese patient with CD19 deficiency. Genes Immun. 2007;8:663–670. doi: 10.1038/sj.gene.6364431. [DOI] [PubMed] [Google Scholar]
- 11.Kuijpers TW, Bende RJ, Baars PA, Grummels A, Derks IA, Dolman KM, et al. CD20 deficiency in humans results in impaired T cell-independent antibody responses. J Clin Invest. 2010;120:214–222. doi: 10.1172/JCI40231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzer U, Maul-Pavicic A, Cunningham-Rundles C, Urschel S, Belohradsky BH, Litzman J, et al. ICOS deficiency in patients with common variable immunodeficiency. Clin Immunol. 2004;113:234–240. doi: 10.1016/j.clim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 13.van Zelm MC, Reisli I, van der Burg M, Castano D, van Noesel CJ, van Tol MJ, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354:1901–1912. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 14.van Zelm MC, Smet J, Adams B, Mascart F, Schandene L, Janssen F, et al. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J Clin Invest. 2010;120:1265–1274. doi: 10.1172/JCI39748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ombrello MJ, Remmers EF, Sun G, Freeman AF, Datta S, Torabi-Parizi P, et al. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N Engl J Med. 2012;366:330–338. doi: 10.1056/NEJMoa1102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekine H, Ferreira RC, Pan-Hammarstrom Q, Graham RR, Ziemba B, de Vries SS, et al. Role for Msh5 in the regulation of Ig class switch recombination. Proc Natl Acad Sci U S A. 2007;104:7193–7198. doi: 10.1073/pnas.0700815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warnatz K, Salzer U, Rizzi M, Fischer B, Gutenberger S, Bohm J, et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106:13945–13950. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, Coonrod EM, Kumanovics A, Franks ZF, Durtschi JD, Margraf RL, et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. Am J Hum Genet. 2013;93:812–824. doi: 10.1016/j.ajhg.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. 2011;127:1360–1367. e6. doi: 10.1016/j.jaci.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volanakis JE, Zhu ZB, Schaffer FM, Macon KJ, Palermos J, Barger BO, et al. Major histocompatibility complex class III genes and susceptibility to immunoglobulin A deficiency and common variable immunodeficiency. J Clin Invest. 1992;89:1914–1922. doi: 10.1172/JCI115797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 28.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 30.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–1116. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt T, Ogris C, Sonnhammer EL. FunCoup 3.0: database of genome-wide functional coupling networks. Nucleic Acids Res. 2014;42:D380–D388. doi: 10.1093/nar/gkt984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao K, Zheng W, Deng X, Xiong W, Song Z, Yang Y, et al. Genetic analysis of the fused in sarcoma gene in Chinese Han patients with Parkinson's disease. Parkinsonism Relat Disord. 2013 doi: 10.1016/j.parkreldis.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Shelkovnikova TA, Robinson HK, Connor-Robson N, Buchman VL. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle. 2013;12:3194–3202. doi: 10.4161/cc.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iguchi Y, Katsuno M, Ikenaka K, Ishigaki S, Sobue G. Amyotrophic lateral sclerosis: an update on recent genetic insights. J Neurol. 2013;260:2917–2927. doi: 10.1007/s00415-013-7112-y. [DOI] [PubMed] [Google Scholar]
- 43.Baron DM, Kaushansky LJ, Ward CL, Sama RR, Chian RJ, Boggio KJ, et al. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener. 2013;8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, et al. Before It Gets Started: Regulating Translation at the 5' UTR. Comp Funct Genomics. 2012;2012:475–731. doi: 10.1155/2012/475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi CF, Martensson A, Mattioli M, Dalla-Favera R, Lobanenkov VV, Morse HC., 3rd CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc Natl Acad Sci U S A. 2003;100:633–638. doi: 10.1073/pnas.0237127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 48.Goymer P. Synonymous mutations break their silence. Nature Reviews Genetics. 2007;8 [Google Scholar]
- 49.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 50.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai Z, Turtle CJ, Booth GC, Riddell SR, Gooley TA, Stevens AM, et al. Normally occurring NKG2D+CD4+ T cells are immunosuppressive and inversely correlated with disease activity in juvenile-onset lupus. J Exp Med. 2009;206:793–805. doi: 10.1084/jem.20081648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green MR, Kennell AS, Larche MJ, Seifert MH, Isenberg DA, Salaman MR. Natural killer T cells in families of patients with systemic lupus erythematosus: their possible role in regulation of IGG production. Arthritis Rheum. 2007;56:303–310. doi: 10.1002/art.22326. [DOI] [PubMed] [Google Scholar]
- 53.Griffin DO, Rothstein TL. A small CD11b(+) human B1 cell subpopulation stimulates T cells and is expanded in lupus. J Exp Med. 2011;208:2591–2598. doi: 10.1084/jem.20110978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner C, Hansch GM, Stegmaier S, Denefleh B, Hug F, Schoels M. The complement receptor 3, CR3 (CD11b/CD18), on T lymphocytes: activation-dependent up-regulation and regulatory function. Eur J Immunol. 2001;31:1173–1180. doi: 10.1002/1521-4141(200104)31:4<1173::aid-immu1173>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 55.Reed JH, Jain M, Lee K, Kandimalla ER, Faridi MH, Buyon JP, et al. Complement receptor 3 influences toll-like receptor 7/8-dependent inflammation: implications for autoimmune diseases characterized by antibody reactivity to ribonucleoproteins. J Biol Chem. 2013;288:9077–9083. doi: 10.1074/jbc.M112.403303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Escobar D, Pons J, Clemente A, Iglesias J, Regueiro V, Bengoechea JA, et al. Defective B cell response to TLR9 ligand (CpG-ODN), Streptococcus pneumoniae and Haemophilus influenzae extracts in common variable immunodeficiency patients. Cell Immunol. 2010;262:105–111. doi: 10.1016/j.cellimm.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 57.Yu JE, Knight AK, Radigan L, Marron TU, Zhang L, Sanchez-Ramon S, et al. Toll-like receptor 7 and 9 defects in common variable immunodeficiency. J Allergy Clin Immunol. 2009;124:349–356. 56 e1–56 e3. doi: 10.1016/j.jaci.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao C, Gao Y, Li Y, Antalis TM, Castellino FJ, Zhang L. The efficacy of activated protein C in murine endotoxemia is dependent on integrin CD11b. J Clin Invest. 2010;120:1971–1980. doi: 10.1172/JCI40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fossati-Jimack L, Ling GS, Cortini A, Szajna M, Malik TH, McDonald JU, et al. Phagocytosis is the main CR3-mediated function affected by the lupus-associated variant of CD11b in human myeloid cells. PLoS One. 2013;8:e57082. doi: 10.1371/journal.pone.0057082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajishengallis G, Harokopakis E. Porphyromonas gingivalis interactions with complement receptor 3 (CR3): innate immunity or immune evasion? Front Biosci. 2007;12:4547–4557. doi: 10.2741/2409. [DOI] [PubMed] [Google Scholar]
- 61.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11:734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 62.Huynh L, Wang L, Shi C, Park-Min KH, Ivashkiv LB. ITAM-coupled receptors inhibit IFNAR signaling and alter macrophage responses to TLR4 and Listeria monocytogenes. J Immunol. 2012;188:3447–3457. doi: 10.4049/jimmunol.1102211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marth T, Kelsall BL. Regulation of interleukin-12 by complement receptor 3 signaling. J Exp Med. 1997;185:1987–1995. doi: 10.1084/jem.185.11.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida Y, Kang K, Berger M, Chen G, Gilliam AC, Moser A, et al. Monocyte induction of IL-10 and down-regulation of IL-12 by iC3b deposited in ultraviolet-exposed human skin. J Immunol. 1998;161:5873–5879. [PubMed] [Google Scholar]
- 65.Zhou Y, Wu J, Kucik DF, White NB, Redden DT, Szalai AJ, et al. Multiple Lupus-Associated ITGAM Variants Alter Mac-1 Functions on Neutrophils. Arthritis Rheum. 2013;65:2907–2916. doi: 10.1002/art.38117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhodes B, Furnrohr BG, Roberts AL, Tzircotis G, Schett G, Spector TD, et al. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann Rheum Dis. 2012;71:2028–2034. doi: 10.1136/annrheumdis-2012-201390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, et al. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Hum Mol Genet. 2009;18:1171–1180. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 69.International Consortium for Systemic Lupus Erythematosus, G. Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 71.Yang W, Zhao M, Hirankarn N, Lau CS, Mok CC, Chan TM, et al. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum Mol Genet. 2009;18:2063–2070. doi: 10.1093/hmg/ddp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosetti F, Tsuboi N, Chen K, Nishi H, Ernandez T, Sethi S, et al. Human lupus serum induces neutrophil-mediated organ damage in mice that is enabled by Mac-1 deficiency. J Immunol. 2012;189:3714–3723. doi: 10.4049/jimmunol.1201594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anaya JM, Kim-Howard X, Prahalad S, Chernavsky A, Canas C, Rojas-Villarraga A, et al. Evaluation of genetic association between an ITGAM non-synonymous SNP (rs1143679) and multiple autoimmune diseases. Autoimmun Rev. 2012;11:276–280. doi: 10.1016/j.autrev.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.MacPherson M, Lek HS, Prescott A, Fagerholm SC. A systemic lupus erythematosus-associated R77H substitution in the CD11b chain of the Mac-1 integrin compromises leukocyte adhesion and phagocytosis. J Biol Chem. 2011;286:17303–17310. doi: 10.1074/jbc.M110.182998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bai Y, Qian C, Qian L, Ma F, Hou J, Chen Y, et al. Integrin CD11b negatively regulates TLR9-triggered dendritic cell cross-priming by upregulating microRNA-146a. J Immunol. 2012;188:5293–5302. doi: 10.4049/jimmunol.1102371. [DOI] [PubMed] [Google Scholar]
- 76.Skoberne M, Somersan S, Almodovar W, Truong T, Petrova K, Henson PM, et al. The apoptotic-cell receptor CR3, but not alphavbeta5, is a regulator of human dendritic-cell immunostimulatory function. Blood. 2006;108:947–955. doi: 10.1182/blood-2005-12-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Gisbergen KP, Ludwig IS, Geijtenbeek TB, van Kooyk Y. Interactions of DC-SIGN with Mac-1 and CEACAM1 regulate contact between dendritic cells and neutrophils. FEBS Lett. 2005;579:6159–6168. doi: 10.1016/j.febslet.2005.09.089. [DOI] [PubMed] [Google Scholar]
- 78.van Gisbergen KP, Sanchez-Hernandez M, Geijtenbeek TB, van Kooyk Y. Neutrophils mediate immune modulation of dendritic cells through glycosylation-dependent interactions between Mac-1 and DC-SIGN. J Exp Med. 2005;201:1281–1292. doi: 10.1084/jem.20041276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coustet B, Agarwal SK, Gourh P, Guedj M, Mayes MD, Dieude P, et al. Association study of ITGAM, ITGAX, and CD58 autoimmune risk loci in systemic sclerosis: results from 2 large European Caucasian cohorts. J Rheumatol. 2011;38:1033–1038. doi: 10.3899/jrheum.101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diamond MS, Garcia-Aguilar J, Bickford JK, Corbi AL, Springer TA. The I domain is a major recognition site on the leukocyte integrin Mac-1 (CD11b/CD18) for four distinct adhesion ligands. J Cell Biol. 1993;120:1031–1043. doi: 10.1083/jcb.120.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sachs UJ, Chavakis T, Fung L, Lohrenz A, Bux J, Reil A, et al. Human alloantibody anti-Mart interferes with Mac-1-dependent leukocyte adhesion. Blood. 2004;104:727–734. doi: 10.1182/blood-2003-11-3809. [DOI] [PubMed] [Google Scholar]
- 82.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 83.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palmer ND, Hester JM, An SS, Adeyemo A, Rotimi C, Langefeld CD, et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes. 2011;60:662–668. doi: 10.2337/db10-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wellcome Trust Case Control, C. Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.