Abstract
We have recently identified a new gene, involved in DNA replication, at the far 3′ end of the adeno-associated virus type 2 (AAV2) genome. The AAV type 6 (AAV6) genome has a disrupted X open reading frame (ORF) whose two halves, when combined, have full-length homology and comparable size to AAV2 X. Hypothesizing that AAV6 X is inactive, we assessed if AAV2 X augments recombinant (r)AAV2 DNA replication and virion production, but with rep and cap trans-functions of AAV6. Using AAV2 X expressing HEK293 cell lines we show AAV2 X significantly boosts rAAV DNA replication/virion production, driven by AAV6 rep/cap as it does the AAV2 rep/cap system. Protein BLAST search for homology between AAV2 X and various AAV Rep78 proteins suggests that X might be AAV8 Rep78-derived and have some of its activities. These data suggest that AAV2 X, and the corresponding X genes of other AAV types/clades, warrant further study.
Introduction
Now over 100 adeno-associated virus (AAV) types have been isolated. While AAV type 2 (AAV2) was the first AAV type used for gene transfer (Hermonat, 1984, 2014; Hermonat and Muzyczka, 1984; Tratschin et al., 1984A), over time more and more AAV types, each with its own somewhat different cellular tropisms, are coming into use. In general these other AAV types have the same genomic structure as AAV2 (Gao et al, 2005; Srivastava et al., 1983). Analysis of the first cloned adeno-associated virus AAV type 2 (AAV2) genome showed that there were two main open reading frames (ORFs) and mutation within the identified ORFs indicated three trans phenotypes were present (Hermonat, et al., 1984, Tratschin et al., 1984B). Mutations in the left half of the genome were defective in DNA replication and transcription and given the rep phenotype. This region encodes replication / transcription factor proteins Rep78, Rep68, Rep52, and Rep40. Mutations within the right half of the genome were defective in wild type virion production, but the region had two phenotypes. One was given the name lip for the production of viral particles of low infectivity (missing VP1)(also described as inf), while the cap phenotype didn’t produce any viral particles at all (encoding the major structural protein, VP3)(Hermonat et al., 1984; Tratschin et al., 1984B). Additionally, recently, a new fourth trans phenotype, involved in virion maturation, has been identified by Jurgen Kleinschmidt and called the AAP gene (Sonntag et al, 2010).
Recently we discovered a fifth phenotype, a new gene we called X (GenBank KM186843.1), within the AAV2 genome (Cao et al., 2014). The X gene is located at the carboxy-end of the cap gene but in a different translational frame. We have shown that X is needed for maximal wt AAV2 and rAAV2 DNA replication and virion production by several methods. The X gene also has a dedicated promoter located just upstream, called p81 (at map unit 81)(Hermonat et al., 1999). However, the question arises is AAV2 X activity only specific for helping/augmenting AAV2, or is it capable of helping other AAV types? Most other AAV clades also have members with an open reading frame (ORF) in the same position as AAV2 X, but these potential genes are usually smaller than AAV2 X. Other AAVs may have mutated X genes, such as in AAV6 there are two ORFs, divided by a few bases, which take up the position analogous to where the AAV2 X gene is. Here we observed that AAV2 X is able to augment or boost an rAAV production system based exclusively on the AAV6 rep and cap, trans sequences and we find that X is capable of increasing rAAV2 DNA replication and virion production when driven by the AAV6 rep and cap genes. Additionally, we hypothesize that AAV2 X may be derived from a 5′ region of the AAV Rep78/NS1 gene.
RESULTS
AAV6 genome contains an X gene but which is divided into two abutting ORFs
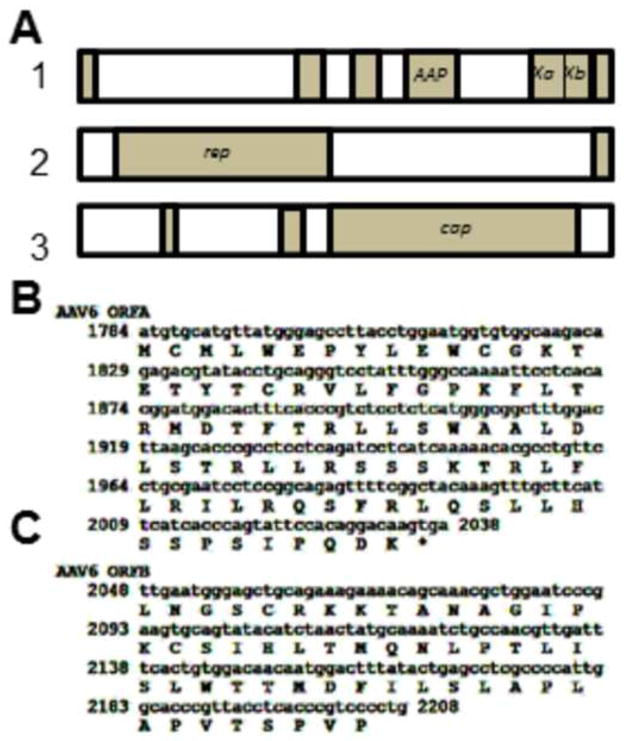
If one observes the open reading frames of the prototype AAV6 genome (Genbank AF028704) it is observed that there are two ORFs, which we refer to as Xa and Xb, which take up the position analogous to where the AAV2 X gene is. There is a small gap between the stop codon of Xa and the initiation codon of Xb. However, analyzing two other AAV6 sequences, specifically Genbank EU368909 and EU36910, there is an even smaller gap between Xa and Xb of only 13 nucleotides, and the Xb ORF encodes a further 22 amino acids (aa) at its amino terminus. Figure 1A shows the gene/ORF organization of AAV6 using largely the AF028704 prototype sequences, but with the X region of EU368909 replacing the analogous sequences of the prototype. Figure 1B and 1C show the DNA and amino acid sequences of Xa and Xb. Figure 2 is a homology analysis by standard NCBI Protein BLAST of the amino acid sequence of AAV2 X versus those of the fused Xa and Xb aa of EU368909. As can be seen there is significant homology between the two X sequences across their length. This extensive homology suggests that AAV6 Xa-b is a homologue of AAV2 X and it has either evolved or mutated at some point in time. Presently, it is unknown if AAV6 Xa and Xb represent two potentially functional proteins or are fully inactive and “broken”. In any case AAV6 appears to have or have had a very AAV2 “X”-like protein(s).
Figure 1. The AAV6 genome showing Xa and Xb.
Shown in A are the ORFs of AAV6 by NIH ORF finder and derived from the Genbank AF028704, but with the AAV6 X region replaced with sequences from EU368909. Note that there are two open reading frames, Xa and Xb, present in the position occupied by AAV2 X. B shows the DNA and amino acid sequences of Xa. C shows the DNA and amino acid sequences of Xb.
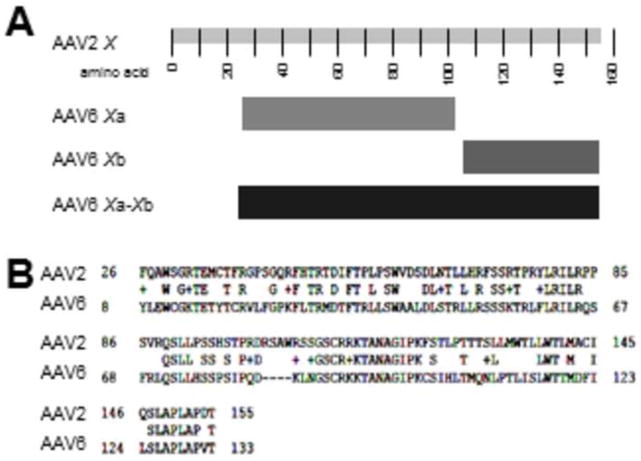
Figure 2. Homology of AAV2 X with AAV6 Xa and Xb.
Shown in A are a more detailed caricature of the X ORFs of AAV6 by NIH ORF finder derived from the Genbank AF028704 plus EU368909. In B is shown an NCBI Protein BLAST analysis of the artificially fused AAV6 Xa-Xb aa sequence (normally two separate ORFs) with that of AAV2 X. Note that the homology of the two X sequences extends the length of fused AAV6 Xa-Xb.
AAV2 X helps rAAV2/6-eGFP DNA replication and virion production
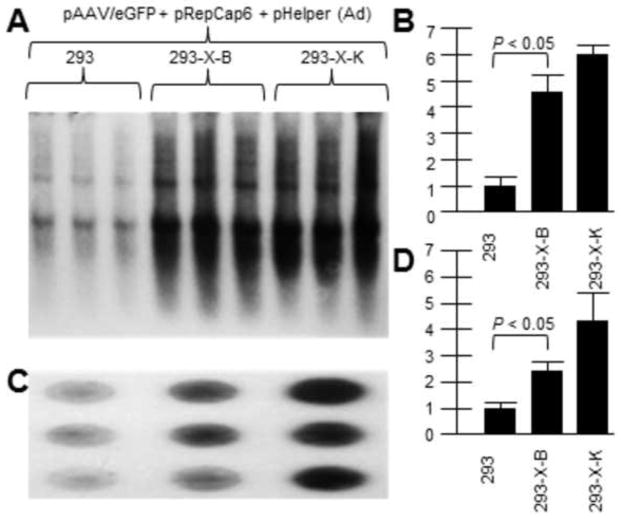
As we know that AAV2 X increases rAAV2 yield, and AAV6 X may be non-functional, we investigated whether AAV2 X might complement AAV6 rep/cap driven rAAV production. Previously we generated HEK293 cell lines containing chromosomal AAV2 X (293-X-B and 293-X-K) and we compared them to parental HEK293 cells for supporting rAAV2 DNA replication and virus production. Shown in Figure 3A is a Southern blot of rAAV2/eGFP DNA replication (probed with32P-eGFP DNA) by transfecting the vector plasmid with AAV6repcap and pHelper (Ad5 helper genes) plasmids. Figure 3B shows a dot blot of DNaseI-resistant virion DNA which shows higher rAAV production in X-positive 293-X-B and 293-X-K than in unaltered 293 cells. Moreover the higher virion production mirrors the higher vector DNA replication levels. As can be seen, the presence of the AAV2 X gene within the B and K cell lines was able to boost rAAV production in based on the AAV6 rep and cap proteins as it did for rAAV based on AAV2 rep and cap driving vector production.
Figure 3. AAV2 X helps rAAV/eGFP DNA and virion production driven by AAV6 rep/cap.
Here we demonstrate that AAV2 X also helps an AAV6-based rep/cap system. We used pRepCap6 which has both the AAV6 rep and cap genes. 293-X-B and 293-X-K are cell lines which contain the X gene. A shows a Southern blot of pAAV/eGFP DNA replication (probed with 32P-eGFP DNA.. B is a densitometric quantification of A. Note that in the presence of AAV2 X, included In 293-X-B and 293-X-K, the level of AAV/eGFP DNA replication was significantly higher compared to 293 cells. C shows a dot blot of DNaseI-resistant pAAV/eGFP virion DNA probed with 32P-eGFP DNA.. D is a densitometric quantification of C. Note that in the presence of AAV2 X, included In 293-X-B and 293-X-K, the level of AAV/eGFP virion DNA was significantly higher compared to 293 cells.
AAV2 X helps rAAV2/6-Foxp3 DNA replication and virion production
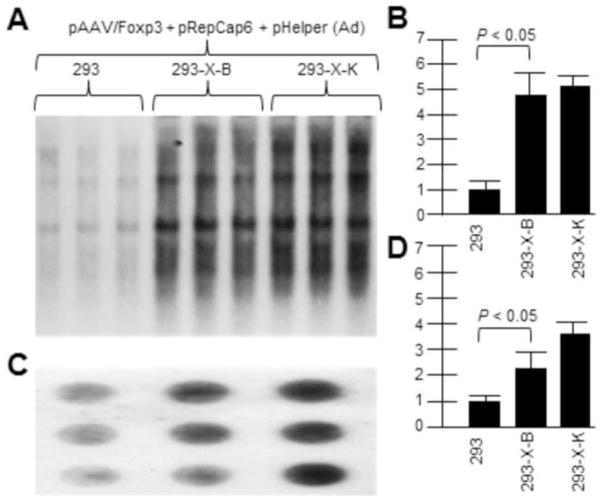
Similar experiments were done with the vector rAAV2/Foxp3 in place of AAV2/eGFP. Figure 4A shows a Southern blot of rAAV2/Foxp3 DNA replication (probed with32P-Foxp3 DNA) by transfecting the vector plasmid with AAV6repcap and pHelper (Ad5 helper genes) plasmids. Figure 4B then shows a dot blot of DNaseI-resistant virion DNA which shows higher rAAV production in X-positive 293-X-B and 293-X-K than in unaltered 293 cells. Again, as with the AAV2/eGFP vector, the presence of AAV2 X in the 293 cells, in conjunction with AAV6 rep and cap proteins, boosted vector rAAV2/Foxp3 DNA replication and virion production as it did for rAAV driven by AAV2 rep and cap.
Figure 4. AAV2 X helps rAAV/Foxp3 DNA and virion production driven by AAV6 rep/cap.
Here we demonstrate that AAV2 X also helps an AAV6-based rep/cap system. We used pRepCap6 which has both the AAV6 rep and cap genes. 293-X-B and 293-X-K are cell lines which contain the X gene. A shows a Southern blot of pAAV/Foxp3 DNA replication (probed with 32P-Foxp3 DNA.. B is a densitometric quantification of A. Note that in the presence of AAV2 X, included In 293-X-B and 293-X-K, the level of AAV/Foxp3 DNA replication was significantly higher compared to 293 cells. C shows a dot blot of DNaseI-resistant pAAV/Foxp3 virion DNA probed with 32P-Foxp3 DNA.. D is a densitometric quantification of C. Note that in the presence of AAV2 X, included In 293-X-B and 293-X-K, the level of AAV/Foxp3 virion DNA was significantly higher compared to 293 cells.
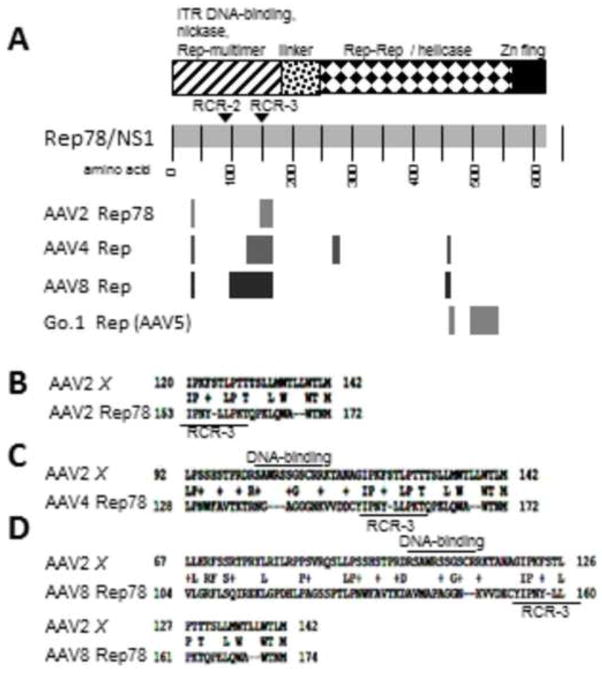
AAV2 X has homology to the Rep78 proteins of various AAVs
It was noticed during various NCBI Protein Blast searches that X showed homology with Rep78/NS1 of AAV2, but also other AAVs as well. Therefore in Figure 5 A, B, C, and D we show the results of homology analyses with AAV2, AAV4, AAV8 and Go.1. The largest region of homology is seen between AAV8 Rep78 and AAV2 X. It can be seen that most homology with X lies in a region from about aa100–200 of the Rep78 protein. However the results were quite different for Go.1, where homology with X was seen at the extreme carboxy-terminus. Clearly this finding is consistent with AAV2 X being involved in AAV DNA replication.
Figure 5. Homology between AAV2 X and various Rep78s.
Shown in A is a map of AAV2 X homologies against the full length Rep78 protein. However, for Go.1, related to AAV5, the homology resides at the extreme carboxy-terminus of Rep78, different from the other AAVs. B, C, and D are NCBI Protein BLAST homology analyses between AAV2 X and AAV type 2, 4, 8, and Go.1 Rep78/NS1 proteins. Note that for most Rep78s the homology with X lies in a region from 100–200 aa. These homologies suggest that Rep78 DNA sequences may have exchanged material with the 3′ end of the AAV genomes. AAV8 may be the most likely source due to its longest length of homology. The putative DNA/RNA binding domain of X and the RCR3 domain of Rep78 are marked.
Discussion
The AAV2 the X ORF (Hermonat et al,1999; Cao et al, 2014) has been largely ignored for over the thirty years since its discovery during the first sequencing of the AAV2 genome (Srivastava et al, 1983). Here, it is demonstrated that the augmentation effect shown for AAV2 X on wild type (wt) and recombinant (r)AAV DNA replication and virion production driven by the AAV2 rep and cap genes/proteins (Hermonat et al, 1984; Hermonat and Muzyczka, 1984), also extends to rAAV these same functions driven by the AAV6 rep and cap proteins. As mentioned earlier, it was observed that there are two AAV6 ORFs, referred to as Xa and Xb, which take up the position analogous to where the AAV2 X gene is. The augmentation of rAAV6-driven functions would seem consistent with AAV6 X being either inactive, or having significantly reduced or different activities than AAV2 X. The possible AAV6 X protein (combined ORFA and B) does have 52% amino acid (aa) homology across its length with AAV2X.
Coupled with the issue of a “broken” AAV6 X gene is a major unanswered question; what is the accuracy of the nucleotide sequence at the 3′ end of the cap gene? This question is not only important for AAV6, but of many of the other AAV types which are less vetted than the AAV2 sequence. The AAV2 sequence has undergone many nucleotide corrections. These nucleotide sequence errors result in altered cap (capsid) encoded VP proteins. In fact, VP1-3 can sometimes be found to have homology with X (National Center for Biotechnology Information [NCBI] Protein BLAST analysis). As X is fully contained within the cap gene a single base addition or subtraction might/fuse or splice X coding sequences into the capsid protein and visa versa. Only continued vetting of AAV genomic sequences can solve this issue.
More certain than “homology” with cap is the homology which can be found between AAV2 X and the rep (replication) encoded protein, Rep78 by Protein BLAST. Selected X-to-Rep78 homologies are shown in Figure 5. As can be seen Figures 5A-C shows Protein BLAST homology with AAV serotypes 2, 4, and 8 Rep78s (NC_001401.2, NC_001829.1, NC_006261.1, respectively). These homologies are present within the amino third of Rep78. AAV8 Rep78 has the most extensive homology, over a 70 aa region, or about half of AAV2 X. These X homologies are shown against the background of a generalized Rep78 protein in Figure 5A. Drawing directly from these homologies, AAV2 X may have been originally derived by some type of non-homologous recombinant exchange from the 5′ half of the rep region and the 3′ portion of the cap region of AAV, possibly to have taken place between the rep gene of AAV8 and the cap gene of AAV2. Moreover, this same region of Rep78, taken from several AAV types, are found to have some homology to AAV2 X (Figure 5).
As Rep78 is a well-studied protein this homology may give suggestions as to what X might do functionally. The Rep78 nickase/helicase requires two Rep-Rep binding sites for hexameric-association on DNA, one in the amino-terminal region (in Rep78/Rep68) and another within the carboxy-two thirds (in all four Rep) (Hermonat and Batchu, 1997; Smith et al., 1997; Zarate-Perez et al., 2012). That X contains significant homology to the amino-hexamer-DNA binding domain of Rep78 (Figure 5) suggests the possibility that X might bind to itself or to Rep78 in the presence of DNA (or without?). A putative DNA binding domain has been identified in X in the region of aa 100–113 by multiple protein-function prediction software (eg. Protein Predict) (Figure 5). Additionally, if X were also able to associate with Rep52/Rep40 the hypothetical X-Rep52 heterodimers might better resemble full length Rep78 and might reconstitute additional biochemistries of that of the full length proteins Rep78/Rep68 proteins. However, likely X cannot augment endonuclease activity as it is deficient in the needed tyrosines residues involved in function (Walker et al., 1997). Yet, many interesting possibilities exist. For example, might X interact with Rep78-ITR complexes and modulate their activities. Additionally, this region of X has homology with the rolling circle replication region 3 (RCR3) of Rep78 which is believed to be involved with cutting and ligating single stranded inverted terminal repeat DNA (Figure 5) (Smith and Kotin, 2000). This is very speculative as the homology with RCR3 is weak. However, if there is a partial activity for this putative RCR of X, it might be involved in the generation of head-to-tail proviral concatemers often seen in integrated AAV provirus (Cheung et al., 1980; Hermonat, 1984; McLaughlin et al., 1988). In any case, these data suggest that AAV2 X is a prototype gene/protein of a type which is significant beyond AAV type 2.
Materials and Methods
Virus and Cells
HEK293 cells were maintained in Dulbecco’s modification of Eagle’s medium with 7% fetal bovine serum and antibiotics. The HEK293 cell lines, 293-X-B and 293-X-K cell lines have been described previously (Cao et al., 2014). AAV/eGFP was generated by ligating the enhanced green fluorescent protein (eGFP) coding sequence into the Xho I site just behind the CMV promoter in dl3-97/CMV. AAV/Foxp3 was generated in a similar manner.
Analysis of AAV2 DNA replication in 293 cells
HEK293, 293-X-B, or 293-X-K cells (6cm plates) at 70% confluence were transfected with 1 μg of the indicated vector plasmid (AAV/eGFP or AAV/Foxp3 plus 1 μg pRepCap6 plus 1 μg of pHelper(Ad) using TransIT according to manufacture’s instructions. For DNA replication analysis cells were lysed with 1.5 ml of 1% SDS, 7.2 pH Tris-HCL, 5mM EDTA, and Pronase K and incubated overnight. The total cellular DNA was then drawn though a 20 gauge needle ten times, phenol extracted, ethanol precipitated twice, and 10 μgs of DNA were agarose gel electrophoresed, Southern blotted and probed with the indicated 32P-labeled DNA probe. After autoradiography densitometric analysis was carried out using the Alpha Imager 2000 with resident software (Alpha Innotech Corporation, San Leandro, CA).
Virion DNA analysis
Six cm plates of transfected HEK 293, 293-X-B and 293-X-K cells, were treated as in the analysis of DNA replication. After three days cells were freeze-thawed three times, cellular debris pelleted by centrifugation at 7,000 rpm for 25 minutes, and the supernatant pushed through a 0.22 μm filter. Three hundred μl of virus stock was treated with 20 units DNase I for 30 minutes at 37°C. After heating the sample for 10 minutes at 100°C, the sample was digested with proteinase K (0.2 μg/ml) for 4 hrs, then phenol extracted and ethanol precipitated (with addition of 10 μg tRNA). The resulting DNA was then dotted blotted onto a nylon membrane and probed with either 32P-eGFP or 32P-Foxp3 DNA, as appropriate, when analyzing for rAAV production.
The AAV6 X gene appears as a disrupted open reading frame (ORF).
The two halves of AAV6 X ORF, when fused, have moderate amino acid homology to AAV2 X across their length.
AAV2 X, expressed in 293-derived cell lines, increases AAV6 rep/cap–driven rAAV production.
AAV2X displays homology to various Rep78s, particularly to that of AAV8.
The X gene of AAV2, and others, warrant further study.
Acknowledgments
This study was funded by a Merit Review award from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development Service and by NIH R56 AI093695 grants to PLH. We thank David Russell for the pRepCap6 plasmid. The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cao M, You H, Hermonat PL. The X gene of adeno-associated virus 2 (AAV2) is involved in viral DNA replication. PLoS ONE. 2014;9:e104596. doi: 10.1371/journal.pone.0104596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AK, Hoggan MD, Hauswirth WW, Berns KI. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5(3):285–97. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- Hermonat PL. Genetic analysis and utilization of adeno-associated virus as a mammalian cloning vector. Univ FL Dissertation. 1984 doi: 10.1073/pnas.81.20.6466. Available at: http://archive.org/details/geneticanalysisu00herm. [DOI] [PMC free article] [PubMed]
- Hermonat PL. First AAV gene transfer experiment, 1983. (2014) Human Gene Therapy. 2014;25:486–487. doi: 10.1089/hum.2014.047. [DOI] [PubMed] [Google Scholar]
- Hermonat PL, Batchu RB. The adeno-associated virus Rep78 major regulatory protein forms multimeric complexes and the domain for this activity is contained within the carboxy-half of the molecule. FEBS Lett. 1997;401(2–3):180–184. doi: 10.1016/s0014-5793(96)01469-x. [DOI] [PubMed] [Google Scholar]
- Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermonat PL, Labow MA, Wright R, Berns KI, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of mutants in adeno-associated virus type 2. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermonat PL, Santin AD, De Greve J, De Rijcke M, Bishop BM, Han L, Mane M, Kokorina N. Chromosomal latency and expression at map unit 96 of a wild-type plus adeno-associated virus (AAV)/Neo vector and identification of p81, a new AAV transcriptional promoter. J Hum Virol. 1999;2(6):359–68. [PubMed] [Google Scholar]
- Kleinschmidt Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SK, Collis P, Hermonat PL, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RH, Spano AJ, Kotin RM. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71(6):4461–71. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RH, Kotin RM. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J Virol. 2000;74(7):3122–9. doi: 10.1128/jvi.74.7.3122-3129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Lusby EW, Berns KI. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratschin JD, West MH, Sandbank T, Carter BJ. A) A human parvovirus, adeno-associated virus, as a eucaryotic vector: transient expression and encapsidation of the procaryotic gene for chloramphenicol acetyltransferase. Mol Cell Biol. 1984;4(10):2072–81. doi: 10.1128/mcb.4.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratschin JD, Miller IL, Carter BJ. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984B;51(3):611–9. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SL, Wonderling RS, Owens RA. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J Virol. 1997;71(4):2722–2730. doi: 10.1128/jvi.71.4.2722-2730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate-Perez F, Bardelli M, Burgner JW, 2nd, Villamil-Jarauta M, Das K, Kekilli D, Mansilla-Soto J, Linden RM, Escalante CR. The interdomain linker of AAV-2 Rep68 is an integral part of its oligomerization domain: role of a conserved SF3 helicase residue in oligomerization. PLoS Pathog 2012. 2012;8(6):e1002764. doi: 10.1371/journal.ppat.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]