Summary
Here we look at modern developmental biology with a focus on the relationship between different approaches of investigation. We argue that direct imaging is a powerful approach not only for obtaining descriptive information but also for model generation and testing that lead to mechanistic insights. Modeling, on the other hand, conceptualizes imaging data and provides guidance to perturbations. The inquiry progresses most efficiently when a trinity of approaches—quantitative imaging (measurement), modeling (theory) and perturbation (test) —are pursued in concert, but not when one approach is dominant. Using recent studies of the zebrafish system, we show how this combination has effectively advanced classic topics in developmental biology compared to a perturbation-centric approach. Finally, we show that interdisciplinary expertise and perhaps specialization are necessary for carrying out a systematic approach, and discuss the technical hurdles.
Introduction
Development of multicellular organisms is a constructive process that relies on integration of interactions across scales. These interactions translate a subtle molecular detail such as a point mutation to a global level phenotype through time. For example, a changed gene product interacts with its cellular partners differently, altering the pathways it participates in. These changes are integrated to modify the behavior of the cell (e.g., reducing the cell’s mobility). The affected cells interact with each other to produce a different collective process (e.g., slowed-down migration of an epithelial sheet). Finally, when the process meets other processes, a developmental phenotype is produced (e.g., a mis-folded tube). While this description sounds quite understandable, the reality is a lot more difficult. The first reason is the sheer number parameters: the “interactions” involve thousands to millions of cells each with their own unique molecular profiles and physical states at micro spatial temporal scales, many of which are either inaccessible by current methods or poorly characterized. Second, along the course of development, evolution has designed multiple regulations and redundancies but also left potentially “odd” relics of its history. Investigators today are looking at a historical complexity, like a several thousand year old city that has been through many rounds of destruction and rebuilding. The difference of this between a well planned de novo city is that it is not based on a rational master plan for accomplishing the city’s current goals, but rather reflects the changing technologies and goals of that city over time. When we ask how development works, we may be confused by the robustness caused by regulation and redundancy. When we ask why development works the way it does, we may be confused because of the historical path from which life evolved.
Are we ever going to understand development then? We likely never have a super microscope to measure all parameters, nor a time machine to watch how developmental processes evolved. But weaker versions of these: modern imaging techniques and imaginative reasoning of how systems interact and were evolved (in other words, modeling), do exist now and bring promise and hope. Systems biology - the arising interdisciplinary field that aims to understand the complexity of dynamic interactions in living systems1,2 - is the ideal field to marry with developmental biology for new conceptual and methodological frameworks. Here interactions across molecular, cellular, tissue and organismal levels are considered together as an integrated whole3. Previously, sitting on the edge of technical limitations, scientists have been speculating and theorizing a lot more beyond available data. Now a new, opposite situation appears thanks to imaging and -omics approaches: having mountains of quantifiable data but not a known framework of theoretic abstraction. Because of this wealth of data, developmental biology is no longer “the last refuge for the mathematically incompetent scientist”4 and becomes approachable using systems thinking. Concepts such as design principles, noise, regulatory networks, and robustness drawn from a range of disciplines to describe biochemical networks are now also being applied to describe developmental processes5. This trend towards systematic models of development is inevitable, as the progress of stem cell sciences and regenerative medicine will ultimately rely on the principles of development to deliver useful applications. Such engineering is impractical (or severely limited, at best) without foundational theory. The quantitative revolution of the field of development will become more evident in the time to come6. Here we explore why and how imaging is one of the major forces driving the transformation and its relationship with modeling and perturbation, review some of the recent works that give the “systems developmental biology” field a solid start, and consider its likely productive future directions.
Under-recognized role of imaging in model generation and testing
Direct observation arguably remains the most powerful approach in biology. The breakthroughs of imaging techniques are often associated with a boost of progress in biological and medical sciences. Unfortunately, careful imaging is often dismissively considered as “descriptive” at best whereas perturbation based approaches are automatically considered more “mechanistic”. The question of how mechanistic an approach is should be addressed by asking how much better a process is understood after the approach is done rather than what approach was used. For example, many genetic perturbations lead to a conclusion “gene X is required for outcome Y” without actually explaining how outcome Y happened. We argue that direct observation not only leads to more comprehensive and accurate description of the subjects and processes, but also allows formulation and testing of specific insightful hypotheses7. On the other hand, perturbations that change one component at a time in a black box are a good way to identify the players but have limited power in unraveling how the game is actually played. The true power of perturbation comes after a specific, testable model is formulated. Overly relying on either imaging or perturbation alone is going to not only be less productive but also may in some cases be misleading.
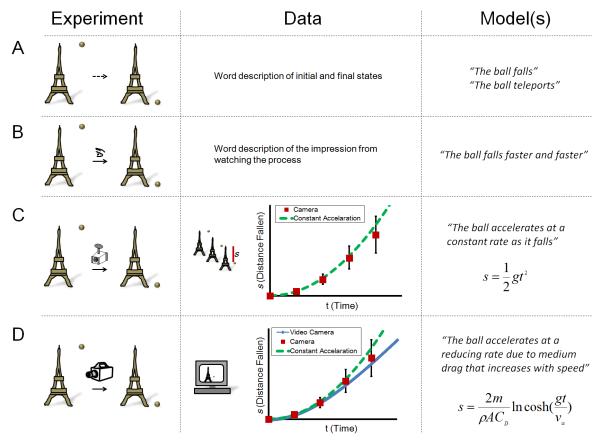
To illustrate this point, imagine a ball is released from the top of a tower (initial state, Figure 1A, experiment). A later observation of the system shows that the ball is on the ground (final state). Without any direct observation of the process, multiple hypotheses that describe what happened cannot be distinguished. A “teleportation model” fits the data as well as a “falling model”. Note that no matter how many different balls and towers are tested (perturbations), without a direct observation of the process between the initial and final states, the models can hardly be distinguished. With very simple observation (watching with the naked eye, Figure 1B), one could immediately develop a more interesting model of the system (the ball falls faster and faster after being dropped). By making the observation quantitative (Figure 1C) using some recording equipment and data analysis (a camera taking photos, a watch recording when the photos were taken, a ruler measuring the distances the ball traveled on the photo), the scientist could take a step further and formulate a mathematical model (the ball accelerates at a constant rate) that adds predictive power to the process general to other similar experiments. At this point, perturbations will be a lot more powerful than before when there was no direct observation, because they now have a specifically stated model (constant acceleration) to falsify. Note that this model will probably stand perturbations (changing balls and towers) and be sufficient until a better direct observation method comes along (Figure 1D). The newer observation (high frame rate video recorder coupled with computer based image analysis) reveals that there are quantitative differences between the model and the experiment. As a result of increased spatial and temporal resolution and coverage, modifications of the model need to be considered. Other parameters such as air resistance, density and cross-sectional area of the ball are then incorporated to form a better model, which can be applied in practical engineering (e.g., aviation). It is worth noting that perturbations (changing the ball to a feather, for example) would be able to identify the parameters in the more advanced model, but are unlikely to quickly lead to precise definitions of the parameters or to reveal their interactions in affecting the ball’s movement. Before a quantitative readout and mathematical modeling become available, dropping different objects from the tower and simply noting that they hit the ground at some point would not be very informative.
Figure 1. Direct observation generates and tests models.
A. The pictures depict a hypothetical experiment where a ball is released from the top of a tower and later found to be on the ground. No direct observation is made here allowing drastically different hypotheses about the process. Note only observations (but not variants of the experiment) can distinguish the given models.
B. Primitive observation with naked eyes generates a new, more specific hypothesis.
C. Measurements using a camera provide quantifiable data, leading to a testable mathematical formulation of the model. s, distance fallen; t, time; g, acceleration.
D. Improved observation using a high frame rate video recorder and analysis using a computer provide more quantitative details, which further improve the model. m, mass of the ball; ρ, density of air; A, projected area of the ball; CD, drag coefficient; v∞, terminal velocity.
Given the large number of genes underlying development, it is not surprising that this field has been dominated for a long time by endpoint analysis of the system when individual players are missing. Overshadowed by the power of developmental genetics in the previous few decades, the role of imaging in generating and testing models for development has been underappreciated. However, as the situation is now changing (imaging technology is advancing and the yield of new interesting terminal null phenotypes is diminishing), it is important to note that poking the black box is not necessarily a more “mechanistic” approach than watching it run. That said, the approach of trying to acquire detailed descriptive data by imaging carries its own risks. Such data might not be comprehensible or it may simply just lend more incremental support or refinement to existing models. Furthermore, the cost and labor of documenting all aspects of a developmental process in detail is high despite technical advances, often putting these approaches under heavy scrutiny. A reviewer may be skeptical of such a proposal if a well-defined question or a foreseeable application is not provided. This is not unjustified: why would one care about how a cell located exactly 100 microns from the midline changes its shape at 8.50 hours by launching sophisticated microscopy, if we already know cells in that area generally become more elongated between 7 and 9 hours from simple, static, low-resolution picture taking? Given that the cost of obtaining data for the former is way out of proportion with the later, it is more than reasonable to question the necessity of the more complex approach.
Here we argue that the above skepticism fails at recognizing the multiple possibilities that the imaging approach may lead to, beyond just a more detailed answer to an already answered question. First, once the proposed movie is taken, one could quantify the dynamics of cell shape changes and formulate models from the data. The data might be argued as just more details, but the models are likely going to contain novel concepts (in a similar way as the ball dropping example - producing a “constant acceleration” idea once the observation of “falling faster and faster” becomes more detailed and quantitative). The perturbations that further arise from these models will surely produce new insights, probably in a specific and resource-saving manner compared to undirected exploratory perturbations. Second, careful imaging often leads to surprises: one may discover previously unknown waves of cell shape change across space and time, one may find novel cell behaviors that drive shape change, one may discover it is in fact not cell shape change but instead cell reorientation, etc. Such unpredicted discoveries may not be immediately comprehensible or modeled and are unlikely to fit in the current picture, but few would deny their potential value and significance. Unfortunately however, these possibilities are difficult to convincingly formulate a priori in the proposal. Imaging’s role in providing good quality data is well appreciated but its roles in model generation, testing and discovery also deserve more recognition.
Quantitative, predictive models of development and the “Approach Trinity”
Developmental biologists have long been keen to apply mathematic formulations to their problems and some specific examples were successful8,9. However, most current models are still qualitative. They describe developmental processes in terms such as one of a few players activating, repressing, inducing, specifying, driving, (and worst of all) mediating a certain process - often without knowing the context, weight or dynamics of these effects. Such “word models”, which are qualitative descriptions or explanations of a process are prone to subjectivity in interpretation, and run the risk of substituting naming with actual mechanistic understanding (“oh, gene X mediates outcome Y…”). It can also be hard to think through the predictions of word models once they contain more than a few non-linearly interacting parts (e.g., A activates B in association with C by mediating D activity). Modeling may help overcome some of these limitations. First, in simply writing down a model as formal mathematical expressions, one must be explicit and precise in terms of what is meant by the words. Some words might have precise mathematical meanings (such as a kinase activating a target through phosphorylation at some rate) while other words may not. The act of converting a sentence to math can shine light on areas in a word model that need more attention. Second, with the aid of computers, a model considers multiple players at the same time while attempting so in experimental perturbations may both be cost-ineffective, technically infeasible, and lead to data that are difficult to interpret. Using modeling, one may also be able to infer and test mechanisms more specifically using a moderate degree of perturbation10 rather than overwhelming the system leading to secondary effects. Whether or not the predictions of a model can be measured using current techniques should not be the sole criteria of judging the model’s value. One should ask if the model advances an intuitive, interesting and generalizable understanding of a process. A good model could help researchers circumvent an area of complexity or parameters that are not measurable to still arrive at a simple prediction that can be tested. Thirdly, models allow a comparison between different interactions and the isolation of important ones among all that have been discovered or all that are theoretically possible (this is where modeling is uniquely powerful amongst the approaches). For example, ‘Core’ motifs of gene regulatory networks are found, and validated, by theoretical analysis and in silico experiments that probe the possible parameter space that could quantitatively describe the interactions between players11. This strategy offers a way to get at the logic of interaction behind the historical complexity of development that has been accumulating since the dawn of multicellular life. Drawing up a huge network diagram of molecules is not really helpful in promoting understanding. To this end, models should only be as complicated as needed to explain the biological question at hand rather than incorporating everything that is known. Finally, when data is of good quality (e.g., high resolution, quantifiable), modeling may be able to predict an exact mechanism, or existence of an unknown player thereby leading to directed discoveries12,13. Such cases are rarer but remain the most exciting aspect/possibility of theoretical work in systems biology. The bottom line is that the models are simplified representations of reality which will only be meaningful when the simplifications have some experimental basis. To do so in practice, one must take caution in not over-simplifying the problem when building a model so that the model is useless, or overly sticking to the specific details of the experimental system (either trying to explain every detail or achieving perfect model-experiment fit) so that the more interesting general principle is masked. This requires insight - which is a different challenge than getting all the quantitative data acquired and analyzed. To improve developmental biologists’ ability to use models efficiently, it is helpful to learn how they are used from other fields such as applied math, physics and engineering.
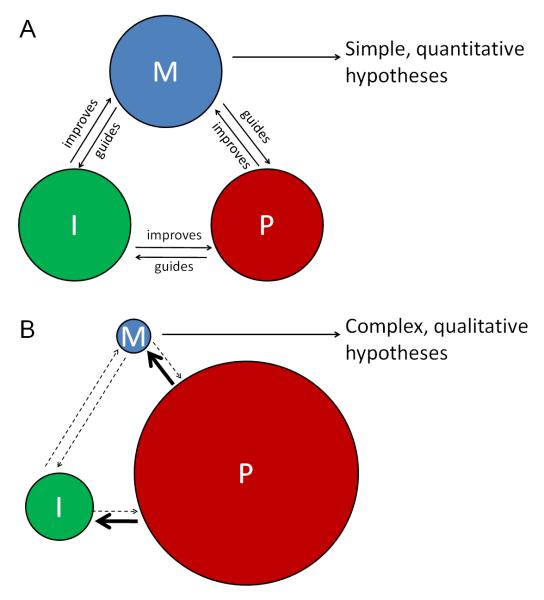
The above discussion shows that modeling can be a lot more useful than an abstractive graphic or word summary of the results. Modeling should be treated seriously as a main branch of the investigative process. Like quantitative imaging, the involvement of modeling feeds back to experiments especially in guiding perturbations along the most effective direction14,15. Together, quantitative imaging, modeling and perturbation form a trinity circle that performs productively when their interactions are matched and balanced (Figure 2A). Questions may remain unanswered or under – answered when this balance is broken e.g. due to perturbation taking dominance (Figure 2B). As illustrated, when a lot of information flows out from perturbations into the absence of a clearly defined theoretic framework or good quality measurement, confusion may increase in the attempt to fit all results into a cohesive picture. The outcome is a collection of statements from which it is hard to extract a generally applicable principle. This situation is unfortunately difficult to overcome, because developmental processes are inherently non-reducible problems. There is no analog of a dropping ball from a tower in development that is immediately feasible for imaging and modeling yet conceptually impacts more complex processes. Therefore, to pursue the balanced “approach trinity”, one needs to seek systems where systematic imaging can be made possible and core parameters can be mathematically defined, then ask questions within the bounds of conceptual and technical limitations.
Figure 2. The relationships between imaging, modeling and perturbation.
A. The circles representing the three basic approaches of investigation: measuring by quantitative imaging (I), theorizing by modeling (M) and testing by perturbation (P). When these approaches are matched, they promote each other to efficiently lead to new understanding.
B. When one of the approaches dominates, such as for developmental biology where perturbation is nearly exclusive, the interactions are out of balance. The models do not have adequate quantitativeness supplied by imaging but have too much information to accommodate from the perturbation side - inevitably leading to complex hypotheses defined by imprecise wording. Without enough independent emphasis on imaging, it became an end point tool directed by perturbations and its role of making the perturbation results more meaningful tends to be diminished.
Zebrafish embryos for systematic imaging
The power of an imaging technique comes from its resolution, coverage and throughput. It is possible to pursue the best result of one of these properties, while forsaking others (e.g., electron microscopy for spatial resolution, but low coverage and temporal resolution; automatic screening scopes for throughput, but low spatial and temporal resolution). For systems biology of development, however, all of them are important. This is especially true when modeling approaches are involved. The quantitative nature of modeling requires correspondingly good data for parameter measurement and confinement (which come in large number and variety in complex developmental processes) and testing of predictions. For example, to use the data for modeling, it is not enough to just know “up-regulated” or “inhibited”. A number must be given which significantly increases the demand on the imaging and image analysis (“representative examples” now must be replaced by a calculated average from a large sample size). In addition, models of development usually do more than predicting the final outcome, they also predict the dynamics leading up to it. This demands imaging to have high temporal coverage (live and continuous if possible). In reality, imaging technique is never ideal, thus the compromises between different technical aspects of imaging thereby define both the restrictions and opportunities at the current moment, although given the fast progress of techniques it is useful to think some time ahead. Fortunately, biological samples come in a huge variety to fill the “niches” allowed by the imaging systems. In vivo, live, developing embryos require non-invasive, high spatial temporal coverage and resolution for “in toto” type of observation16. Conversely, microscopes require samples that are easy to handle, optically accessible, and brightly labeled. Zebrafish, among others, situates comfortably at this enlarging intersection.
Zebrafish has emerged as a low cost vertebrate system for studies of development and disease in the past 30 years. The small size, fast development, and transparency of zebrafish embryos are ideal for pushing imaging to very high coverage and resolution. In addition, the toolkit of genetic manipulation and embryology for this system has been rapidly expanding (zfin.org)17. It would be no surprise, in the next 10 years to see zebrafish as the first vertebrate whose early developmental lineages are mapped to a level of precision as the C. elegans18, or to see gene expression dynamics registered to a zebrafish developmental atlas. Recently, gene expression patterns have been documented in great detail for zebrafish19. Beside the molecular profiles, the cellular and tissue-level dynamics during development are more poorly documented. For example, the changes of cell shapes, the trajectories of cell migration and the mechanical property changes of tissues are only superficially described. Studies aiming to generate a digital, quantified representation of these processes are emerging recently.
Below we review some recent works in zebrafish that employ high-resolution imaging and attempt to derive quantitative models from the data. These examples provide in-action showcases of the points we discussed above.
Case 1: Early lineage and divisions in zebrafish embryos
Zebrafish embryos are most accessible for observation and perturbation during the cleavage and gastrulation stages20. The embryo undergoes incomplete cleavage, separating the transparent blastomeres from the more obscure yolk cell. This allows video recording of cellular events for cells not only on the surface but also in deeper layers. In addition, the cells are large and robust enough so that by mosaic labeling using acute injections the lineage and fates of the progeny of a blastomere can be determined later21,22. Using these approaches earlier investigations revealed timing of cell cycles, different mitotic domains, first differentiation events and cell movements that reorganize the embryo. These results provide a framework of concepts that enable characterization of different mutant phenotypes20,23.
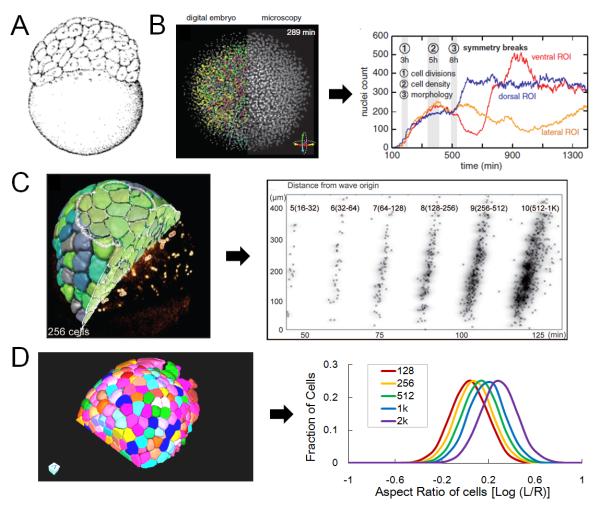
Recent development of imaging of the same early stages brings new insights. Keller and colleagues used an impressive digital scanning light sheet microscope (DSLM) to capture zebrafish early embryonic development in toto24. The authors segmented and tracked the nucleus of all the early embryo cells. The data provide high temporal resolution information of cell number, cell division times and cell movements, revealing the cell cycle synchrony break at the 10th cell cycle (Figure 3B). Moreover, the authors were able to pinpoint exactly what went wrong in a mutant embryo on the cellular level. In a later study, Olivier and colleagues implemented second and third harmonic generation in the imaging to capture cell membranes and cells that are very deep inside25 (Figure 3C). In addition to performing division and lineage analysis, with the membrane data the authors were able to calculate the volumes of the cells and potentially cell shapes. We performed a systematic analysis of the surface cell shapes in order to reveal the interaction logic of different processes that are happening during this time15. Note that this is only possible when the relevant parameters are measurable. Using imaging and cell tracking we found that surface cells follow a “division rule” that determines their division orientation (and lineage outcome) as a function of its shape. By having this link, the cells move steadily towards a flattened shape robust to initial conditions and perturbations (Figure 3D). Moreover, by tuning this link one can obtain the full range of different epithelial cell shapes, thus explaining one way by which local cell behaviors could lead a global process on the tissue level. In this example, we see that imaging not only improves the resolution and coverage of a well characterized process during zebrafish embryonic development, but also allows quantitative models to be formulated and tested, leading to interesting new insights. The modeling in return directly points to the perturbations that would test the hypothesis. Completion of the approach trinity here brings the understanding of the process to a new level.
Figure 3. Imaging and modeling of the early zebrafish embryo.
A. Sketch of 128-cell stage zebrafish embryo, a model system for early developmental stages accessible for imaging and perturbation. Image reprinted with permission from ref. 20.
B. Microscopy data and digital embryo allowing tracking of all cells in the early embryo. The nuclei are labeled. The plots show nuclei counts over time. This analysis pinpoints the symmetry breaking events of the whole embryo which marks transitions between different developmental stages. Image reprinted with permission from ref. 24.
C. Reconstruction of 256-cell stage embryos using second and third harmonic generation imaging data. These quantitative data allow analysis such as seen in the scatter plot showing individual mitosis time as a function of distance to a mitosis pseudo-wave origin. Image reprinted with permission from ref. 25.
D. Membrane segmentation using ACME42 on early embryo imaging data. The data allow cell shape measurements as plotted by the aspect ratio of cells on the surface where L means the width on the surface and R means the depth perpendicular to the surface. The distributions of the aspect ratio were then used to model a new feedback mechanism of cell shape control. Images adapted from ref. 15.
Case 2: Mechanically regulated tissue morphogenesis
Morphogenesis refers to the processes of forming tissue shapes and structures. This is another key component of development and is much less understood compared to the molecular signaling networks that benefited from the progresses in genetics and biochemistry. This domain requires input from physics and material sciences in the context of developmental genetics. To apply concepts from these fields to development, careful measurements of the dynamic changes of the tissue undergoing morphogenesis and relevant cellular and molecular behaviors must be made. In zebrafish embryos, the epiboly process moves cells around the yolk to transform a hemisphere of cells into a more flattened sheet several layers thick20 (Figure 4A, compare to Figure 3A). This process is essential for setting up the geometry for the gastrulating embryo. What are the forces driving this drastic morphogenetic movement and how the forces are generated have been long-standing, puzzling questions. Mutation of the epithelial adhesion molecule E-cadherin, which is highly expressed in the enveloping layer (EVL), prevents completion of epiboly of the deep cells26, raising the model that EVL cells are driving epiboly and drag deep cells along via adhesive junctions with the deep cells. Subsequent studies identified a contracting ring of actomyosin on the marching frontier of the EVL thus revealing where the force might come from27 (Figure 4B).
Figure 4. Using imaging to measure forces driving morphogenesis.
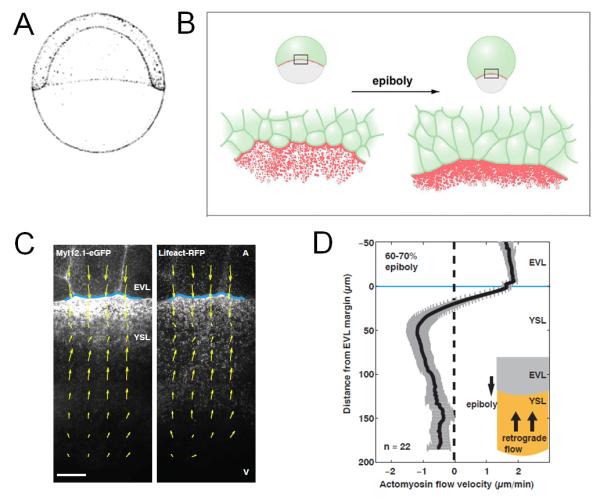
A. Sketch of 50% epiboly stage zebrafish embryo, a model system for collective cell movement during morphogenesis. Image reprinted with permission from ref. 20.
B. During epiboly, enveloping layer (EVL) cells (green) become more elongated as an ‘Actin ring’ forms as a result of Actin (Red) accumulation at the marching frontier. Image reprinted with permission from ref. 27.
C. Cortical flows highlighted by labeled Myosin-2 (left) and F-actin (right) at the EVL margin (blue line). Image reprinted with permission from ref. 28.
D. The plot shows the average cortical flow revealing retrograde flow (negative velocities) toward the animal pole. Image reprinted with permission from ref. 28.
Imaging played a crucial role in promoting the physical understanding of this morphogenetic process. Behrndt and colleagues imaged epiboly at high resolution at the marching frontier and analyzed the dynamics of local myosin flow28 (Figure 4C). By quantifying the amount of tension in the tissue they were able to distinguish two force-generating mechanisms driving the spreading. Importantly, the movies allowed the calculation of flow rate of myosin and estimation of the friction force the flow generates on the tissue. This example shows how a mechanical hypothesis of morphogenesis became testable and distinguishable once the process is translated into quantitative descriptions, and imaging is the required tool for the translation and testing. Here the trinity is complete as imaging and modeling grow stronger to match a “well-perturbed” process.
Case 3: Developmental patterning in a morphogen gradient
An important concept in development is patterning, which means the formation of spatial organizations of different cells. The patterns carry out important functions and can be characterized clearly with differential gene expressions in different cells. Patterns can emerge automatically as a result of chemical interactions in a reaction-diffusion system29 or under the instruction of spatial signals called morphogens30. In these mechanisms, the diffusivity of morphogen molecules is predicted to be essential for shaping the profile of morphogen distribution, which in turn explains many characteristics of the pattern such as sizes of different domains and boundaries between neighboring domains.
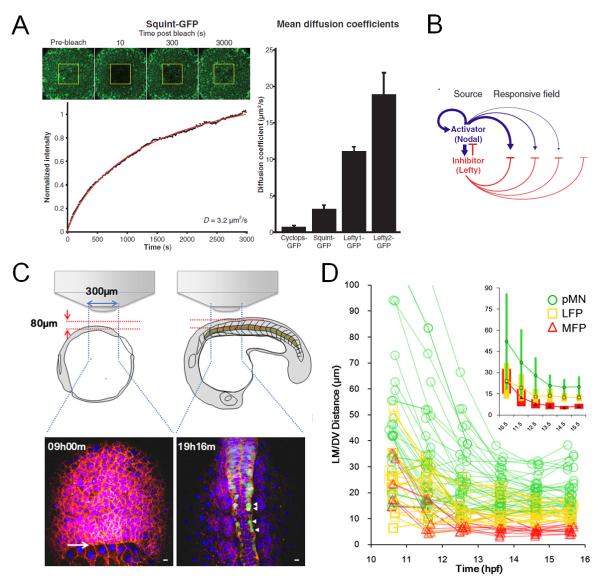
To directly validate and test these models, it is essential to watch the morphogen’s action and the cell’s responses. Perturbations that remove or over-express the morphogens are not sufficient by themselves without imaging that capture the dynamics leading up to the phenotypes. A recent study using zebrafish embryos and fluorescent fusion proteins provides the key evidence of different diffusivity of morphogen molecules setting up the pattern31. By live imaging of the fluorescence recovery after photobleaching (FRAP) of the tagged morphogen molecules expressed by injected cells, the authors were able to measure the ability of the molecules to move in an in vivo context (Figure 5A). Coupling with the interaction between these molecules with a model, they were able to explain how mesoderm patterning works as a polarized reaction-diffusion network (Figure 5B). In this example quantification by imaging and conceptualization by modeling completes the approach trinity which leads to a definitive result. We used single cell tracking based analysis in the captured patterning process of the zebrafish neural tube32 (Figure 5C). The progenitors respond to Sonic Hedgehog (Shh) morphogen in a time and concentration dependent manner to acquire different fates33,34. We found that cells move a lot during this process and mix extensively with other cell types. The ability to track single cells allows us to define a lot more precisely the timing of specification vs. localization, revealing that specified cells of different types initially are spatially mixed but continue to move and rearrange (Figure 5D). Therefore previously unknown processes of noise in morphogen specification and patterning by cell sorting were uncovered. In this example, imaging provides cellular dynamics of signaling and position that can be compared with theoretic models that were built from lower resolution, perturbation data. This again promotes the interaction of the approach trinity, leading to a challenge of previous models. Moreover, just like the high frame rate camera filming the falling ball (Figure 1D), direct imaging of the morphogens and their recipient cells also goes beyond better description and re-validation of previous studies to generate novel mechanistic insights.
Figure 5. Morphogen patterning dynamics studied using live imaging.
A. Using Fluorescence recovery after photobleaching (FRAP) to measure the diffusion coefficients of tagged morphogen molecules (Squint as example here). The recovery profile (black) was fit with simulated recovery curves (red). The plot shows a summary of measured diffusion coefficients. Image reprinted with permission from ref. 31.
B. Model diagram of the activator/inhibitor reaction-diffusion system for Nodal/Lefty morphogens, the diffusivities of activator and inhibitors are essential for the patterning system. Image reprinted with permission from ref. 31.
C. Schematic illustration of imaging setup for neural tube live imaging and sample data time points. Image adapted from ref. 32.
D. Positional trajectories of cells of different fates. Inset shows population average position ± SD (colored bars) by cell type plotted on the same axes. Cells show rearrangement of positions during the patterning process, an observation made possible by live imaging based cell tracking. Image adapted from ref. 32.
Outlook
The examples we discussed suggest that a new chapter has begun in the pursuit of understanding development using mathematical, predictive models. Zebrafish serves as an excellent system for imaging allowing it to be a key model system leading the way into the quantitative era of developmental biology. The field will benefit from improvement in the following areas that together constitute a systematic approach. First, more innovations in imaging techniques need to be developed, especially ones that increase the access to larger, deeper samples and that are faster and more sensitive. There has been great progresses in imaging innovations in recent years and this progress promises to carry on35. Second, samples with enhanced transparency and brighter labeling need to be prepared to improve the accessibility of imaging. Zebrafish has some in-born advantage here but others are coming along. For example, several approaches have recently been developed for clearing large samples such as a mouse brain or whole mouse embryos albeit only on fixed specimens36,37,38. New fluorescent proteins and other labeling agents are also flourishing39. Besides pushing the limitation of in vivo experimental models, the use of in vitro models will also continue to play an important role. In vitro systems such as embryonic stem (ES) cell cultures offer simpler, more accessible models by compromising the need of quantitativeness and the difficulty of achieving it in vivo. In vitro models have served as a main force in reducing the complexity of developing systems and revealed many principles that can be applied to understand the in vivo processes. In some cases, the in vitro systems have directly provided useful applications mimicking the in vivo counterparts40,41. Third, programmable or even automated standard imaging pipelines that increase throughput need to be created. Variation and flexibility is a major part of developmental processes, whose underlying mechanisms have deep implications in understanding evolution of development and ensuring robustness to reduce developmental defects. The developmental lineage tree of one zebrafish embryo is probably quite different from another. Standardized imaging that collects more individual datasets and lends more confidence for the comparison between them is essential. Here it is possible that specialization will occur, for example, imaging cores with personnel highly experienced with the equipment may collaborate with researchers to design efficient, reproducible methods of data acquisition. Still, developmental biologists should be encouraged to learn more about imaging systems especially how they fit with the model systems and scientific questions at hand. Fourth, imaging analysis tools that efficiently translate data into easily shared quantitative information need to be developed42,43. It is not surprising that classically trained developmental biologists start to find the large datasets difficult to handle. The challenges range from data storage to analysis to sharing analysis results. Even with great efforts underway in the imaging analysis community, the hurdles are already extending significantly the time between conducting an experiment and having results on a figure. Now there are situations where a developmental biologist will have the data and know the interesting results must be in there, for quite a while, without an idea of what they actually are. This is akin to recording a great diffraction pattern for a crystalized protein but not yet knowing its structure. Again, while we may see more specialization in this area dividing data acquisition and analysis, it is advisable for a researcher to learn more about the concepts and techniques behind imaging analysis to better guide experiments to produce data suitable for analysis. Fifth, new conceptual frameworks that handle the complex molecular networks and physical properties of cells and tissues need to be proposed and debated. There is a lot to borrow already from different fields, such as feedback, robustness, optimality, visco-elasticity etc. But development has its uniqueness and confinements: it operates robustly in narrow ranges of physical parameters, bio-molecules are limited in the type of functions they can achieve, individual cells are error prone, and the system relies on self assembly with limited capacity of information exchange. Developmental mechanisms we observe may be one of a few possible solutions evolution has found under these many constraints, and should therefore be solvable as equations.
Taken together, one could probably see that only when multiple disciplines join forces with a common goal of better understanding development is ‘systems biology in vivo’ possible. Similar to when systems biology allied biochemistry, microbiology and cell biology, imaging and modeling will play a central role in the systematic revisiting of multicellular development. The exciting upcoming questions enabled by systems biology are demanding a lot more in the training of today’s developmental biologists, who should take the initiative to step out of the “refuge” and adopt the mathematical thinking and tools that will drive the field towards its quantitative future.
Acknowledgements
The authors are supported by NIH grant R01 DC010791.
Footnotes
The authors declare no competing financial interest.
References
- 1.Kirschner MW. The meaning of systems biology. Cell. 2005;121:503–504. doi: 10.1016/j.cell.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Megason SG, Fraser SE. Imaging in systems biology. Cell. 2007;130:784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 3.Megason SG, Srinivas S, Dickinson ME, Hadjantonakis AK. Microscopy to mechanism across the scales of development. Curr. Opin. Genet. Dev. 2011;21:519–522. doi: 10.1016/j.gde.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert SF. Mathematical Modeling of Development. 6th edition. Sinauer Associates; Sunderland (MA): 2000. Developmental Biology. http://www.ncbi.nlm.nih.gov/books/NBK10126/ [Google Scholar]
- 5.Davidson LA, Baum B. Making waves: the rise and fall and rise of quantitative developmental biology. Development. 2012;139:3065–3069. doi: 10.1242/dev.080093. [DOI] [PubMed] [Google Scholar]
- 6.Pantazis P, Supatto W. Advances in whole-embryo imaging: a quantitative transition is underway. Nat. Rev. Mol. Cell Biol. 2014;15:327–339. doi: 10.1038/nrm3786. [DOI] [PubMed] [Google Scholar]
- 7.Sharpe J. Two ways to use imaging: focusing directly on mechanism, or indirectly via behaviour? Curr Opin Genet Dev. 2011;21:523–9. doi: 10.1016/j.gde.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Thompson DW. On Growth and Form. Cambridge University Press; Cambridge: 1942. [Google Scholar]
- 9.Odell G, Oster G, Alberch P, Burnside B. The mechanical basis of morphogenesis. I. Epithelial folding and invagination. Dev. Biol. 1981;85:446–462. doi: 10.1016/0012-1606(81)90276-1. [DOI] [PubMed] [Google Scholar]
- 10.Barkoulas M, van Zon JS, Milloz J, van Oudenaarden A, Félix MA. Robustness and epistasis in the C. elegans vulval signaling network revealed by pathway dosage modulation. Dev Cell. 2013;24:64–75. doi: 10.1016/j.devcel.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 1976;260:799–802. doi: 10.1038/260799a0. [DOI] [PubMed] [Google Scholar]
- 14.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–8. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- 15.Xiong F, Ma W, Hiscock TW, Mosaliganti KR, Tentner AR, Brakke KA, Rannou N, Gelas A, Souhait L, Swinburne IA, Obholzer ND, Megason SG. Interplay of Cell Shape and Division Orientation Promotes Robust Morphogenesis of Developing Epithelia. Cell. 2014;159:415–427. doi: 10.1016/j.cell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megason SG, Fraser SE. Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech. Dev. 2003;120:1407–1420. doi: 10.1016/j.mod.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Westerfield M, Doerry E, Kirkpatrick AE, Douglas SA. Zebrafish informatics and the ZFIN database. Methods Cell Biol. 1999;60:339–55. doi: 10.1016/s0091-679x(08)61909-3. [DOI] [PubMed] [Google Scholar]
- 18.Sulston J, Schierenberg E, White J, Thomson J. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–6119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 19.Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159:662–75. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 21.Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres. III. Clonal analyses of the blastula and gastrula stages. Dev. Biol. 1985;108:94–101. doi: 10.1016/0012-1606(85)90012-0. [DOI] [PubMed] [Google Scholar]
- 22.Strehlow D, Heinrich G, Gilbert W. The fates of the blastomeres of the 16-cell zebrafish embryo. Development. 1994;120:1791–1798. doi: 10.1242/dev.120.7.1791. [DOI] [PubMed] [Google Scholar]
- 23.Kane D, Hammerschmidt M, Mullins M, Maischein H, Brand M, van Eeden F, Furutani-Seiki M, Granato M, Haffter P, Heisenberg C, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nüsslein-Volhard C. The zebrafish epiboly mutants. Development. 1996;123:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- 24.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 25.Olivier N, Luengo-Oroz MA, Duloquin L, Faure E, Savy T, Veilleux I, Solinas X, Debarre D, Bourgine P, Santos A, Peyrieras N, Beaurepaire E. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science. 2010;329:967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- 26.Kane DA, McFarland KN, Warga RM. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development. 2000;132:1105–16. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- 27.Köppen M, Fernández BG, Carvalho L, Jacinto A, Heisenberg CP. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 2006;133:2671–81. doi: 10.1242/dev.02439. [DOI] [PubMed] [Google Scholar]
- 28.Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, Grill SW, Heisenberg CP. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338:257–260. doi: 10.1126/science.1224143. [DOI] [PubMed] [Google Scholar]
- 29.Turing A. The chemical basis of morphogenesis. Bull. Math. Biol. 1953;52:153–97. doi: 10.1007/BF02459572. [DOI] [PubMed] [Google Scholar]
- 30.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 31.Muller P, Rogers KW, Jordan BM, Lee JS, Robson D, Ramanathan S, Schier AF. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science. 2012;336:721–724. doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong F, Tentner AR, Huang P, Gelas A, Mosaliganti KR, Souhait L, Rannou N, Swinburne IA, Obholzer ND, Cowgill PD, Schier AF, Megason SG. Specified neural progenitors sort to form sharp domains after noisy shh signaling. Cell. 2013;153:550–561. doi: 10.1016/j.cell.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 35.Keller PJ. Imaging morphogenesis: technological advances and biological insights. Science. 2013;340:1234168. doi: 10.1126/science.1234168. [DOI] [PubMed] [Google Scholar]
- 36.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, Ueda HR. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 39.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10:512–23. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 41.Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 42.Mosaliganti KR, Noche RR, Xiong F, Swinburne IA, Megason SG. ACME: automated cell morphology extractor for comprehensive reconstruction of cell membranes. PLoS Comput. Biol. 2012;8:e1002780. doi: 10.1371/journal.pcbi.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amat F, Lemon W, Mossing DP, McDole K, Wan Y, Branson K, Myers EW, Keller PJ. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat Methods. 2014;11:951–8. doi: 10.1038/nmeth.3036. [DOI] [PubMed] [Google Scholar]