Abstract
The HIV vaccine-induced neutralizing antibodies (Abs) display low rates of mutation in their variable regions. To determine the range of neutralization mediated by similar human monoclonal Abs (mAbs) but derived from unselected chronically HIV-1 infected subjects, we tested a panel of 66 mAbs specific to V3, CD4 binding site (CD4bs) and V2 regions. The mAbs were tested against 41 pseudoviruses, including 15 tier 1 and 26 tier 2, 3 viruses, showing that the neutralization potency and breadth of anti-V3 mAbs were significantly higher than those of the anti-CD4bs and anti-V2 mAbs, and only anti-V3 mAbs were able to neutralize some tier 2, 3 viruses. The percentage of mutations in the variable regions of the heavy (VH) and light (VL) chains varied broadly in a range from 2% to 18% and correlated moderately with the neutralization breadth of tier 2, 3 viruses. There was no correlation with neutralization of tier 1 viruses as some mAbs with low and high percentages of mutations neutralized the same number of viruses. The electrostatic interactions between anti-V3 mAbs and the charged V3 region may contribute to their neutralization because the isoelectric points of the VH CDR3 of 48 anti-V3 mAbs were inversely correlated with the neutralization breadth of tier 2, 3 viruses. The results demonstrate that infection-induced antibodies to CD4bs, V3 and V2 regions can mediate cross-clade neutralization despite low levels of mutations which can be achieved by HIV-1 vaccine-induced antibodies.
Keywords: HIV-1, V3 region, CD4 binding site, V2 region, Human monoclonal antibodies, HIV neutralizing antibodies, Somatic mutation
1. Introduction
The identification of anti-HIV-1 broadly neutralizing antibodies (bnAbs) suggests the possibility of designing immunogens that can induce potent and cross-reactive antibodies (Abs) in HIV vaccinees. Although this approach is very attractive, it faces several major challenges including immunogen design, an increased level of somatic mutations (15–36%) in bnAbs, and the fact that the induction of bnAbs by a HIV vaccine has not been achieved in any animal model (reviewed in (van Gils and Sanders, 2013; West et al., 2014).
In contrast to the concept that bnAbs need to be induced to reduce infection by HIV-1, are the results of the recent RV144 clinical vaccine efficacy trial, which showed a reduction in HIV-1 infection of 31.2% in vaccinees (Haynes et al., 2012). This vaccine used a prime and boost regimen with a recombinant HIV-avian pox virus and two different recombinant gp120 proteins which induced a broad range of anti-gp120 Abs, including three types of neutralizing Abs against CD4-binding site (CD4bs), V3 and V2 regions; however, bnAbs were not detected (Gottardo et al., 2013; Haynes et al., 2012). Data analysis showed that reduced infection was inversely correlated with levels of anti-V2 plasma Abs (Haynes et al., 2012; Zolla-Pazner et al., 2013). The anti-V3 Abs were also correlated with infection risk but only in vaccinees with lower levels of gp120-specific plasma IgA Abs (Gottardo et al., 2013).
The plasma Abs from recipients of the RV144 neutralized tier 1 pseudoviruses and presence of neutralizing anti-V3 Abs was determined based on peptide blocking assays which does not exclude that other, conformation-dependent neutralizing Abs, were involved (Haynes et al., 2012; Montefiori et al., 2012). In addition, two anti-V3 mAbs – CH22 and CH23 – derived from recipients of the vaccine displayed weak neutralizing activity which could be related to a low level of mutations, 3.7% and 4.5%, respectively, in their variable regions (Montefiori et al., 2012). This is comparable with the low percentage of mutations observed in other vaccine-induced anti-V2 and anti-gp120 mAbs (Liao et al., 2013; Moody et al., 2012). It is possible that during several months of vaccination, responding Abs are characterized by a limited percentage of mutations, but the range of their neutralization potency and breadth is unknown due to the existence of only several such mAbs (Liao et al., 2013; Montefiori et al., 2012).
To address this issue we analyzed the neutralization potency and breadth as well as the percentage of mutations in 66 human mAbs against CD4bs, V3 and V2 regions of HIV-1 gp120 which were derived from chronically infected individuals. These three types of neutralizing Abs, anti-CD4bs, anti-V3 and anti-V2, are commonly present in the plasma of HIV-1 infected individuals (Kayman et al., 1994; Lynch et al., 2012; Vogel et al., 1994) and corresponds to HIV vaccine induced neutralizing Abs which can be classified as conventional Abs in contrast to bnAbs (Zolla-Pazner, 2014).
This study showed that anti-V3 mAbs neutralized tier 1 and some tier 2, 3 viruses from diverse HIV-1 subtypes, while anti-CD4bs and anti-V2 mAbs neutralized only tier 1 viruses. The percentage of mutations was not related to neutralization breadth of tier 1 viruses but moderately correlated with the neutralization breadth of tier 2, 3 viruses which was mediated by anti-V3 mAbs. The electrostatic interactions between anti-V3 mAbs and V3 may contribute to neutralization of tier 2, 3 viruses which inversely correlated with low isoelectric points (pI) of the CDR domain in the heavy chains.
2. Materials and methods
2.1. Human monoclonal antibodies
Sixty-six human mAbs specific to gp120 HIV-1 were studied, including 48 anti-V3, 12 anti-CD4bs and six anti-V2 mAbs (Table 1, Supplementary Table 1). All mAbs were produced in our laboratory and those which displayed the ability to neutralize HIV-1 pseudoviruses were selected for this study (Table 1). One human mAb, 1418 against parvovirus B19, was used as negative control (Gigler et al., 1999). The anti-HIV-1 mAbs were derived from 58 unselected and chronically HIV-1 infected individuals and except for two clonal anti-V2 mAbs (1393A and 1361), each mAb is unique with different VH CDR3 sequences (Supplementary Table 1). Forty mAbs were derived from individuals living primarily in the New York City area whom were presumably infected with subtype B viruses (Table 1). Twenty-six mAbs originated from individuals infected with non-clade B viruses and living in Cameroon, India, Ivory Coast and Thailand; the blood donors were infected with subtypes C, G, H, F2, CRF01_AE, CRF02_AG, CRF09_cpx, CRF13_cpx viruses or undetermined (Table 1). The subtypes of plasma viruses were determined by sequencing the envelope proteins in our lab as previously described (Zhong et al., 2002). Immunoglobulin (IG) gene usage (IGHV, IGKV and IGLV) and alleles evaluated by IMGT/V-QUEST are shown in Supplementary Table 1.
Table 1.
Human anti-HIV-1 gp120 monoclonal antibodies used in the study.
| # | mAb | Site | Virus subtype1 |
VH, V % mut2 |
VL, V % mut2 |
Neutral. breadth3 |
References |
|---|---|---|---|---|---|---|---|
| 1 | 3791 | V3 | C | 12.15 | 4.3 | 5 | (Gorny et al., 2009) |
| 2 | 4121 | V3 | C | 5.21 | 1.4 | 7 | (Gorny et al., 2009) |
| 3 | 391/95-D | V3 | B | 12.5 | 7.53 | 7 | (Gorny et al., 1993) |
| 4 | 311-11D | V3 | B | 7.99 | 2.87 | 3 | (Gorny et al., 1993) |
| 5 | 1334 | V3 | B | 10.07 | 3.58 | 7 | (Gorny et al., 2000) |
| 6 | 4139 | V3 | C | 6.6 | 6.6 | 5 | (Andrabi et al., 2013) |
| 7 | 2191 | V3 | B | 17.36 | 11.58 | 16 | (Gorny et al., 2002) |
| 8 | 4591 | V3 | non-B | 6.6 | 6.32 | 10 | This study |
| 9 | 3869 | V3 | non-B | 8.33 | 8.6 | 13 | (Gorny et al., 2009) |
| 10 | 4210 | V3 | non-B | 5.56 | 4.3 | 8 | (Andrabi et al., 2013) |
| 11 | 3697 | V3 | G | 10.07 | 5.5 | 9 | (Gorny et al., 2009) |
| 12 | 694/98-D | V3 | B | 5.84 | 4.66 | 8 | (Gorny et al., 1992) |
| 13 | 2412 | V3 | B | 6.53 | 6.45 | 6 | (Gorny et al., 2002) |
| 14 | 447-52D | V3 | B | 5.44 | 2.46 | 13 | (Gorny et al., 1993) |
| 15 | 2601 | V3 | CRF13 cpx | 5.21 | 3.23 | 4 | (Gorny et al., 2006) |
| 16 | 1324E | V3 | AE | 6.6 | 2.51 | 4 | (Gorny et al., 1998) |
| 17 | 3904 | V3 | C | 5.9 | 5.02 | 3 | (Gorny et al., 2009) |
| 18 | 3b54 | V3 | B | 14.24 | 7.72 | 7 | (Li et al., 2012) |
| 19 | 2424 | V3 | B | 13.33 | 5.1 | 5 | (Gorny et al., 2004) |
| 20 | 3881 | V3 | CRF02_AG | 11.23 | 5.38 | 3 | (Gorny et al., 2009) |
| 21 | 4508 | V3 | non-B | 4.56 | 4.71 | 5 | (Andrabi et al., 2013) |
| 22 | 2442 | V3 | B | 9.82 | 1.4 | 11 | (Gorny et al., 2002) |
| 23 | 3074 | V3 | CRF02_AG | 5.96 | 5.26 | 12 | (Gorny et al., 2006) |
| 24 | 268-D | V3 | B | 14.03 | 8.24 | 4 | (Gorny et al., 1991) |
| 25 | 386-D | V3 | B | 10.18 | 6.09 | 5 | (Gorny et al., 1993) |
| 26 | 2182 | V3 | CRF02_AG | 11.93 | 9.68 | 5 | (Gorny et al., 2002) |
| 27 | 782-D | V3 | B | 6.6 | 4.21 | 4 | (Gorny et al., 1997) |
| 28 | 908-D | V3 | B | 6.94 | 5.26 | 7 | (Gorny et al., 1997) |
| 29 | 1006-15D | V3 | B | 8.33 | 5.61 | 9 | (Gorny et al., 1997) |
| 30 | 2219 | V3 | B | 10.76 | 8.73 | 11 | (Gorny et al., 2002) |
| 31 | 2483 | V3 | B | 8.33 | 4.21 | 6 | (Gorny et al., 2004) |
| 32 | 4085 | V3 | non-B | 10.076 | 8.07 | 7 | (Gorny et al., 2011) |
| 33 | 4487 | V3 | non-B | 8.33 | 5.61 | 7 | (Andrabi et al., 2013) |
| 34 | 838-D | V3 | B | 7.29 | 6.45 | 10 | (Gorny et al., 1997) |
| 35 | 2557 | V3 | CRF02_AG | 14.24 | 13.98 | 12 | (Gorny et al., 2004) |
| 36 | 3906 | V3 | C | 7.29 | 9.68 | 8 | (Gorny et al., 2009) |
| 37 | 4490 | V3 | non-B | 4.51 | 5.02 | 6 | (Andrabi et al., 2013) |
| 38 | 4682 | V3 | non-B | 6.6 | 10.04 | 11 | This study |
| 39 | 2558 | V3 | CRF02 AG | 7.29 | 5.73 | 14 | (Gorny et al., 2004) |
| 40 | 3019 | V3 | CRF02 AG | 5.9 | 9.68 | 8 | (Gorny et al., 2006) |
| 41 | 3694 | V3 | H | 18.06 | 11.47 | 8 | (Gorny et al., 2009) |
| 42 | 4022 | V3 | C | 11.81 | 7.89 | 7 | (Gorny et al., 2009) |
| 43 | 4025 | V3 | non-B | 3.82 | 4.66 | 5 | (Gorny et al., 2009) |
| 44 | 4415 | V3 | B | 9.72 | 6.45 | 6 | This study |
| 45 | 4647 | V3 | non-B | 7.64 | 6.09 | 7 | This study |
| 46 | 2456 | V3 | B | 3.47 | 1.79 | 7 | (Gorny et al., 2002) |
| 47 | 419-D | V3 | B | 14.24 | 6.19 | 8 | (Gorny et al., 1993) |
| 48 | 3b384 | V3 | B | 6.326 | 5.84 | 9 | (Li et al., 2012) |
| 49 | 448-D | CD4bs | B | 7.64 | 6.25 | 6 | (Karwowska et al., |
| 50 | 3c504 | CD4bs | B | 9.03 | 5.38 | 6 | (Li et al., 2012) |
| 51 | 3c814 | CD4bs | B | 4.86 | 3.23 | 3 | (Li et al., 2012) |
| 52 | 9CL | CD4bs | B | 5.9 | 4.81 | 5 | (Jeffs et al., 2001) |
| 53 | 1027-30D | CD4bs | B | 4.08 | 1.77 | 5 | (Jeffs et al., 2001) |
| 54 | 654-D | CD4bs | B | 13.61 | 6.6 | 9 | (Nyambi et al., 1998) |
| 55 | 1570 | CD4bs | B | 6.6 | 3.4 | 7 | (Jeffs et al., 2001) |
| 56 | 729-D | CD4bs | B | 14.24 | 8.5 | 7 | (Zolla-Pazner et al., |
| 57 | 1202 | CD4bs | B | 9.62 | 4.76 | 7 | (Nyambi et al.., 1998) |
| 58 | 1263 | CD4bs | B | 17.19 | 11.46 | 3 | (Jeffs et al., 2001) |
| 59 | 559/64-D | CD4bs | B | 8.93 | 8.24 | 6 | (Karwowska et al., |
| 60 | 1331-E | CD4bs | B | 2.78 | 2.84 | 6 | (Gorny et al., 2000) |
| 61 | 2158 | V2 | B | 8.68 | 3.94 | 7 | (Pinter et al., 2004) |
| 62 | 1393A5 | V2 | B | 2.43 | 0.35 | 8 | (Nyambi et al., 2000) |
| 63 | 13615 | V2 | B | 4.51 | 1.796 | 8 | (Gorny et al., 2000) |
| 64 | 697-D | V2 | B | 5.9 | 1.04 | 6 | (Gorny et al., 1994) |
| 65 | 1357 | V2 | B | 5.9 | 2.87 | 6 | (Gorny et al., 2000) |
| 66 | 830A | V2 | B | 12.98 | 8.96 | 4 | (Nyambi et al., 2000) |
Virus subtype which infected the blood donors; it was determined in our laboratory either by sequencing the envelope fragment (C2-V5) from the donor plasma virus or by presumption based on donor’s location (subtype B); non-B – plasma virus originated from either Cameroon or India but its subtype was not determined.
Percentage of mutations in nucleotide sequences of the V region of VH and VL is deduced from the V-REGION identity % which is evaluated by IMGT/V-QUEST up to C104 for IGH sequences (which corresponds to FR1 to FR3), and up to codon 109 for IGK and codon 110 for IGL sequences.
Neutral. breadth indicates the number of viruses neutralized, out of 41 tested, by a particular mAb.
Four mAbs produced from single B cells using molecular methods, derived from the same donor.
Two clonally related mAbs derived from the same donor.
The V has insertions (4085 and 3b38) or deletion (1361) which was counted as one point mutation.
All but four mAbs were generated using the hybridoma technology based on fusion of Epstein-Barr virus (EBV)-transformed lymphocytes with the heteromyeloma cell line SHM-D33 (Gorny et al., 1991). Four mAbs were produced using the single B cell method (Li et al., 2012). Sixty-two mAbs have been described in various papers (see references in Table 1), while four mAbs (4415, 4591, 4647 and 4682) were produced for this study using the hybridoma method. Briefly, these latter mAbs were generated from peripheral blood mononuclear cells (PBMC) which were transformed by EBV and screened by ELISA using V3-cholera toxin B (CTB)-fusion proteins containing the V3 consensus sequences from clades A and C (Totrov et al., 2010). The V3-CTB reactive cells were fused with SHM-D33 heteromyeloma, and the resulting hybridoma cells were cloned by limiting dilutions until monoclonality was achieved.
Blood samples were obtained from infected donors who signed informed consent forms, which were approved by the New York University (NYU) and Veteran Affairs (VA) Institutional Review Boards, the Ethics Committee of All India Institute of Medical Sciences, New Delhi, and the National Ethical Committee of Cameroon.
All sequences of the VH and VL regions were deposited in GenBank and the accession numbers are shown in Supplementary Table 1.
2.2. Amplification and nucleotide sequencing of the mAb variable domains
The nucleotide sequences of the heavy (VH) and light (VL) chain genes of the variable domains of mAbs produced by hybridoma technology were determined as described (Gorny et al., 2011; Gorny et al., 2009). Briefly, messenger RNA was extracted from the hybridoma cell lines producing anti-HIV-1 Env mAbs and reverse transcribed into cDNA using oligo dT primer. A homopolymeric tail was added to the 3’ end of cDNA by the terminal deoxynucleotidyl transferase (TdT). The IG genes coding for the VH and VL were amplified from poly-C tailed cDNA by PCR using the forward primer containing anchored tail (Invitrogen) and the reverse primer specific for the constant region encoded by IGHG, IGKC or IGLC genes. PCR amplification was performed using a cycling program, and ethidium bromide-stained 0.8% agarose gels were used to visualize the PCR products. The bands of appropriate size were excised, purified and cloned into the 2.1-TOPO TA cloning vector (Invitrogen); plasmids were transformed into Top10 competent cells. For each heavy and/or light chain, 6 to 12 independent clones were screened. The plasmids with the appropriate inserts were sequenced in both directions using M13 primers. All sequencing reactions were performed at Macrogen, Rockville, MD.
2.3. Analysis of immunoglobulin gene sequences
The nucleotide sequence data of the 66 mAbs were analyzed using Pregap4 and BioEdit software. The percentage of mutations was determined by two methods: a) in the variable (V) regions and b) in the V plus CDR3 (V+CDR3) using IMGT/V-QUEST (Brochet et al., 2008; Giudicelli et al., 2011) and IMGT/JunctionAnalysis (Giudicelli and Lefranc, 2011; Yousfi Monod et al., 2004) from the international ImMunoGeneTics information system® (http://www.imgt.org ) (Lefranc et al., 2014). The V region includes nucleotides up to 2nd-CYS 104 codon which delimits the end of FR3-IMGT of all functional IG V genes of the VH and VL (V-KAPPA and V-LAMBDA) (Lefranc, 2014). This system provides the data about percentage of identical nucleotides in the V region of antibody which allows calculating the percentage of mutations. In addition, we calculated the percentage of mutations in the CDR3 domains which was added to mutations in the V region (V+CDR3); this analysis included the regions of CDR3 which have the corresponding germ line sequences, 5’V-REGION, D-REGION and 3’J-REGION, but not the palindromic (P) and non-templated (N) nucleotides regions.
2.4. Neutralization assay
All 66 anti-HIV-1 Env-specific mAbs and one negative control mAb 1418 were tested in the same laboratory (M. S. Seaman) for neutralizing activities against a standard panel of 41 Env-pseudotyped viruses using the TZM-bl cell line (Li et al., 2005; Seaman et al., 2010). Briefly, this standardized assay consists of seven two-fold serial dilutions of mAbs which were tested at IgG concentration in a range of 50 to 0.4 µg/ml, while four anti-V2 mAbs were tested at 100 to 0.4 µg/ml. The highest IgG concentration used limits the measurement of neutralizing activities of mAbs to 50 µg/ml or for four anti-V2 mAbs to 100 µg/ml. The mAbs were pre-incubated with the virion-containing culture supernatants for one hour. The virus/mAb mixtures were then incubated for 48 hrs with TZM-bl cells expressing CD4, CCR5 and CXCR4. Virus infectivity was determined by measuring luciferase activity in cell lysates. The 50% inhibitory concentration (IC50) was determined as the mAb IgG concentration (μg/ml) that resulted in a 50% reduction in relative light units (RLU) compared to wells with the virus only, after the subtraction of cell control RLUs (Hioe et al., 2010; Seaman et al., 2010).
2.5. Statistical analysis
The neutralizing activity represented by IC50 values was compared between three groups of mAbs (anti-V3, anti-CD4bs and anti-V2) using the Haenszel-Mantel log-rank test as applied to Kaplan-Meier curves, and the neutralizing IC50 data was treated as censored data. Unpaired t test was used for analysis of mutation frequency because the data follow a Gaussian distribution. The breadth-potency (B-P) scores were calculated as described (Zolla-Pazner et al., 2011) with some modifications. Briefly, the reciprocal IC50 values (Fig. 1) were scaled up by 100, and the score based on the log10 of that value. The calculation approximates the area under the curve (AUC) by the intuitive product formula: B-P score = (fraction neutralized) x (average of mAbs which neutralized). The relationship between the percentage of mutations and neutralization breadth was determined by linear regression and by use of the Pearson correlation coefficient (r) with P values. To account for multiple comparisons, the collection of p-values were processed to choose significant comparisons by the false discovery rate (FDR) criterion via the method of Benjami and Hochberg using the R statistical system. Storey’s q-values were computed using the statistical software (http://genomics.princeton.edu/storeylab/qvalue) which also utilizes R (R Foundation for Statistical Computing, Vienna, Austria). Graphs of the data were made using GraphPad Prism version 5 (GraphPad Software, La Jolla, CA).
Figure 1. Neutralizing activity of 66 human anti-HIV-1 gp120 (anti-V3, anti-CD4bs and anti-V2) monoclonal antibodies against a panel of 41 pseudoviruses determined by the TZM-bl cell assay.
All antibodies were tested at IgG concentation in a range of 50 to 0.4 µg/ml, while four anti-V2 mAbs were tested at 100 to 0.4 µg/ml. The IC50 values are shown in each cell, with numbers rounded and color-coded: IC50<1 µg/ml (red); 1-50 µg/ml (yellow). An empty cell indicates that the antibody did not reach 50% neutralization at the maximal concentration (50 or 100 µg/ml). Monoclonal Ab 1418 (designated as “C”) is specific to parvovirus B19 and was used as a negative control. A double horizontal line separates tier 1 from tier 2 and 3 viruses. Breadth (%) indicates the percentage of pseudoviruses in the panel neutralized by a particular mAb. nt – not tested.
3. Results
3.1. Neutralization of pseudoviruses
Three panels of anti-HIV-1 Env mAbs, including 48 anti-V3, 12 anti-CD4bs, and 6 anti-V2 mAbs, were tested to compare their neutralizing potency and breadth against a panel of 41 envelope-pseudotyped viruses. The neutralization potency was defined by IC50 values, which indicate the dose of mAbs needed for 50% neutralization. The neutralization breadth corresponds to the number of viruses neutralized by a particular mAb.
All 66 mAbs mediated neutralization but varied in their potency and breadth of neutralization (Fig. 1). Comparing the IC50 values for neutralization of all 41 viruses (tier 1, 2 and 3), the anti-V3 mAbs were significantly more potent than the anti-CD4bs mAbs (p=0.02) and had a tendency to be stronger than anti-V2 mAbs (p=0.18), while anti-CD4bs and anti-V2 mAbs had similar neutralizing activities (p=0.74) (Fig. 1). The IC50 values were comparable for three panels of mAbs in neutralization of 15 sensitive tier 1 viruses; in contrast, the tier 2, 3 viruses were neutralized, although sporadically, by anti-V3 mAbs but not by anti-CD4bs and anti-V2 mAbs (Fig. 1).
Neutralization breadth was also larger for anti-V3 mAbs, which neutralized between three and 16 of the 41 pseudoviruses (7–39%) while the anti-CD4bs and anti-V2 mAbs neutralized between three and nine pseudoviruses (7–22%) (Fig. 1). The four mAbs with the greatest breadth (>30% of the 41 viruses neutralized) were all V3-specific: mAbs 2191, 2558, 447, and 3869 (Fig. 1). The large neutralization breadth of the anti-V3 mAbs partly depended on their ability to neutralize tier 2, 3 viruses as 27 of 48 (56%) anti-V3 mAbs neutralized one to seven of the 26 tier 2, 3 viruses (Fig. 1).
Neutralization potency strongly correlated with breadth for all 66 mAbs (p<0.0001, Fig. 2) indicating that the more potent antibodies (lower IC50) neutralize a greater range of viruses. Therefore, the breadth-potency (B-P) scores as previously described (Zolla-Pazner et al., 2011) were examined for each mAb, revealing that anti-V3 mAbs have significantly higher B-P scores than anti-CD4bs and anti-V2 mAbs (p=0.02 and p=0.01, respectively) (Fig. 3). The B-P scores for anti-CD4bs and anti-V2 mAbs were comparable without a significant difference (p=0.11) (Fig. 3). Thus, in this panel of conventional mAbs, the anti-V3 mAbs appeared to be more efficient in neutralization compared to anti-CD4bs and anti-V2 mAbs.
Figure 2. Neutralization potency and breadth of all 66 human neutralizing anti-HIV-1 gp120 mAbs correlated.
The neutralization potency of 66 mAbs, represented by the reciprocal average of IC50 (y-axis), is strongly correlated with neutralization breadth expressed as the number of neutralized pseudoviruses (x-axis) of the total of 41 viruses tested. r - Pearson correlation coefficient.
Figure 3. The breadth-potency scores (B-P scores) of anti-HIV-1 mAbs.
The B-P score for each mAb accounts for both the number of neutralized pseudoviruses (breadth) and its neutralization potency (IC50) and was calculated as described (Zolla-Pazner et al., 2011). The anti-V3 mAbs have significantly higher B-P scores compared to anti-CD4bs and anti-V2 mAbs.
Our panel of 66 mAbs were derived from individuals infected with different viruses. The anti-CD4bs and anti-V2 mAbs were derived from subtype B viruses, while 21 and 27 anti-V3 mAbs were derived from subtypes B and non-B infected donors, respectively. The neutralization breadth and potency were comparable between both groups of anti-V3 mAbs, suggesting that the infecting virus of different subtypes induced anti-V3 antibodies with comparable neutralizing activities (data not shown).
3.2. Somatic mutation analysis
As the recognition function of antibodies depends on affinity maturation which is driven by somatic mutations, we analyzed the percentage of mutations in the 66 mAbs using IMGT/V-QUEST which identifies nucleotides not identical to the closest germline sequence (Brochet et al., 2008; Giudicelli et al., 2011). The percentage of mutations in the V region ranged between 2.43% to 18.06% (mean 8.57%) for VH and 0.35% to 13.98% (mean 5.74%) for VL (Table 1). In the V+CDR3 the range of the percentage of mutations was very similar: 2.4% to 18.6% (mean 8.78%) for VH and 0.4% to 13.9% (mean 5.78%) for VL (Supplementary Table 1), and the mutations in both analysis correlated very strongly (r=0.98, p<0.0001) (data not shown). When the three panels of mAbs, anti-V3, anti-CD4bs and anti-V2, were compared to each other, no significant difference in the percentage of mutations was found except that of VL mutations for anti-V2 mAbs (Fig. 4A, 4B, 4D, 4E). In the three mAbs the nucleotide insertions were found in VH (4085 and 3b38) or deletion in VL (1361) which were counted as one point mutation (Table 1).
Figure 4. Percentage of point mutations in the nucleotide sequence of the variable region of mAbs.
The percentage of mutations in the V (A, B) and V+CDR3 regions (D, E) of the VH and VL of the 66 mAbs as determined by IMGT/V-QUEST and IMGT/JunctionAnalysis. Statistical significance was determined by t test. The percentage of mutations in the VH and VL analyzed in the V (C ) and V+CDR3 (F) are strongly correlated; r – Pearson correlation coefficient.
For an individual antibody, the percentage of mutations is usually higher in VH than in VL, and this was observed in our panel of mAbs as well, except for three mAbs – 3019, 3906 and 4682 – in which the percentage of mutations was higher in VL than VH by more than 2.0% (Table 1). Despite this small difference, there was a strong correlation between the percentage of mutations in the VH and VL (Fig. 4C, 4F) confirming that mutations in both chains proceed in parallel.
Thus, the percentage of mutations was very broad in both VH and VL and very similar when calculated in the V and V+CDR3 regions of IG genes, moreover, very similar for each type of mAbs except of anti-V2 mAbs possibly due to a smaller size of this panel.
3.3. Study of correlation between neutralization breadth and somatic mutations
We further analyzed the relationship between the neutralization breadth and the percentage of mutations in the V region of the VH and VL by linear regression and by use of the Pearson correlation coefficient (r) with P values (Fig. 5A, 5B). The correlation was moderate (low r) and was found between the percentage of mutations in the VL, but not in the VH, and the neutralization breadth (r=0.25, p=0.04), Fig. 5B and 5A, respectively.
Figure 5. Correlation between the neutralization breadth and percentage of mutations in the variable (V) region or in V+CDR3 of 66 anti-HIV-1 mAbs.
The percentage of mutations is shown for the VH and VL in the V region only (A, B) and in the V+CDR3 for VH and VL (C, D); this was determined using IMGT/V-QUEST. The mutations in the CDR3 were determined only in the regions corresponding to germline genes (5’V-REGION, D-REGION and 3’J-REGION) and were analyzed using IMGT/JunctionAnalysis integrated in IMGT/V-QUEST. The two methods showed that the percentage of mutations correlated moderately with neutralization breadth when determined in the VL domains (B and D) but not in the VH domains (A and C); r – Pearson correlation coefficient.
The correlation limited only to the VL region was surprising because the VH usually dominates the interaction between an antibody and its antigen epitope. As the VH CDR3 is the most involved from the six CDRs in the interaction with the antigens, we tested whether mutations in CDR3 may have an impact on the correlation with neutralization breadth. The results of analysis in the V+CDR3 were similar to those in Fig. 5A and 5B showing moderate correlation in the VL (r=0.24, p=0.04; Fig. 5D), but not in the VH (Fig. 5C).
We continued the analysis using the percentage of mutations in the V+CDR3 of the VH and VL of all 66 mAbs and compared it with the neutralization of two panels of pseudoviruses: 15 sensitive to neutralization tier 1 and 26 more resistant tier 2/3 viruses (Fig. 6). The percentage of mutations in the VH and VL was not correlated with neutralization of tier 1 viruses (Fig. 6A, 6B), but a moderate correlation was found between the percentage of mutations in VL (r=0.31, p=0.01), and some tendency for the mutations in VH, and neutralization of tier 2, 3 viruses (r=0.24, p=0.05), Fig.6D and 6C, respectively.
Figure 6. Correlation between the percentage of mutations and neutralization of tier 1 and tier 2, 3 pseudoviruses.
The percentage of mutations in the V+CDR3 of the VH (A, C) and VL (B, D) of 66 human mAbs is compared to neutralization breadth for tier 1 viruses (A, B), and tier 2, 3 viruses (C, D). Tendency to ( C) or moderate correlation (D) was determined between percentage of mutations and neutralization breadth of tier 2, 3 viruses but not tier 1 viruses; r – Pearson correlation coefficient.
3.4. The VH CDR3 features that may influence the antibody neutralizing function
Lack of correlation between the percentage of mutations in V+CDR3 in the VH (p=0.05) and neutralization breadth may result from an incomplete analysis of mutations in the CDR3, in contrast to the VL (p=0.01) (Fig. 6). The VH CDR3 contains an increased number of P and N nucleotides compared to VL CDR3, mean 16.6 versus 2.6, respectively (p<0.0001), which have no corresponding germline gene sequences (Lefranc et al., 2014) and the percentage of mutations cannot be determined (Fig. 7).
Figure 7. Number of palindromic (P-REGION) and non-templated nucleotides (N1-REGION and N2-REGION for VH, N-REGION for VL) in the VH CDR3 and VL CDR3 of 66 mAbs.
The number of nucleotides (P and N) was determined using IMGT/JunctionAnalysis integrated in IMGT/V-QUEST (http://www.imgt.org). These nucleotide sequences have no corresponding germline genomic sequences.
To determine if other features of the VH CDR3 may have some relationship with neutralization breadth, the amino acid (AA) length and isoelectric point (pI) were evaluated using IMGT/JunctionAnalysis (Giudicelli and Lefranc, 2011; Yousfi Monod et al., 2004) and the hydropathy value evaluated using GRAVY calculator (www.gravy-calculator.de)(Fig. 8). None of these features, the VH CDR3 length, hydropathy and pI correlated with the neutralization breadth (data not shown).
Figure 8. Analysis of three features of VH CDR3 of 66 mAbs against V3, CD4bs and V2 and their relationships with neutralization breadth.
The VH CDR3 length (A) and isoelectric point (pI) (C) were determined using IMGT/JunctionAnalysis while hydropathy (B) was determined using GRAVY calculator. The VH CDR3 length and hydropathy were comparable for mAbs that neutralized tier 1 only and tier 1, 2, 3 viruses (A, B) but the pI was significantly lower for mAbs that neutralized tier 1, 2, 3 (C). The statistical difference was determined using t tests.
Since the percentage of mutations in the V region of the VL domain correlated with neutralization breadth of tier 2, 3 viruses but not with tier 1 viruses, we also analyzed VH CDR3 features in two groups of mAbs, those neutralizing tier 1 viruses only and those neutralizing tier 1, 2, 3 viruses (Fig. 8). It was found that the VH CDR3 length and hydropathy was comparable for mAbs neutralizing the two panels of pseudoviruses (Fig. 8A, 8B), while pI was significantly lower (p<0.0001) for mAbs neutralizing tier 1, 2, 3 viruses versus mAbs neutralizing tier 1 viruses (Fig. 8C).
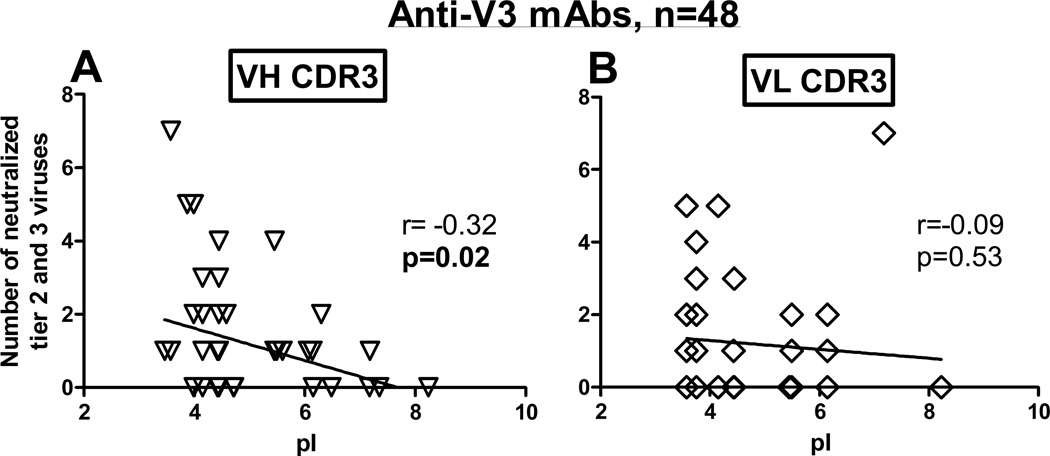
Because the tier 2, 3 viruses were neutralized only by anti-V3 mAbs, we tested whether the pI of the CDR3s of 48 mAbs have any association with neutralization breadth. It was found that the pI of the VH CDR3, but not the pI of the VL CDR3, inversely correlated with the number of neutralized tier 2, 3 viruses (r=−0.32, p=0.02) (Fig. 9A, 9B). Thus, the lower pI of the VH CDR3 of anti-V3 mAbs correlated with more tier 2, 3 viruses neutralized. These data suggest that the pI of the VH CDR3 may play role in the neutralizing activity of anti-V3 mAbs against tier 2, 3 viruses.
Figure 9. Isoelectric point (pI) of the VH CDR3 in anti-V3 mAbs inversely correlates with the number of neutralized tier 2, 3 pseudoviruses.
The pI of the VH CDR3 (A) and VL CDR3 (B) were determined using IMGT/JunctionAnalysis. The pI of the VH CDR3 (A) inversely correlated with the increasing number of neutralized tier 2, 3 pseudoviruses; r – Pearson correlation coefficient.
The collection of 27 p-values in the study was additionally tested to exclude false positives via the method of Benjamini and Hochberg methodology and Storey q-values and the primary results were confirmed.
4. Discussion
The two objectives of this study, the comparative analysis of the neutralizing activities of human anti-V3, anti-CD4bs and anti-V2 mAbs derived from chronically HIV-infected subjects, and the relationships between neutralization breadth and the percentage of mutations, are relevant to the development of an HIV-1 vaccine. First, the mAbs tested in this study may correspond to antibodies elicited by vaccines as they are not selected for elite neutralizing activity in serum, and represent the commonly induced antibodies in HIV-1 infected individuals. Second, the rate of mutations in some mAbs displaying neutralization breadth was relatively low and comparable to mutations in mAbs derived from recipients of candidate HIV-1 vaccines (Liao et al., 2013; Montefiori et al., 2012; Moody et al., 2012).
This comparative study of three types of antibodies revealed significantly higher neutralization potency and breadth of anti-V3 mAbs over the anti-CD4bs and anti-V2 mAbs. This depended mainly on the ability of anti-V3 mAbs to neutralize, along with tier 1, some tier 2, 3 viruses while anti-CD4bs and anti-V2 mAbs neutralized sensitive tier 1 viruses only; neutralization of tier 1 viruses was comparable for all three panels of mAbs. Anti-V3 mAbs are well known for their ability to cross-neutralize (Andrabi et al., 2013; Corti et al., 2010; Hioe et al., 2010; Mouquet et al., 2011; Pantophlet et al., 2007; Scheid et al., 2009), and in our study, they neutralized in a range from 7% to 39% of the 41 viruses representing the sequences of subtypes A, AG, B and C. Nine of these anti-V3 mAbs (Table 1) can be listed as bnAbs based on two reviews which classify bnAbs as those exhibiting neutralization breadth in the range of 25–98% and 32–98% (van Gils and Sanders, 2013; Verkoczy et al., 2011).
The mAbs against CD4bs and V2 region displayed lower neutralization breadth in a range from 7% to 22% and were not able to neutralize tier 2, 3 viruses which might be related to limited accessibility of the corresponding epitopes on native virions. Notably, anti-CD4bs and anti-V2 compared to anti-V3 mAbs were much less efficient in binding to intact virions, virus-infected and envelope-transfected cells (Liu et al., 2013; Nyambi et al., 2000; Zolla-Pazner et al., 1995). This difference between 3 types of mAbs was well illustrated by the results of neutralization of the JRFL virus which is classified as tier 2 and has a cryptic V3 region due to masking by the V2 region (Bou-Habib et al., 1994; Pinter et al., 2004). In our panel of pseudoviruses, the JRFL.JB was neutralized by 12 out of 48 anti-V3 mAbs in a range of IC50 from 5 to 50 µg/ml, while being completely resistant to neutralization by anti-CD4bs and anti-V2 mAbs (Table 1, Fig.1).
The recent structural studies of an HIV-1 constrained gp140 trimer, SOSIP.664, revealed a close association between V1V2 and V3; access to V3 appeared to be restricted by association with V1V2, while V1V2 was itself shielded by N-glycosylation (Julien et al., 2013). Access to the CD4bs is also restricted by spatial arrangements of V1V2 and V3 and also by glycans (Lyumkis et al., 2013). The accessibility of the V3 region for antibody binding is increased by binding of soluble CD4 and several anti-CD4bs antibodies which results in conformational changes in gp120 leading to enhanced exposure of the V3 loop (Mbah et al., 2001; Thali et al., 1992; Upadhyay et al., 2014; Verrier et al., 2001). These structural requirements in the gp120 trimer can decrease the binding affinity of anti-V3, anti-CD4bs and anti-V2 antibodies to unliganded virus resulting in their limited neutralization potency and breadth, especially in tier 2, 3 viruses.
The percentage of mutations, which we used in this study to assess antibody maturation and relationship with neutralization, contributes to binding affinity but is not directly related. This may explain the only moderate correlation between the percentage of mutations and neutralization of tier 2, 3 viruses, but not tier 1 viruses, which possibly requires higher antibody affinity. It is in part confirmed by our results which showed that the relative affinity binding to a biotinylated cyclic V3A244 peptide of a selected group of 22 anti-V3 mAbs encoded by the IGHV5–51/IGLV lambda genes (Gorny et al., 2011; Jiang et al., 2010), and which are included in this study, inversely correlated with the neutralization breadth of tier 1, 2, 3 viruses but not with the percentage of mutations (Supplementary Fig. 1). Moreover, the increased percentage of mutations in mAbs is not strictly related to neutralization breadth because mAbs with both a low (2.4% to 8.7%) and high rate of somatic mutations (8.8% to 18.6%) can neutralize a similar number of viruses (Supplementary Figure 2).
The broad range of the percentage of mutations between 2% and 18% was found (mean 8.8%), both in the V and V+CDR3 regions of the heavy chains. A similarly low percentage of somatic hypermutations in the VH, mean 7.9%, was observed in another study of 25 neutralizing mAbs against V3, CD4bs and CD4-induced epitopes which were isolated from one chronically HIV-1 infected individual (Ramirez Valdez et al., 2014). However, only a moderate correlation between the percentage of mutations and neutralization of tier 2, 3, but not tier 1 viruses, was determined. The mAbs with a minimal and high percentage of mutations displayed cross-neutralizing activities, for example, a mAb with 2.4% mutations was able to cross-clade neutralize 8 pseudoviruses (Table 1). The tier 1 viruses can even be neutralized by mAbs without somatic mutations in the V region as it was shown recently that the three germline-reverted anti-V3 mAbs, which retained the VH CDR3 sequence, neutralized several tier 1 viruses (McGuire et al., 2014).
The tier 2, 3 viruses were neutralized sporadically by some anti-V3 mAbs only and their percentage of mutations, particularly in the VL regions, was moderately correlated with neutralization breadth. This indicated that access to the V3 epitopes on tier 2, 3 viruses is very limited and possible for mAbs with higher percentage of mutations. Since the VH CDR3 is the main domain of each antibody interacting with the antigen some of its features may have an impact on neutralization and it was found that the isoelectric points, but not the length and hydrophobicity, were inversely correlated with the neutralization breadth. These results underline the role of the electrostatic interactions of the anti-V3 mAbs with the V3 loop which has the overall positive charge in the range from +2 to +10 (Kwong et al., 2000).
In conclusion, the results of this study revealed the significantly higher neutralizing activity for anti-V3 mAbs over the anti-CD4bs and anti-V2 mAbs which were all generated from randomly selected HIV-1 infected individuals. This dominant neutralization breadth of anti-V3 mAbs was dependent on their ability to neutralize tier 1 along with some tier 2, 3 viruses, while anti-CD4bs and anti-V2 mAbs neutralized only tier 1 viruses. While the percentage of mutations in the V region of the VH and VL including CDR3 was not correlated with the neutralization breadth of tier 1 viruses, a moderate correlation was found with neutralization of tier 2, 3 viruses mediated by anti-V3 mAbs. The isoelectric points of the VH CDR3 of anti-V3 mAbs inversely correlated with the neutralization of tier 2, 3 viruses suggesting that the electrostatic interactions between anti-V3 mAbs and the V3 crown of the virus which is positively charged may contribute to their neutralizing activity against these viruses.
Supplementary Material
Highlights.
-
▶
Sixty-six human mAbs against V3, CD4bs and V2 were studied.
-
▶
Anti-V3 mAbs have higher neutralizing potential than anti-CD4bs and anti-V2 mAbs.
-
▶
The majority of anti-V3 mAbs neutralized some tier 2 and 3 viruses.
-
▶
The percentage of mutations of mAbs partly correlated with neutralization breadth.
-
▶
A lower pI of the VH CDR3 inversely correlated with neutralization
Acknowledgements
This study was supported in part by NIH grants AI091543, AI096977 (MKG), HL59725, AI36085 (SZP), P01 AI100151 (SZP and X-P Kong) and research funds from the Department of Veterans Affairs. Monoclonal antibodies were supplied with support from the NIH AIDS Research and Reference Reagent Program. The authors thank Drs. Alok Choudhary and Raiees Andrabi for their assistance in conducting these studies. We thank Drs. Susanne Tranguch and Michelle Ryndak for critical review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure
The authors declare no competing financial interests.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version, at
References
- Andrabi R, Williams C, Wang XH, Li L, Choudhary AK, Wig N, Biswas A, Luthra K, Nadas A, Seaman MS, Nyambi P, Zolla-Pazner S, Gorny MK. Cross-neutralizing activity of human anti-V3 monoclonal antibodies derived from non-B clade HIV-1 infected individuals. Virology. 2013;439:81–88. doi: 10.1016/j.virol.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, Norcross MA. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O’Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PloS one. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigler A, Dorsch S, Hemauer A, Williams C, Kim S, Young NS, Zolla-Pazner S, Wolf H, Gorny MK, Modrow S. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J. Virol. 1999;73:1974–1979. doi: 10.1128/jvi.73.3.1974-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harbor protocols. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- Giudicelli V, Lefranc MP. IMGT/junctionanalysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR) Cold Spring Harbor protocols. 2011;2011:716–725. doi: 10.1101/pdb.prot5634. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse HIV-1 variants by an anti-V3 human monoclonal antibody. J. Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Mascola JR, Israel ZR, VanCott TC, Williams C, Balfe P, Hioe C, Brodine S, Burda S, Zolla-Pazner S. A human monoclonal antibody specific for the V3 loop of HIV type 1 clade E cross-reacts with other HIV type 1 clades. AIDS Res. Hum. Retroviruses. 1998;14:213–221. doi: 10.1089/aid.1998.14.213. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of HIV-1. J. Virol. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov CP, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 2004;78:2394–2404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang X-H, Williams C, O’Neal T, Volsky B, Li L, Cardozo T, Nyambi P, Zolla-Pazner S, Kong X-P. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PloS one. 2011;6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, 2nd, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Williams C, Revesz K, Zolla-Pazner S. Effects of oligomerization on the epitopes of the Human Immunodeficiency Virus Type 1 envelope glycoproteins. Virology. 2000;267:220–228. doi: 10.1006/viro.1999.0095. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Wang XH, Williams C, Volsky B, Revesz K, Witover B, Burda S, Urbanski M, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S, Nadas A. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol. Immunol. 2009;46:917–926. doi: 10.1016/j.molimm.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov CP, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize HIV-1 primary isolates from various clades. J. Virol. 2002;76:9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Koning FA, Nadas A, Anyangwe C, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of HIV-1. J. Virol. 2006;80:6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu J-Y, Gianakakos V, Karwowska S, Williams C, Sheppard HW, Hanson CV, Zolla-Pazner S. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3238–3242. doi: 10.1073/pnas.88.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu J-Y, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PloS one. 2013;8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. New Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PloS one. 2010;5:e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs SA, Gorny MK, Williams C, Revesz K, Volsky B, Burda S, Wang XH, Bandres J, Zolla-Pazner S, Holmes H. Characterization of human monoclonal antibodies selected with a hypervariable loop-deleted recombinant HIV-1(IIIB) gp120l. Immuno. Letters. 2001;79:209–213. doi: 10.1016/s0165-2478(01)00289-9. [DOI] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong X-P. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 2010;17:955–961. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwowska S, Gorny MK, Buchbinder A, Gianakakos V, Williams C, Fuerst T, Zolla-Pazner S. Production of human monoclonal antibodies specific for conformational and linear non-V3 epitopes of gp120. AIDS Res. Hum. Retroviruses. 1992;8:1099–1106. doi: 10.1089/aid.1992.8.1099. [DOI] [PubMed] [Google Scholar]
- Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 1994;68:400–410. doi: 10.1128/jvi.68.1.400-410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP. Immunoglobulin and T Cell Receptor Genes: IMGT((R)) and the Birth and Rise of Immunoinformatics. Front. Immunol. 2014;5:22. doi: 10.3389/fimmu.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, Hadi-Saljoqi S, Sasorith S, Lefranc G, Kossida S. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015;43:D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang X-H, Banerjee S, Volsky B, Williams C, Virland D, Nadas A, Seaman MS, Chen X, Spearman P, Zolla-Pazner S, Gorny MK. Different pattern of immunoglobulin gene usage by HIV-1 compared to non-HIV-1 antibodies derived from the same infecgted subject.e. PloS one. 2012;7:e39534. doi: 10.1371/journal.pone.0039534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J. Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Yates NL, Shen X, Bonsignori M, Moody MA, Liao HX, Fong Y, Alam SM, Overman RG, Denny T, Ferrari G, Ochsenbauer C, Kappes JC, Polonis VR, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Montefiori DC, Gilbert P, Michael NL, Kim JH, Haynes BF, Tomaras GD. Infectious virion capture by HIV-1 gp120-specific IgG from RV144 vaccinees. J. Virol. 2013;87:7828–7836. doi: 10.1128/JVI.02737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Members CCT, Dersimonian R, Euler Z, Gray ES, Abdool Karim S, Kirchherr J, Montefiori DC, Sibeko S, Soderberg K, Tomaras G, Yang ZY, Nabel GJ, Schuitemaker H, Morris L, Haynes BF, Mascola JR. The development of CD4 binding site antibodies during HIV-1 infection. J. Virol. 2012;86:7588–7595. doi: 10.1128/JVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbah HA, Burda S, Gorny MK, Williams C, Revesz K, Zolla-Pazner S, Nyambi PN. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 2001;75:7785–7788. doi: 10.1128/JVI.75.16.7785-7788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J. Infect. Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, Yu JS, Zhang R, Meyerhoff RR, Parks R, Scull JC, Wang L, Vandergrift NA, Pickeral J, Pollara J, Kelsoe G, Alam SM, Ferrari G, Montefiori DC, Voss G, Liao HX, Tomaras GD, Haynes BF. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J. Virol. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PloS one. 2011;6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 1998;72:9384–9391. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi PN, Mbah HA, Burda S, Williams C, Gorny MK, Nadas A, Zolla-Pazner S. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 2000;74:7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Aguilar-Sino RO, Wrin T, Cavacini LA, Burton DR. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology. 2007;364:441–453. doi: 10.1016/j.virol.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 2004;78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez Valdez KP, Kuwata T, Maruta Y, Tanaka K, Alam M, Yoshimura K, Matsushita S. Complementary and synergistic activities of anti-V3, CD4bs and CD4i antibodies derived from a single individual can cover a wide range of HIV-1 strains. Virology. 2014;475C:187–203. doi: 10.1016/j.virol.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Furman C, Wahren B, Posner M, Ho DD, Robinson J, Sodroski J. Cooperativity of neutralizing antibodies directed against the V3 and CD4 binding regions of the human immunodeficiency virus gp120 envelope glycoprotein. J. Acquir. Immun. Defic. Syndrome. 1992;5:591–599. [PubMed] [Google Scholar]
- Totrov M, Jiang X, Kong XP, Cohen S, Krachmarov C, Salomon A, Williams C, Seaman MS, Cardozo T, Gorny MK, Wang S, Lu S, Pinter A, Zolla-Pazner S. Structure-guided design and immunological characterization of immunogens presenting the HIV-1 gp120 V3 loop on a CTB scaffold. Virology. 2010;405:513–523. doi: 10.1016/j.virol.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay C, Mayr LM, Zhang J, Kumar R, Gorny MK, Nadas A, Zolla-Pazner S, Hioe CE. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J. Virol. 2014;88:12853–12865. doi: 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, Sanders RW. Broadly neutralizing antibodies against HIV-1: templates for a vaccine. Virology. 2013;435:46–56. doi: 10.1016/j.virol.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Current opinion in immunology. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier F, Nadas A, Gorny MK, Zolla-Pazner S. Additive effects characterize the interaction of antibodies involved in neutralization of the primary dualtropic human immunodeficiency virus type 1 isolate 89.6. J. Virol. 2001;75:9177–9186. doi: 10.1128/JVI.75.19.9177-9186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Kurth R, Norley S. The majority of neutralizing Abs in HIV-1-infected patients recognize linear V3 loop sequences. Studies using HIV-1MN multiple antigenic peptides. J. Immunol. 1994;153:1895–1904. [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J Junctions. Bioinformatics. 2004;20(Suppl 1):379–385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- Zhong P, Burda S, Urbanski M, Kenfack H, Tongo M, Heyndrickx L, Nanfack A, Shang J, Agyingi L, Zolla-Pazner S, Zekeng L, Nyambi P. HIV Type 1 Group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J. Acquir. Immune Defic. Syndromes. 2002;31:495–505. doi: 10.1097/00126334-200212150-00007. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S. A critical question for HIV vaccine development: which antibodies to induce? Science. 2014;345:167–168. doi: 10.1126/science.1256526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, deCamp AC, Cardozo T, Karasavvas N, Gottardo R, Williams C, Morris DE, Tomaras G, Rao M, Billings E, Berman P, Shen X, Andrews C, O’Connell RJ, Ngauy V, Nitayaphan S, de Souza M, Korber B, Koup R, Bailer RT, Mascola JR, Pinter A, Montefiori D, Haynes BF, Robb ML, Rerks-Ngarm S, Michael NL, Gilbert PB, Kim JH. Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PloS one. 2013;8:e53629. doi: 10.1371/journal.pone.0053629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Kong X, Jiang X, Cardozo T, Nadas A, Cohen S, Totrov M, Seaman MS, Wang S, Lu S. Cross-clade HIV-1 neutralizing antibodies induced with V3-scaffold protein immunogens following priming with gp120 DNA. J. Virol. 2011;85:9887–9898. doi: 10.1128/JVI.05086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, O’Leary J, Burda S, Gorny MK, Kim M, Mascola J, McCutchan FE. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J. Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.