Abstract
Objectives
Pharmacogenetic testing is projected to improve health outcomes and reduce the cost of care by increasing therapeutic efficacy and minimizing drug toxicity. American Indian and Alaska Native (AI/AN) people historically have been excluded from pharmacogenetic research and its potential benefits, a deficiency we sought to address. The vitamin K antagonist warfarin is prescribed for prevention of thromboembolic events, although its narrow therapeutic index and wide inter-individual variability necessitate close monitoring of drug response. Therefore, we were interested in variation in CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX, which encode enzymes important for the activity of warfarin and synthesis of vitamin K dependent blood clotting factors.
Methods
We resequenced these genes in 188 AI/AN people in partnership with Southcentral Foundation (SCF) in Anchorage, AK and 94 Yup'ik people living in the Yukon-Kuskokwim Delta of southwest Alaska to identify known or novel function-disrupting variation. We conducted genotyping for specific SNPs in larger cohorts of each study population (380 and 350, respectively).
Results
We identified high frequencies of the lower-warfarin dose VKORC1 haplotype (−1639G>A and 1173C>T) and the higher-warfarin dose CYP4F2*3 variant. We also identified two relatively common, novel, and potentially function-disrupting variants in CYP2C9 (M1L and N218I), which, along with CYP2C9*3, CYP2C9*2 and CYP2C9*29, predict that a significant proportion of AI/AN people will have decreased CYP2C9 activity.
Conclusions
Overall, we predict a lower average warfarin dose requirement in AI/AN populations in Alaska than that seen in non-AI/AN populations of the US, a finding consistent with clinical experience in Alaska.
Keywords: indigenous populations, underserved populations, Coumadin, cytochrome P450, individualized therapy, personalized medicine
INTRODUCTION
Genetic variation affects the pharmacokinetics and pharmacodynamics of medical drugs by altering enzyme and transporter expression and function, resulting in differences in drug efficacy and safety between individuals [1-5]. The prevalence and frequency of genetic variation affecting pharmacologic response is diverse across racial and ethnic populations [6-8]. As a result, populations that are rarely included in medical research, such as American Indian and Alaska Native (AI/AN) people, are less likely to benefit from genome-based, personalized, drug therapy [6-8]. Thus, pharmacogenetic research in these understudied populations is needed to improve both the selection of drugs and their dosages, and to reduce the number of adverse effects [6-8].
The incidence of stroke is disproportionately high in AN communities, with AN people more likely to be affected by stroke at an earlier age than other populations [9-11] and death rates due to stroke being approximately 20% greater than for individuals in other racial and ethnic groups in the United States [9]. An oral vitamin K antagonist, warfarin, is used to prevent thromboembolic events, but it has a narrow therapeutic index and is affected by wide inter-individual and inter-ethnic dose variability (up to 20-fold), requiring intensive dose management to prevent adverse bleeding events, which can be difficult when patients live in remote communities [12-15].
Over the last 7 years, the average stable anticoagulation dose used in long-term care (> 22 weeks of treatment) for the AI/AN people receiving drug therapy at Southcentral Foundation (SCF) in Anchorage, AK was found to be lower than the average for other sites in North America and Europe, using the same DAWN AC Anticoagulation Management Software (4S Information Systems Ltd., Cumbria, England). For a target INR of 2.5, with a mean INR of 2.0 to 3.0, the average dose at SCF was 4.5 mg/day, with a percent time within INR of 69.7%. For all other locations, the average dose was 4.9 mg/day, with a percent time within INR of 73.0% [16]. These differences in dose may be attributed to inter-individual and inter-ethnic differences, including differences in dietary vitamin K consumption, drug-drug interactions, age, body surface area, gender, concurrent health conditions (e.g., diabetes), and genetic polymorphisms [17-20].
To explore the sources of this reported lower dose among AI/AN people, we characterized the variation in 5 genes that have been associated with altered warfarin response. These include the genes that encode the following enzymes: 1) the pharmacological target of warfarin (VKOR), which reduces vitamin K-epoxide to its active form; 2) the major cytochrome P450 enzyme that clears (S)-warfarin (CYP2C9); 3) two enzymes that catabolize vitamin K (CYP4F2 and CYP4F11); and 4) the γ-carboxylase that activates vitamin K-dependent clotting factors (e.g., Factors II, VII IX and X) (GGCX). It is estimated that by combining knowledge of VKORC1, CYP2C9, and CYP4F2 genotypes with readily accessible clinical factors, including age, gender, and body mass index (BMI), more than 60% of the variance in warfarin dosage can be explained in European-American populations [21].
To assess novel variation in CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX, each gene was resequenced in a sample of AI/AN study participants. Population frequencies were determined for alleles that had been previously associated with the dose of warfarin in other populations and for novel non-synonymous alleles that were discovered during resequencing. We hypothesized that there could be novel, function-disrupting variation or higher frequencies of known gene variants in this population that could reduce warfarin dose requirement and impact bleeding/thrombotic risk.
METHODS
Setting
As of 2012, 106,260 AI/AN people live in Alaska, with approximately 1/3 living in more densely populated areas such as Anchorage, Fairbanks, and Juneau, and 2/3 living primarily in rural communities with populations of 50 to 1000 people, with many of the communities accessible only by air or water [22]. Geographic isolation leads to substantial portions of AI/AN people in Alaska being medically under-served and having considerable health disparities compared to other populations [22].
Southcentral Foundation (SCF), a tribally owned and operated regional health corporation, provides pre-paid healthcare services to 58,000 AI/AN patients, who are considered “customer-owners” of SCF. The Anchorage Service Unit (ASU) served by SCF is comprised of both urban and rural areas, including Anchorage, the Matanuska-Susitna Borough, and 60 outlying villages (most with fewer than 500 residents). It provides primary care services to ~55% of the total AN population at 6 SCF primary care clinics on the Alaska Native Medical Center (ANMC) campus. Tertiary care is provided at the 150-bed ANMC hospital, which is co-owned and co-managed by SCF and Alaska Native Tribal Health Consortium (ANTHC).
The Center for Alaska Native Health Research (CANHR) is based at the University of Alaska Fairbanks. CANHR has an ongoing genetic research partnership with 11 of the 58 rural communities in the Yukon-Kuskokwim River Delta (Y-K Delta) that are served by the Yukon-Kuskokwim Health Corporation (YKHC). The YKHC is based in Bethel and provides healthcare to about 23,000 Yup'ik people. CANHR collaborates with Yup'ik communities in community-based participatory research focused on understanding, preventing, and reducing health disparities.
Institutional Review Board (IRB) Approval
The Alaska Area Institutional Review Board (IRB), and the SCF and ANTHC tribal review boards approved work conducted at SCF on the ANMC campus. The YKHC Executive Board of Directors and the University of Alaska Fairbanks IRB approved the work conducted in the Y-K Delta by CANHR. The University of Washington (UW) IRB approved the overall research project, as UW is the academic home of the grant funding this research (Pharmacogenetics in Rural and Underserved Populations) and its principal investigators. The National Institute of General Medical Sciences and the Indian Health Service granted a Certificate of Confidentiality for protection of participant information, and the respective Alaska IRBs approved forms for written consent prior to initiating research. Research questions were developed through community based participatory research at SCF and CANHR.
Study Participants
A convenience sample of study participants (n = 380) was obtained through recruitment by research staff members at SCF's primary care clinics. Any AI/AN person ≥18 years of age and receiving care at SCF was eligible to participate in the study. Surveys collected self-reported gender, date of birth, and tribal affiliation. A representative subset (n=188) was used for resequencing of CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX genes.
A convenience sample of 350 residents of the Y-K Delta, ≥18 years of age, was recruited using written and oral advertisement during research-focused community visits by the CANHR research personnel. All CANHR participants self-identified as Yup'ik. A subset of 94 individuals was chosen for targeted resequencing of CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX; because all participants were recruited from the same communities, these 94 individuals were selected from the set of 350 individuals on the basis of being unrelated according to either pedigree-based kinship coefficients obtained from available genealogical information [23] as well as empirical kinship coefficients calculated using the KING (kinship-based inference for GWASs)-robust method [24] for sample individuals with genome-wide SNP genotyping data available.
Specimen Processing and Storage
Buffy coats were extracted from blood that was collected into EDTA-coated tubes (BD Vacutainer® CPT™), centrifuged (900 × g, 15 min) at room temperature, incubated with Puregene RBC Lysis Solution for 10 minutes, and centrifuged again (1800 × g, 10 min) at room temperature. White blood cells from CANHR samples were then re-suspended in 10 mL Puregene Cell Lysis Solution until DNA purification. At CANHR, genomic DNA was isolated using the Gentra Puregene kit (Qiagen, Valencia, California, USA) prior to shipment to UW investigators. White blood cells from SCF samples were washed in phosphate-buffer saline (1X PBS), centrifuged again (800 × g, 6 min) at room temperature, then re-suspended in PBS and frozen (−80°C) until shipped to UW investigators for DNA isolation. Genomic DNA from the samples of SCF participants was isolated using a QIAamp DNA Blood Midi/Maxi kit (Qiagen, Valencia, California, USA). Quality and concentration of DNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware, USA).
Gene Resequencing Methods
Exons, 50-100 base pairs into each adjacent intronic region, 1000-4000 base pairs in the 5’ flanking region, and 150-300 base pairs into 3’ flanking regions were resequenced using PCR amplicons ~500–600 bp in size, with amplicons containing overlapping segments of ~150 bp to validate primer binding sites and to prevent allele-specific amplification [25]. The PCR primers were standardized with a universal M13-tailed PCR sequence, and used in conventional Sanger sequencing reactions using BigDye chemistry under standard conditions and separated on an ABI 3730 DNA analyzer (Life Technologies, Grand Island, NY, USA). Chromatograms were analyzed using Phrap software (UW, Seattle, Washington, USA) for base calling and quality assignment, and Consed software (UW) was used for assembly and editing [26]. Single nucleotide polymorphisms (SNPs) and small insertions/deletions were identified through pairwise comparison of chromatogram peak heights/intensities using the PolyPhred program (version 5.0; UW) [27, 28], which produces chromatograms averaging greater than 500 bp and averaging a Phred quality greater than 40 (corresponding to a 1/10,000 probability of incorrect base assignment) [29]. Data were verified using second-strand confirmation. Automated scripts were used to map variants onto the intron–exon gene structure. For the CYP2C9 and VKORC1 haplotype analysis, sites were based on human reference sequence AL359672.
Coding variants CYP2C9 M1L, N218I, and P279T (CYP2C9*29) were analyzed for PolyPhen2 and Grantham scores to predict the phenotypic effect of the amino acid change on enzyme function [30, 31].
Genotyping Methods
We genotyped DNA samples from all study participants for novel coding variants identified through resequencing and for those variants, both intronic and coding, that have published phenotypes. This included 9 SNPs in CYP2C9, 3 SNPs in VKORC1, 6 SNPs in CYP4F2, 2 SNPs in CYP4F11, and 3 SNPs in GGCX. DNA samples were pre-amplified following Fluidigm's (South San Francisco, CA) Specific Target Amplification protocol to increase available template DNA. TaqMan SNP Genotyping Assays (Applied Biosystems, Inc.) were run on 96.96 Dynamic Genotyping Arrays (Fluidigm) according to the BioMark™ 96.96 Genotyping protocol. Dynamic Arrays were primed and loaded on the Fluidigm HX and thermal cycled on the Fluidigm FC1 controller following pre-set programs in the instruments. End-point fluorescence was read on a BioMark™ Real-Time PCR System (Fluidigm) and analyzed using SNP Genotyping Analysis software (Fluidigm). Samples with overall call rates below 95% were removed from further analysis. Of DNA samples selected for genotyping, 22 from the SCF cohort were excluded due to call rates below 95%. For samples from participants included in both sequencing and genotyping, concordance between calls for the 2 methods was over 99.5%.
Population Substructure Analysis
Genealogical information for the participants from the CANHR sample set (Yup'ik residents of the Y-K Delta in western Alaska) were used to calculate pairwise kinship coefficients between each participant [23]. Using these pedigree relationships, allele frequencies and confidence intervals in the CANHR dataset were calculated according to the best linear unbiased estimator (BLUE) of allele frequency [32] to account for the non-independence of these samples resulting from family structure. This adjustment appropriately weights correlated genotypes based on kinship coefficients.
For participants from SCF, neither pedigree nor genome-wide marker information was collected, so this kinship adjustment could not be calculated. To account for population substructure within the SCF cohort, participants were asked for self-reported tribal affiliation. Participants were grouped based on geographic and language similarities of these affiliations, clustered based on linguistic studies by Krauss [33, 34]. These regions are Northern (Inupiaq), Interior (Athabascan subgroups), Southeastern (Tlingit, Tsimshian, Haida, Eyak), Southwestern (Aleut/Unangan), and Western (Central Yup'ik, Cup'ik, Sugpiaq/Alutiiq). Participants also were given the option of choosing affiliation with multiple groups and affiliation with tribes in the lower 48 states of the US, which resulted in 7 total subgroups of participants. All subgroups are represented in the SCF cohort, with each individual subgroup comprising no more than 17% of the total. Between subgroups of study participants at SCF, Analysis of Variance (ANOVA) was performed at SNPs with known phenotypic effects, with a significance threshold of 0.05.
Comparisons with Other Populations
Population frequencies were compared to the 1000 Genomes Database, which documents the distribution of common genetic variants in geographically and historically diverse populations [35]. The populations used for comparison were Admixed American (AMR), African (AFR), Asian (ASN), and European (EUR). Confidence intervals for allele frequencies were calculated based on the number of individuals included in the 1000 Genomes Database for each SNP, as accessed on September 17, 2014.
Statistical Analysis
Allele frequencies were compared using RStudio version 0.97.551 (RStudio, Inc., Boston, MA) and Haploview 4.2 software [36]. All SNPs identified were tested for deviations from Hardy–Weinberg equilibrium using a χ2-test. Pairwise linkage disequilibrium (LD) was calculated using Haploview 4.2 software [36]. The r2 values were used to determine the LD between all non-monomorphic SNPs. The LD display was generated using Haploview 4.2 software [36].
RESULTS
Resequencing for SNP Identification
The exons and bordering intronic regions of the 5 genes CYP2C9, VKORC1, CYP4F2, CYP4F11, and GGCX were resequenced in 94 CANHR participants and 188 SCF participants to identify any novel population-specific variation. All SNPs identified in the SCF and CANHR samples are listed in Supplemental Table 1. Novel SNPs not found in the 1000 genomes database as of November 5, 2014 are labeled rsNA, as they do not have rs numbers.
For CYP2C9, 33 SNPs were identified in the samples from SCF participants, including 2 novel SNPs. In the samples from CANHR participants, 25 SNPs were identified, including the same 2 novel SNPs. One novel SNP predicted a non-synonymous change from asparagine to isoleucine at amino acid 218 (N218I allele). The other was discovered in the first codon, resulting in a change from methionine to leucine (M1L allele). The sequencing chromatograms identifying N218I are found in Supplemental Figure 1 and those for M1L are found in Supplemental Figure 2. This M1L SNP was found at frequency of 9.7% (+/− 4.3%) of chromosomes in the 94 CANHR samples subjected to resequencing. M1L was also identified in the samples from SCF participants, though at a lower frequency of 1.0% (+/− 0.7%). A known SNP, rs182132442, resulting in a proline to threonine substitution at amino acid 279 (CYP2C9*29) is not well characterized and was found in the CANHR cohort only [37]. PolyPhen and Grantham scores predicted a deleterious effect on protein function for all 3 variants [30, 31, 38]. The M1L variant had a PolyPhen score of 0.904, predicting a severe effect on protein function based on likely truncation. The N218I variant had a Grantham score of 149 and the CYP2C9*29 variant had a Grantham score of 38, predicting severe effects due to chemical dissimilarities of the affected amino acids.
For VKORC1, 10 SNPs were identified in resequencing the samples from SCF participants. Of these, 3 were novel synonymous changes. Only 2 SNPs were identified in the CANHR participants, neither of them novel. While the −1639 SNP differentiating the major VKORC1 haplotypes was assessed, the 1173 base was outside of the sequencing range, though both sites were assessed in subsequent genotyping.
For CYP4F2, 34 SNPs were identified in the samples from SCF participants, with 4 of those being novel. One of these novel SNPs changed the splice site of exon 1 (exon+splice allele). Within the CANHR participants, 22 SNPs were identified, with the only novel SNP being the exon+splice.
For CYP4F11, 28 SNPs were identified in the samples from SCF participants, including 5 novel SNPs. One SNP that was novel at the time of sequencing, but has since been named rs199657164, predicted a glycine to arginine change at amino acid 12 (G12R allele). One of these five novel SNPs found in the samples from SCF participants predicted a coding change from asparagine to aspartic acid at amino acid 285. In the CANHR participants, 25 SNPs were identified, including 4 novel SNPs, 3 of which were also in with the samples from SCF participants.
Resequencing of GGCX identified 21 SNPs in the samples from SCF participants. These SNPs included 3 novel SNPs, including a predicted alanine to glycine change at amino acid 421 (A421G allele). Of the SNPs identified in the samples from SCF participants, 11 of those were identified in the samples from CANHR participants, including 1 of the novel SNPs. No unique SNPs were identified in the CANHR cohort that were not found in the SCF cohort.
Genotyping for Population Frequencies
A summary of the characteristics of study participants for whom we recovered DNA producing ≥ 95% genotyping call rates is presented in Table 1. Genotyping at specific SNPs was performed to verify the findings from resequencing and to establish better estimates of population frequencies (Table 2). The SNPs chosen for genotyping either are SNPs that have a published phenotype, or are non-synonymous SNPs that were discovered during resequencing. Allele frequencies of the samples from the CANHR cohort were adjusted for the kinship between study participants using BLUE [32]. All SNPs were in Hardy-Weinberg equilibrium.
Table 1.
Demographic characteristics of genotyped study cohorts. SCF participants were classified by self-reported tribal affiliation, clustered by geographic region and linguistic similarities. Only participants for whom genotyping reached ≥ 95% call rate for all alleles tested were included.
| Population | Subpopulation | Males | Females | Total |
|---|---|---|---|---|
| SCF | 125 | 234 | 359 | |
| Interior | 17 | 20 | 37 | |
| Northern | 30 | 55 | 85 | |
| Southeastern | 16 | 26 | 42 | |
| Southwestern | 23 | 37 | 60 | |
| Western | 8 | 41 | 49 | |
| Multiple | 21 | 35 | 56 | |
| Lower 48 | 10 | 20 | 30 | |
| CANHR | 165 | 185 | 350 |
Table 2.
Prevalence of CYP2C9, CYP4F2, VKORC1, and GGCX variant alleles in the SCF and CANHR AI/AN cohorts of Alaska, as determined using the Fluidigm genotyping platform. The SCF sample participants are presented in total (column 6) and divided into regional subgroups (columns 7-13). Reference allele (Ref) obtained from dbSNP1. Reported frequency is of the variant allele (Var) listed. Frequencies are reported in percentages, with 95% confidence intervals for the true population allele frequency in parentheses.
| CYP2C9 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Ref | Var | CANHR (n=350) | SCF (n=359) | Interior (n=37) | Northern (n=85) | South eastern (n=42) | South western (n=60) | Western (n=49) | Multiple (n=56) | Lower 48 (n=30) |
|
*13 L90P |
72558187 | T | C | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 (0 - 3.5) |
0.0 | 0.0 | 0.0 | 0.0 |
|
*14 R125H |
72558189 | G | A | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 (0 - 3.5) |
0.0 | 0.0 | 0.0 | 0.0 |
|
*2 R144C |
1799853 | C | T | 0.3 (0 - 0.7) |
5.2 (3.6 - 6.8) |
5.4 (0.3 - 10.5) |
4.1 (1.1 - 7.1) |
7.1 (1.6 - 12.6) |
5.8 (1.6 - 10.0) |
6.1 (1.4 - 10.8) |
1.8 (0 - 4.3) |
8.3 (1.3 - 15.3) |
|
*8 R150L |
7900194 | G | A | 0.0 | 0.1 (0 - 0.3) |
0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.7 (0 - 5.0) |
|
*11 R355W |
28371685 | C | T | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| *3 I359L | 1057910 | A | C | 2.1 (1.0 - 3.2) |
3.4 (2.1 - 4.7) |
4.1 (0 - 8.6) |
1.2 (0 - 2.8) |
3.6 (0 - 7.6) |
5.9 (1.7 - 10.1) |
0.0 | 3.6 (0.1 - 7.1) |
8.3 (1.3 - 15.3) |
| M1L | NA | T | A | 6.3 (4.5 - 8.1) |
1.0 (0.3 - 1.7) |
1.4 (0 - 4.1) |
1.8 (0 - 3.8) |
0.0 | 0.8 (0 - 2.4) |
1.0 (0 - 3.0) |
0.9 (0 - 2.6) |
0.0 |
| N218I | NA | A | T | 3.8 (2.4 - 5.3) |
1.4 (0.5 - 2.3) |
5.4 (0.3 - 10.5) |
2.4 (0.1 - 4.7) |
0.0 | 0.8 (0 - 2.4) |
1.0 (0 - 3.0) |
0.0 | 0.0 |
|
*29 P279T |
182132442 | C | A | 2.1 (1.0 - 3.2) |
0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| VKORC1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Ref | Var | CANHR (n=350) | SCF (n=359) | Interior (n=37) | Northern (n=85) | South eastern (n=42) | South western (n=60) | Western (n=49) | Multiple (n=56) | Lower 48 (n=30) |
| V29L | 28940302 | G | T | 0.0 | 0.1 (0 - 0.3) |
0.0 | 0.0 | 1.2 (0.0 - 3.6) |
0.0 | 0.0 | 0.0 | 0.0 |
| 1173 | 9934438 | G | A | 77.5 (74.4 - 80.7) |
59.7 (56.1 - 63.3) |
52.7 (41.3 - 64.1) |
64.7 (57.5 - 71.9) |
53.6 (42.9 - 64.3) |
55.0 (46.1 - 63.9) |
68.4 (59.2 - 77.6) |
64.3 (55.4 - 73.2) |
50.0 (37.3 - 62.7) |
| −1639 | 9923231 | C | T | 77.8 (74.6 - 80.9) |
59.7 (56.1 - 63.3) |
54.1 (42.7 - 65.5) |
64.1 (56.9 - 71.3) |
53.6 (42.9 - 64.3) |
54.2 (45.3 - 63.1) |
68.4 (59.2 - 77.6) |
64.3 (55.4 - 73.2) |
50.0 (37.3 - 62.7) |
| CYP4F | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Ref | Var | CANHR (n=350) | SCF (n=359) | Interior (n=37) | Northern (n=85) | South eastern (n=42) | South western (n=60) | Western (n=49) | Multiple (n=56) | Lower 48 (n=30) |
| 4F2 | 2189784 | G | A | 39.6 (35.9 - 43.2) |
31.0 (27.6 - 34.4) |
29.7 (19.3 - 40.1) |
34.1 (27.0 - 41.2) |
32.1 (22.1 - 42.1) |
28.0 (20.0 - 36.0) |
34.7 (25.3 - 44.1) |
27.7 (19.4 - 36.0) |
30.0 (18.4 - 41.6) |
|
4F2 M519L |
3093200 | C | A | 0.0 | 2.7 (1.5 - 3.9) |
2.7 (0 - 6.4) |
1.8 (0 - 3.8) |
2.4 (0 - 5.7) |
0.8 (0 - 2.4) |
1.1 (0 - 3.2) |
5.4 (1.2 - 9.6) |
6.7 (0.4 - 13.0) |
|
4F2*3 V433M |
2108622 | C | T | 50.9 (47.2 - 54.7) |
31.5 (28.1 - 34.9) |
31.1 (20.6 - 41.6) |
36.5 (29.3 - 43.7) |
39.3 (28.9 - 49.7) |
22.9 (15.4 - 30.4) |
35.7 (26.2 - 45.2) |
32.1 (23.5 - 40.7) |
16.7 (7.3 - 26.1) |
|
4F2 G185V |
3093153 | G | T | 0.3 (0 - 0.6) |
2.2 (1.1 - 3.3) |
4.1 (0 - 8.6) |
1.8 (0 - 3.8) |
0.0 | 4.2 (0.6 - 7.8) |
1.0 (0 - 3.0) |
1.8 (0 - 4.3) |
3.3 (0 - 7.8) |
|
4F2*2 W12G |
3093105 | T | G | 3.7 (2.3 - 5.1) |
11.0 (8.7 - 13.3) |
16.2 (7.8 - 24.6) |
10.6 (6.0 - 15.2) |
22.6 (13.7 - 31.5) |
7.5 (2.8 - 12.2) |
6.1 (1.4 - 10.8) |
8.0 (3.0 - 13.0) |
10.0 (2.4 - 17.6) |
|
4F2 spIiceCG |
NA | C | G | 0.7 (0.1 - 1.4) |
1.4 (0.5 - 2.3) |
9.7 (3.0 - 16.4) |
1.2 (0 - 2.8) |
1.2 (0 - 3.5) |
0.0 | 1.0 (0 - 3.0) |
0.9 (0 - 2.6) |
0.0 |
|
4F11 R276C |
8104361 | G | A | 0.3 (0 - 0.7) |
9.1 (7.0 - 11.2) |
6.8 (1.1 - 12.5) |
7.6 (3.6 - 11.6) |
8.3 (2.4 - 14.2) |
10.8 (5.2 - 16.4) |
7.1 (2.0 - 12.2) |
3.6 (0.1 - 7.1) |
23.3 (12.6 - 34.0) |
|
4F11 G12R |
NA | C | G | 1.7 (0.7 - 2.6) |
0.8 (0.2 - 1.5) |
1.4 (0 - 4.1) |
0.0 | 2.4 (0 - 5.7) |
0.8 (0 - 2.4) |
1.0 (0 - 3.0) |
0.9 (0 - 2.6) |
0.0 |
| GGCX | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Ref | Var | CANHR (n=350) | SCF (n=359) | Interior (n=37) | Northern (n=85) | South eastern (n=42) | South western (n=60) | Western (n=49) | Multiple (n=56) | Lower 48 (n=30) |
| 11676382 | C | G | 0.3 (0 - 0.7) |
3.8 (2.4 - 5.2) |
5.4 (0.3 - 10.5) |
2.4 (0.1 - 4.7) |
2.4 (0 - 5.7) |
4.2 (0.6 - 7.8) |
4.1 (1.7 - 8.0) |
2.7 (0 - 5.7) |
8.3 (1.3 - 15.3) |
|
| G421A | NA | G | C | 0.0 | 0.6 (0 - 1.2) |
0.0 | 0.0 | 2.4 (0 - 5.7) |
0.0 | 0.0 | 1.8 (0 - 4.3) |
0.0 |
| R325Q | 699664 | G | A | 49.1 (45.3 - 52.9) |
35.9 (32.4 - 39.4) |
31.1 (20.6 - 41.6) |
38.2 (30.9 - 45.5) |
23.8 (14.7 - 32.9) |
29.2 (21.1 - 37.3) |
49.0 (39.1 - 58.9) |
43.8 (34.6 - 53.0) |
30.0 (18.4 - 41.6) |
Of the 9 SNPs genotyped in CYP2C9, 6 were previously known alleles (*2, *3, *8, *11, *13, *14, and *29) and 2 were the M1L and N218I novel non-synonymous SNPs identified in resequencing. The frequencies of the *29 allele and both novel variants M1L and N218I were significantly higher (p < 0.05) in the CANHR cohort, and the frequency of CYP2C9*2 was higher in samples from SCF participants. All other SNPs, with the exception of the CYP2C9*3 allele, were found at frequencies below 1% of alleles in both cohorts.
Of the 3 SNPs related to VKORC1 activity, 2 are the SNPs that differentiate the major haplotype groups and typically are seen in complete linkage disequilibrium [39, 40]. The other (rs28940302) predicts a substitution of leucine for valine at amino acid 29 (V29L) and has been associated with warfarin resistance [41]. The V29L variant was found at less than 1% in both cohorts. Both SNPs designating the major VKORC1 haplotype associated with lower warfarin dose were found at significantly higher frequencies in the CANHR cohort than in the SCF cohort (p < 0.05). The VKORC1 and CYP2C9 diplotypes of the CANHR and SCF cohorts are reported in Table 3 and predict phenotypes for warfarin metabolism.
Table 3.
Frequencies of predicted diplotypes known to affect warfarin dose in the CANHR and SCF datasets.
| CANHR Diplotype Frequencies | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VKORC1 diplotype | CYP2C9 diplotype | |||||||||
| *1/*1 | *1/*2 | *1/*3 | *2/*2 | *1/M1L | *1/N218I | *1/*29 | M1L/M1L | *3/M1L | Total | |
| Low dose homozygous (AT/AT) | 153 | 1 | 6 | 0 | 25 | 20 | 12 | 2 | 3 | 222 |
| Low dose heterozygous (AT/GC) | 77 | 0 | 3 | 1 | 13 | 6 | 1 | 1 | 0 | 102 |
| High dose homozygous (GC/GC) | 17 | 1 | 2 | 0 | 4 | 1 | 0 | 1 | 0 | 26 |
| Total | 247 | 2 | 11 | 1 | 42 | 27 | 13 | 4 | 3 | 350 |
| SCF Diplotype Frequencies | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VKORC1 diplotype | CYP2C9 diplotype | |||||||||
| *1/*1 | *1/*2 | *1/*3 | *2/*2 | *1/M1L | *1/N218I | *2/N218I | *2/*3 | *3/*3 | Total | |
| Low dose homozygous (AT/AT) | 101 | 10 | 5 | 0 | 4 | 3 | 0 | 1 | 0 | 124 |
| Low dose heterozygous (AT/GC) | 145 | 15 | 10 | 1 | 1 | 5 | 1 | 0 | 1 | 179 |
| High dose homozygous (GC/GC) | 41 | 7 | 5 | 0 | 1 | 1 | 0 | 1 | 0 | 56 |
| Total | 287 | 32 | 20 | 1 | 6 | 9 | 1 | 2 | 1 | 359 |
Diplotypes of VKORC1 and CYP2C9 were calculated from observed 1173 and -1639-containing high and low dose VKORC1 haplotypes, and CYP2C9 *1, *2, *3 and the M1L, N218I, and P279T genotypes, assuming no LD between CYP2C9 variants.
At the CYP4F locus, 6 previously identified alleles were assessed, as well as 2 novel non-synonymous changes. The CYP4F2*3 (V433M rs2108622) allele was found at significantly lower frequency in the SCF cohort than in the CANHR cohort. However, the frequency of this variant was high in both populations and the frequency in the CANHR cohort was one of the highest reported for a population to date – similar to the 53.2% reported in the Saudi Arabian tribal subgroup of the Kuwaiti population [42]. Samples from SCF participants had a significantly higher frequency of the CYP4F2*2 allele (W12G rs3093105) than the samples from CANHR participants. The same was true of the CYP4F2 M519L (rs3093200) substitution, the CYP4F2 amino acid 185 change from glycine to valine (G185V rs3093153), and the CYP4F11 amino acid 276 change from arginine to cysteine (R276C rs8104361).
The 2 GGCX SNPs described previously [43-45] were both found at significantly different frequencies in the SCF and CANHR cohorts (p < 0.05). The SNP rs11676382 was less common in the samples from CANHR participants, and the missense SNP (rs699664) was less common in the samples from SCF participants.
Linkage Disequilibrium
Linkage disequilibrium (LD) was calculated between every non-monomorphic SNP in each gene. For CYP2C9, LD was low between all SNPs, in both the SCF and CANHR cohorts. In the VKORC1 gene, the 1173C>T (rs9934438) and −1639G>A (rs9923231) SNPs, which differentiate the 2 major haplotypes of VKORC1, were in tight LD in both the samples from SCF participants (r2 = 0.96) and the samples from CANHR participants (r2 = 0.97).
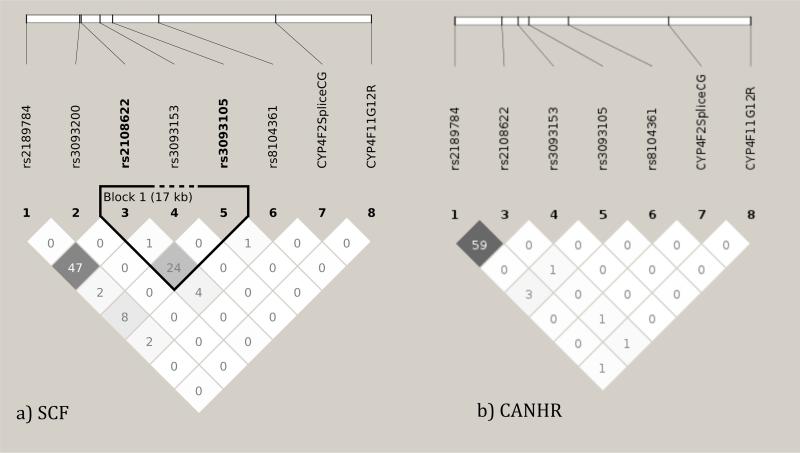
At the CYP4F locus (Figure 1), the samples from both SCF and CANHR participants show moderate levels of LD between the rs2108622 and rs2189784 SNPs. The samples from SCF participants show moderate LD between the rs2108622 (*3 allele) and the rs3093105 (*2 allele). LD was low between all other SNPs.
Figure 1. Linkage Disequilibrium (LD) in the CYP4F locus for all non-monomorphic SNPs in the participants from a) SCF and b) CANHR sample sets, by r2 measure.
Pairwise comparisons illustrate low LD between most SNPs. The rs2189784 and rs2108622 SNPs are found in tighter LD in the samples from CANHR participants than in the samples from SCF participants. These patterns illustrate the effects of potential founder effects and population isolation, resulting in differences in genetic patterns found in regional subgroups of Alaska. Variant pairs with LD scores closer to 100 were more often inherited together than not. Scores were calculated with Haploview version 4.2. Haplotype blocks determined by confidence intervals.
The SNPs in GGCX were not in LD.
Allele Frequency Comparison Between AN Regional Subgroups
While the samples in the CANHR cohort all were collected from participants living in the Y-K Delta and self-reporting Yup'ik ancestry, many of the participants in the SCF cohort self-identify with tribes historically distributed throughout Alaska but now live in Anchorage or make visits there for healthcare. The allele frequencies of the genotyped SNPs were determined for each of the self-identified regional subgroups of Alaska (Table 2). Because subgroup frequencies could not be adjusted for population substructure, the frequency variance is expected to be slightly larger than presented here, but the frequency estimate should not be greatly affected. Based on ANOVA results, SCF subgroups had significantly different allele frequencies at the VKORC1 −1639, VKORC1 1173, and CYP4F2*3 loci.
Allele Frequency Comparison to Other Populations
Comparisons to the 4 main continental groups of the 1000 Genomes Database are presented in Table 4 [35]. Generally, the frequencies of variant alleles CYP2C9*2 and CYP2C9*3 were low in the samples from SCF participants by global comparison and even lower in the samples from CANHR participants. For VKORC1, the frequency of the lower warfarin dose associated AT haplotype (A at rs9934438 and T at rs9923231) was high in the SCF cohort (~60%) and higher in the CANHR cohort (~80%), though both frequencies were higher than in the EUR and AMR and lower than in the ASN populations. At the CYP4F2 locus, the frequency of the CYP4F2*3 SNP was higher in both the SCF and CANHR cohorts than in any of the 1000 Genomes Database populations. The allele frequency of CYP4F2*2 in the CANHR cohort was low compared to the 1000 Genomes Database populations, whereas that of the SCF cohort was similar to the EUR, AMR, and ASN populations. For GGCX, the variant allele frequencies in the SCF and CANHR cohorts were similar to the 1000 Genomes Database populations.
Table 4.
Comparison of allele frequencies of SCF and CANHR cohorts to global populations from the 1000 genomes database2.
| CYP2C9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Reference allele | Variant allele | SCF | CANHR | AMR | AFR | ASN | EUR |
| Number of alleles | 718 | 700 | 362 | 492 | 572 | 758 | |||
| *2 R144C | 1799853 | C | T | 5.2 (3.6 - 6.8) | 0.3 (0 - 0.7) | 12.4 (9.0 - 15.8) | 1.8 (0.6 - 3.0) | 0.3 (0 - 0.7) | 12.3 (10.0 - 14.6) |
| *3 I359L | 1057910 | A | C | 3.4 (2.1 - 4.7) | 2.1 (1.0 - 3.2) | 5.8 (3.4 - 8.2) | 0.6 (0 - 1.3) | 4.0 (2.4 - 5.6) | 6.1 (4.4 - 7.8) |
| VKORC1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Reference Allele | Variant allele | SCF | CANHR | AMR | AFR | ASN | EUR |
| Number of alleles | 718 | 700 | 362 | 492 | 572 | 758 | |||
| 1173 | 9934438 | G | A | 59.7 (56.1 - 63.3) | 77.5 (74.4 - 80.7) | 43.9 (38.8 - 49.0) | 6.5 (4.3 - 8.7) | 91.8 (89.6 - 94.0) | 40.1 (36.6 - 43.6) |
| −1639 | 9923231 | C | T | 59.7 (56.1 - 63.3) | 77.8 (74.6- 80.9) | 43.9 (38.8 - 49.0) | 6.5 (4.3 - 8.7) | 91.8 (89.6 - 94.0) | 40.1 (36.6 - 43.6) |
| CYP4F | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Reference Allele | Variant Allele | SCF | CANHR | AMR | AFR | ASN | EUR |
| Number of alleles | 718 | 700 | 362 | 492 | 572 | 758 | |||
| 4F2 | 2189784 | G | A | 31.0 (27.6 - 34.4) | 39.6 (35.9 - 43.2) | 41.7 (36.6 - 46.8) | 28.7 (24.7 - 32.7) | 26.6 (23.0 - 30.2) | 43.0 (39.5 - 46.5) |
| 4F2*3 V433M | 2108622 | C | T | 31.5 (28.1 - 34.9) | 50.9 (47.2 - 54.7) | 28.5 (23.8 - 33.2) | 8.5 (6.0 - 11.0) | 20.6 (17.3 - 23.9) | 27.3 (24.1 - 30.5) |
| 4F2 G185V | 3093153 | C | A | 2.2 (1.1 - 3.3) | 0.3 (0 - 0.6) | 4.4 (2.3 - 6.5) | 2.6 (1.2 - 4.0) | 0 | 7.5 (5.6 - 9.4) |
| 4F2*2 W12G | 3093105 | A | C | 11.0 (8.7 - 13.3) | 3.7 (2.3 - 5.1) | 17.7 (13.8 - 21.6) | 24.0 (20.2 - 27.8) | 7.2 (5.1 - 9.3) | 15.8 (13.2 - 18.4) |
| 4F11 R276C | 8104361 | G | A | 9.1 (7.0 - 11.2) | 0.3 (0 - 0.7) | 22.4 (18.1 - 26.7) | 29.3 (25.3 - 33.3) | 7.2 (5.1 - 9.3) | 26.5 (23.4 - 30.0) |
| GGCX | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | rs number | Reference Allele | Variant Allele | SCF | CANHR | AMR | AFR | ASN | EUR |
| Number of alleles | 718 | 700 | 362 | 492 | 572 | 758 | |||
| 11676382 | C | G | 3.8 (2.4 - 5.2) | 0.3 (0 - 0.7) | 3.3 (1.5 - 5.1) | 0.6 (0 - 1.3) | 0 | 8.7 (6.7 - 10.7) | |
| R325Q | 699664 | C | T | 35.9 (32.4 - 39.4) | 49.1 (45.3 - 52.9) | 29.3 (24.6 - 34.0) | 69.1 (65.0 - 73.2) | 32.9 (29.0 - 36.8) | 35.9 (32.5 - 39.3) |
Frequencies are reported as percentage of the variant allele, including 95% confidence interval. The populations used for comparison were Admixed American (AMR), African (AFR), Asian (ASN), and European (EUR) from the 1000 genomes database.
1. Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308-11.
Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56-65.
DISCUSSION
The most significant new findings in the AN subpopulations studied were (1) the presence of two, previously unreported, relatively high frequency coding variants in the CYP2C9 gene (M1L and N218I) in most, but not all, regional subgroups; (2) a high frequency of the low warfarin dose associated non-coding variants in the VKORC1 gene, especially in the Yup'ik population living in the Y-K Delta; and 3) a relatively high frequency of the higher warfarin dose associated CYP4F2*3 variant in some, but not all, regional subgroups. These results are generally consistent with an observed requirement of lower warfarin doses to achieve target INR values in an AI/AN population receiving healthcare in Anchorage, in comparison to that for the non-indigenous population of the US [16].
The identification of relatively common, novel, potentially function-disrupting variants in CYP2C9 illustrates how pharmacogenetic discoveries made from studies of “representative” world populations do not always capture variation that could be important for other, historically geographically isolated populations, such as the AN people. Population-specific pharmacogenetic studies are necessary to guide anticoagulation therapy in the AI/AN community if clinical testing becomes standard of care and is to be implemented effectively. A prime example is the ATG to TTG change in the first codon of the CYP2C9 gene (resulting in a predicted methionine to leucine substitution at amino acid position 1) that would be expected to adversely affect mRNA translation and protein synthesis. Although we have not yet confirmed the phenotype of this variant, M1L is predicted to disrupt mRNA translation, by alteration of the first codon. Similar disruption of codon 1 (M1V or M1L) in other genes is associated with a highly penetrant, loss of function phenotype [46-49]. Indeed, the extremely rare M1V variant of CYP2C9 (CYP2C9*36) was recently described in a Chinese population, and its recombinant expression was found to result in low accumulation of the variant enzyme relative to wild-type in COS cells [50].
The novel asparagine to leucine substitution at amino acid 218 was also predicted to have a deleterious effect on CYP2C9 enzyme function. N218I is located between helices F and G in an area of the enzyme known to be important for catalytic activity [51]. To our knowledge, no protein variant at this position has been prepared and tested for function, but the neighboring, inter-helical, Q214L variant is CYP2C9*28, which expressed well in COS-7 cells, but exhibited no detectable S-warfarin 7-hydroxylation activity [52].
Another known, but little studied variant, a proline to threonine change at position 279 is predicted to reduce the catalytic function of CYP2C9. The CYP2C9 P279T variant (CYP2C9*29) resides between the H and I helices of CYP2C9 [51], and has been found at low frequencies in Japanese and Chinese populations [37, 50]. The recombinant enzyme expressed well in mammalian cells and exhibited a ~30% reduction in tolbutamide hydroxylation activity relative to wild-type [50]. A more detailed kinetic analysis of S-warfarin 7-hydroxylation found no difference in Km compared to wild-type CYP2C9 and a ~50% decrease in Vmax, similar to that found for CYP2C9*2 [52], which is an established risk factor for warfarin sensitivity.
Combined with the known, functionally important CYP2C9 loss of function coding variants (*2 and *3 alleles), these novel potentially loss of function variants were found at a frequency that ranged from 9.5% in the Northern to 14.8% in the Interior AN subgroups. Importantly, individuals carrying CYP2C9*2 and *3 alleles are at increased risk of major bleeding events following the initiation for warfarin therapy [53]. If relying on variants identified in European populations, the much lower frequencies of *2 and *3 in AN populations, especially in the Yup'ik population, compared to European populations would underestimate overall CYP2C9 coding variation. Novel CYP2C9 variants (M1L and N218I) first identified in this paper, and a relatively new variant (P279T), may confer a similar risk in AN subgroups. Thus, while the exact composition of CYP2C9 coding variation differs between the AN regional subgroups and other populations globally, the overall impact of these changes on warfarin dose requirement and bleeding risk could be the same.
Variation seen in the VKORC1 gene is also likely to have a significant effect on warfarin dose requirement in regional AN subgroups. This is especially true for the Yup'ik people of the Y-K Delta, for whom we observed the highest frequency of the low dose haplotype. Although the specific mechanism is unknown, these non-coding SNPs are consistently associated with reduced gene transcription and protein expression, and with lower warfarin dose requirements in other populations [54, 55].
Although CYP4F2 variation is thought to contribute less to inter-individual differences in warfarin dose requirement than do variation in CYP2C9 and VKORC1, it may have a greater generalized impact on warfarin dose requirements in several AN regional subpopulations because of the observed relatively high frequency of the *3 allele. The CYP4F2*3 variant is associated with reduced metabolic clearance of vitamin K, increased vitamin K levels, and increased warfarin dose [56, 57]. It would be expected to counteract to some degree the effect of the reduced function CYP2C9 and VKORC1 variants on average dose requirement, although a given individual could fall anywhere along a wide continuum of warfarin dose sensitivity based on overall genetic constitution. Selective pressure may have acted on the CYP4F2 gene to conserve vitamin K as a result of inconsistent access to greens (e.g., traditional tundra greens, beach greens) throughout the year. Alternatively, it could be the result of a founder effect, with ancestral blocks of DNA preserved over time. Of particular relevance is the recent report of reduced bleeding risk following variation in vitamin K consumption in individuals carrying the CYP4F2*3 allele who receive long-term warfarin therapy [53]. If changes in vitamin K consumption and accumulation in the body affect the risk of a major bleeding event in individuals receiving warfarin, the relatively high frequency of the CYP4F2*3 allele may provide some resiliency against that adverse event. Indeed, in a recent study of vitamin K homeostasis in Yup'ik people of the Y-K Delta, we found significantly higher levels of plasma vitamin K1 in individuals carrying 1 or 2 copies of the CYP4F2*3 allele, in comparison to those homozygous for the reference CYP4F2*1 allele (unpublished observations).
Variation in GGCX and CYP4F11 is not expected to have implications for warfarin therapy in the AN subpopulations. While genes were sequenced to look for novel SNPs, none were discovered at a frequency expected to affect enzyme function at a population level. Known variation in these genes has not been associated with warfarin dose or response.
We sought to identify and characterize variation in 5 genes associated with warfarin dose requirement and drug response in two cohorts of AN people. This study is the first to partner with a diverse population to systematically sequence these genes to identify population-specific variation. Furthermore, by genotyping these and other SNPs relevant for warfarin response in cohorts of AN subgroups, we have established robust population frequencies that can be used to guide patient care. Although this study does not have information on drug dose or bleeding events for participants or functional studies on novel variation, based on known genotype-phenotype relationships, our phenotypic predictions of these genetic findings are well supported.
The presence of novel CYP2C9 gene variants and relatively high frequencies of variant alleles in the VKORC1 and CYP4F2 genes support our hypothesis that pharmacogenetic research in understudied populations is needed and suggests the possibility of significant associations with warfarin dose requirement and bleeding risk in the AN subpopulations. Warfarin is still the drug of choice for chronic therapy in the prevention of thromboembolic events for at-risk individuals receiving health care at SCF and YKHC. Although treatment is challenging for all of the reasons that apply to other populations [12-15], major bleeding risk associated with alternative direct thrombin inhibitors and the absence of an antidote for those drugs is a deterrent to changing treatment in patients who often have restricted access to emergency care because of geographic isolation. Thus, the findings we report may advance the development of genetic tests for optimizing the initiation of warfarin therapy and for identifying individuals at increased risk of major bleeding events during chronic therapy.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Barbara Kavanaugh, Program Manager for the NWA-PGRN, for directing programmatic collaborations, and LT Catherine Arnatt, PharmD, LT Christopher Chong, PharmD, and Mr. Austen Rodgers, who collected blood samples at SCF. They sincerely thank customer-owners of the Southcentral Foundation and community members of the Yukon-Kuskokwim River Delta for their input, guidance, and study participation.
Funding: National Institutes of Health (U01 GM092676, P30 ES007033, P20 RR16430, U19 HL069757, R01 DK074842, P30 GM103325, and GM109743)
Abbreviations
- AI/AN
American Indian / Alaska Native
- CYP2C9
Cytochrome P450, family 2, subfamily C, polypeptide 9
- CYP4F2
Cytochrome P450, family 4, subfamily F, polypeptide 2
- CYP4F11
Cytochrome P450, family 4, subfamily F, polypeptide 11
- VKORC1
Vitamin K epoxide oxidase reductase complex subunit 1
- GGCX
gamma-glutamyl carboxylase
- BMI
Body Mass Index
- SCF
Southcentral Foundation
- ANMC
Alaska Native Medical Center
- ANTHC
Alaska Native Tribal Health Consortium
- CANHR
Center for Alaska Native Health Research
- LD
Linkage Disequilibrium
- YKHC
Yukon-Kuskokwim Health Corporation
- SNP
Single Nucleotide Polymorphism
- ASU
Anchorage Service Unit
Footnotes
Conflict of Interest: No conflicts of interest were declared by the authors.
REFERENCES
- 1.Shin J. Clinical pharmacogenomics of warfarin and clopidogrel. J Pharm Pract. 2012;25(4):428–38. doi: 10.1177/0897190012448310. [DOI] [PubMed] [Google Scholar]
- 2.Bhathena A, Spear BB. Pharmacogenetics: improving drug and dose selection. Curr Opin Pharmacol. 2008;8(5):639–46. doi: 10.1016/j.coph.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Thummel KE, Lin YS. Sources of interindividual variability. Methods Mol Biol. 2014;1113:363–415. doi: 10.1007/978-1-62703-758-7_17. [DOI] [PubMed] [Google Scholar]
- 4.Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and Individualized Drug Therapy. Annual Review of Medicine. 2006;57(1) doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 5.Smart A, Martin P. The promise of pharmacogenetics: assessing the prospects for disease and patient stratification. Studies in history and philosophy of biological and biomedical sciences. 2006;37(3):583–601. doi: 10.1016/j.shpsc.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Boyer B, Dillard D, Woodahl EL, Whitener R, Thummel KE, Burke W. Ethical issues in developing pharmacogenetic research partnerships with American Indigenous communities. Clinical pharmacology and therapeutics. 2011;89(3):343–5. doi: 10.1038/clpt.2010.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fohner A, Muzquiz LI, Austin MA, Gaedigk A, Gordon A, Thornton T, et al. Pharmacogenetics in American Indian populations: analysis of CYP2D6, CYP3A4, CYP3A5, and CYP2C9 in the Confederated Salish and Kootenai Tribes. Pharmacogenetics and genomics. 2013;23(8):403–14. doi: 10.1097/FPC.0b013e3283629ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw JL, Robinson R, Starks H, Burke W, Dillard DA. Risk, reward, and the double-edged sword: perspectives on pharmacogenetic research and clinical testing among Alaska Native people. American journal of public health. 2013;103(12):2220–5. doi: 10.2105/AJPH.2013.301596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horner RD, Day GM, Lanier AP, Provost EM, Hamel RD, Trimble BA. Stroke mortality among Alaska Native people. American journal of public health. 2009;99(11):1996–2000. doi: 10.2105/AJPH.2008.148221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129(3):e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard BV, Metzger JS, Killer KR, Jolly SE, Asay ED, Wang H, et al. All-cause, cardiovascular, and cancer mortality in western Alaska Native people: Westen Alaska Tribal Collaborative for Health (WATCH). American journal of public health. 2014;104(7):1334, 40. doi: 10.2105/AJPH.2013.301614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stafford RS, Singer DE. Recent national patterns of warfarin use in atrial fibrillation. Circulation. 1998;97(13):1231–3. doi: 10.1161/01.cir.97.13.1231. [DOI] [PubMed] [Google Scholar]
- 13.Birman-Deych E, Radford MJ, Nilasena DS, Gage BF. Use and effectiveness of warfarin in Medicare beneficiaries with atrial fibrillation. Stroke. 2006;37(4):1070–4. doi: 10.1161/01.STR.0000208294.46968.a4. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro SS. Treating thrombosis in the 21st century. The New England journal of medicine. 2003;349(18):1762–4. doi: 10.1056/NEJMe038152. [DOI] [PubMed] [Google Scholar]
- 15.Hart RG, Pearce LA, Aguilar MI. Meta-analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med. 2007;146(12):857–67. doi: 10.7326/0003-4819-146-12-200706190-00007. [DOI] [PubMed] [Google Scholar]
- 16.Schilling B. Anticoagulation Care for Alaska Native Customer-Owners within the Nuka Model of Care.. 7th Dawn AC Anticoagulation Management Software North American User Group Meeting; La Jolla, CA. 2013. [Google Scholar]
- 17.Daly AK. Pharmacogenetics of the major polymorphic metabolizing enzymes. Fundam Clin Pharmacol. 2003;17(1):27–41. doi: 10.1046/j.1472-8206.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- 18.Daly AK. Pharmacogenetics of the cytochromes P450. Curr Top Med Chem. 2004;4(16):1733–44. doi: 10.2174/1568026043387070. [DOI] [PubMed] [Google Scholar]
- 19.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacology & therapeutics. 2007;116(3):496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Zanger UM, Turpeinen M, Klein K, Schwab M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Analytical and bioanalytical chemistry. 2008;392(6):1093–108. doi: 10.1007/s00216-008-2291-6. [DOI] [PubMed] [Google Scholar]
- 21.McDonagh E, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomarkers in medicine. 2011;5(6):795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alaska Department of Labor and Workforce Development RaAS . Alaska Population Overivew: 2012 Estimates. 2013. p. 128. [Google Scholar]
- 23.Bourgain C, Zhang Q. Kinship and Inbreeding coefficients computation in general pedigrees. 1.1 ed. Free Software Foundation, Inc.; Boston, MA: 2009. [Google Scholar]
- 24.Manachaikul A, Mychaleckyi JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wise associaiton studies. Bioinformatics. 2010;26(22):2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieder MJ, L. TS, G. CA, A. ND. Sequence variation in the human angiotensin converting enzyme. Nature genetics. 1999;22(1):59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- 26.Gordon D, Abajian C, Green P. Consed: A Graphical Tool for Sequence Finishing. Genome Research. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 27.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic acids research. 1997;25(14):2745. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nature genetics. 2006;38(3):375–81. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- 29.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome research. 1998;8(3):186–94. [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grantham R. Amino Acid Difference Formula to Help Explain Protein Evolution. Science. 1974;185(4154):862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 32.McPeek MS, Wu X, Ober C. Best Linear Unbiased Allele-Frequency Estimation in Complex Pedigrees. Biometrics. 2004;60:359–67. doi: 10.1111/j.0006-341X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- 33.Krauss ME. Alaska Native Languages: Past, Present, and Future. Alaska Native Language Center; Fairbanks, AK: 1980. [Google Scholar]
- 34.Krauss ME, Alaska ANLCUo. Research AIoSE . Indigenous peoples and languages of Alaska. Alaska Native Language Center, University of Alaska Fairbanks; Fairbanks: 2011. [Google Scholar]
- 35.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett J, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Maekawa K, Fukushima-Uesaka H, Tohkin M, Hasegawa R, Kayjio H, Kuzuya N, et al. Four novel defective alleles and comprehensive haplotype analysis of CYP2C9 in Japanese. Pharmacogenetics and genomics. 2006;16:497–514. doi: 10.1097/01.fpc.0000215069.14095.c6. [DOI] [PubMed] [Google Scholar]
- 38.Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D, et al. Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5902–7. doi: 10.1073/pnas.0730858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Limdi NA, Wiener H, Goldstein JA, Acton RT, Beasley TM. Influence of CYP2C9 and VKORC1 on warfarin response during initiation of therapy. Blood cells, molecules & diseases. 2009;43(1):119–28. doi: 10.1016/j.bcmd.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Limdi NA, Wadelius M, Cavallari L, Eriksson N, Crawford DC, Lee MT, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115(18):3827–34. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet. 2008;82(2):495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alsmadi O, John SE, Thareja G, Hebbar P, Antony D, Behbehani K, et al. Genome at juncture of early human migration: a systematic analysis of two whole genomes and thirteen exomes from Kwaiti Population subgroup of inferred Saudi Arabian tribe ancestry. PLOS ONE. 2014;9(6):e99069. doi: 10.1371/journal.pone.0099069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. The pharmacogenomics journal. 2005;5(4):262–70. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 44.King CR, Deych E, Milligan P, Eby C, Lenzini P, Grice G, et al. Gamma-glutamyl carboxylase and its influence on warfarin dose. Thrombosis and haemostasis. 2010;104(4):750–4. doi: 10.1160/TH09-11-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, et al. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thrombosis research. 2007;120(2):181–6. doi: 10.1016/j.thromres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Vijayasarathy C, Sui R, Zeng Y, Yang G, Xu F, Caruso RC, et al. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Human mutation. 2010;31(11):1251–60. doi: 10.1002/humu.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gannage-Yared MH, Makrythanasis P, Chouery E, Sobacchi C, Mehawej C, Santoni FA, et al. Exome sequencing reveals a mutation in DMP1 in a family with familial sclerosing bone dysplasia. Bone. 2014;68:142–5. doi: 10.1016/j.bone.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Farrow EG, Davis SI, Ward LM, Summers LJ, Bubbear JS, Keen R, et al. Molecular analysis of DMP1 mutants causing autosomal recessive hypophosphatemic rickets. Bone. 2009;44(2):287–94. doi: 10.1016/j.bone.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caridi G, Dagnino M, Lugani F, Shalev SA, Campagnoli M, Galliano M, et al. A novel mutation in the albumin gene (c.1A>C) resulting in analbuminemia. European journal of clinical investigation. 2013;43(1):72–8. doi: 10.1111/eci.12019. [DOI] [PubMed] [Google Scholar]
- 50.Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, et al. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. The pharmacogenomics journal. 2014;14(1):85–92. doi: 10.1038/tpj.2013.2. [DOI] [PubMed] [Google Scholar]
- 51.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, et al. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. The Journal of biological chemistry. 2004;279(34):35630–7. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 52.Niinuma Y, Saito T, Takahashi M, Tsukada C, Ito M, Hirasawa N, et al. Functional characterization of 32 CYP2C9 allelic variants. The pharmacogenomics journal. 2014;14(2):107–14. doi: 10.1038/tpj.2013.22. [DOI] [PubMed] [Google Scholar]
- 53.Roth JA, Boudreau D, Fujii MM, Farin FM, Rettie AE, Thummel KE, et al. Genetic risk factors for major bleeding in patients treated with warfarin in a community setting. Clin Pharmacol Ther. 2014;95(6):636–43. doi: 10.1038/clpt.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium IWP. Estimation of the warfarin dose with clinical and pharmacogenetic data. The New England journal of medicine. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 Haplotypes on Transcriptional Regulation and Warfarin Dose. New England Journal of Medicine. 2005;352(22) doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 56.Nahar R, Saxena R, Deb R, Parakh R, Shad S, Sethi PK, et al. CYP2C9, VKORC1, CYP4F2, ABCB1 and F5 variants: influence on quality of long-term anticoagulation. Pharmacological reports : PR. 2014;66(2):243–9. doi: 10.1016/j.pharep.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Edson KZ, Prasad B, Unadkat JD, Suhara Y, Okano T, Guengerich FP, et al. Cytochrome P450-dependent catabolism of vitamin K: omega-hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry. 2013;52(46):8276–85. doi: 10.1021/bi401208m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.