Abstract
ABCC2 is a transporter with key influence on liver and kidney pharmacokinetics. In order to explore the structure–activity relationships of compounds that modulate ABCC2, and by doing so gain insights into drug–drug interactions, we screened a library of 432 compounds for modulators of radiolabeled β-estradiol 17-(β-D-glucuronide) (EG) and fluorescent 2′,7′-dichlorofluorescin transport (CDCF) in membrane vesicles. Following the primary screen at 80 μM, dose–response curves were used to investigate in detail 86 compounds, identifying 16 low μM inhibitors and providing data about the structure–activity relationships in four series containing 19, 24, 10, and eight analogues. Measurements with the CDCF probe were consistently more robust than for the EG probe. Only one compound was clearly probe-selective with 50-fold difference in the IC50s obtained by the two assays. We built 24 classification models using the SVM and fused-XY Kohonen methods, revealing molecular descriptors related to number of rings, solubility and lipophilicity as important to distinguish inhibitors from inactive compounds. This study is to our best knowledge the first to provide details about structure–activity relationships in ABCC2 modulation.
Keywords: ABCC2, ABC transporter, CDCF, Estradiol glucuronide, Efflux, Inhibitor, Membrane vesicles, Modulator, MRP2, Sf9, Stimulator, Structure–activity relationships, SVM modelling, Transport, Transport assay
1. Introduction
The ATP Binding Cassette family C2 (ABCC2, also known as Multidrug Resistance-associated Protein 2, MRP2, or canalicular multispecific organic anion transporter 1, cMOAT), is a member of the ATP-binding cassette protein family. ABCC2 is expressed at the apical side of polarized cells in the liver, kidney, small intestine, and placenta. This localization is fundamental to its role in the elimination of endogenous compounds as well as xenobiotics.
ABCC2 has been classified as a Multi-Drug Resistance Protein family member following its identification as overexpressed in cisplatin-resistant cancer cell lines.1 ABCC2 expression has since then been associated with intrinsic as well as probably acquired resistance to chemotherapy in patients, for example, with oesophageal squamous cell carcinoma.2 ABCC2 indeed transports many chemotherapeutic agents such as vinblastine,3 vincristine,4 epirubicin4 and chlorambucil.5 The main physiological role for ABCC2 identified to date is to efflux organic anionic metabolites and conjugates including leukotriene-C4,4 glutathione,6 bilirubin as mono or bi-glucuronide,7 estradiol glucuronide4 and p-aminohippurate.8 There is evidence that a majority but not all ABCC2 substrates have an anionic character.9–12
While no clinically relevant drug–drug interactions affecting ABCC2 have been identified to date, there is a high risk that these would occur.13 An impaired activity of ABCC2 due to genetic factors or through inhibition (drug–drug interaction) can lead to conjugated hyperbilirubinemia. The importance of examining ABCC2 inhibition was recently underlined by the International Transporter Consortium for potential drug candidates that induce conjugated hyperbilirubenemia.13 Compounds or metabolites that would inhibit ABCC2, as well as other transporters that are present in the hepatocytes such as MRP3 and MRP4, may cause hepatotoxicity. 13 Similarly, blocking ABCC22 with small molecules, typically chemotherapeutic agents, may cause the accumulation of organic anions inside renal proximal tubule cells; inhibition of mitochondrial DNA synthesis by these organic anions could subsequently cause the iatrogenic Fanconi Syndrome.14 In this context, potent inhibitors of ABCC2 are of interest for the study of drug–drug interactions. In addition, there is interest in developing drug molecules able to evade or circumvent transporters such as ABCC2.
ABCC2 is a large membrane-embedded protein (1545 amino acids, 17 transmembrane segments) that folds into five domains: the ABCC2 sequence contains a duplicated unit composed of one transmembrane domain (six transmembrane segments) and one intracellular nucleotide binding domain. This unit, duplicated or not, is found in most ABC systems. Characteristic of the ABCC family is an extra domain, composed of five transmembrane segments of unknown function. Upon folding, the two domains with six transmembrane segments would arrange in parallel to form a central binding cavity in ABC transporters, as is seen in the crystal structures of the mouse MDR1A (PDB code 4M1M),15,16 human ABCB10 (4AYT),17 as well as in bacterial transporters such as Sav 1866 (2HYD)18 and MsbA lipid flippase (3B60).19 This central binding cavity located mid-way to the membrane is an essential component of the alternate access model proposed by Jadzerky in 1966, which postulates that the transporter ‘oscillates’ between two conformations during transport cycle.20 All known transport proteins crystallized to date, including the ABC transporters, present characteristics compatible with the alternate access model.21–23 ABCC2-mediated transport is complex, as demonstrated by the sigmoidal plot of transport, characteristic of self-stimulation, of the endogenous molecule EG with human (but not rat) ABCC2.24–26
The interaction of compounds with ABCC2 is often studied using the vesicular transport assay, where compounds are assessed for their modulatory effect (stimulation, inhibition) on the transport of a detectable probe. Transport of the tested compounds in itself is not seen in this assay, but most of the transported substrates should be detected along with other non-transported inhibitors (transported substrates are likely to behave as competitive inhibitors). In the framework of the ABCC2 three-dimensional structure (Fig. 1), modulators may exert their action either directly by binding to the central cavity and competing with the transported substrate (modulators are then not necessarily transported themselves) or binding to either of the two nucleotide binding domains, uncoupling the transporter from its energy source. For substrates, the modulatory effect is a consequence not only of the affinity towards the central binding cavity, but also of the capacity to trigger transport, which affect the residence time of the probe competitor and therefore the assay readout. Substrates that differ in their transport capacity are for example CDCF and LTC4.27 More subtly modulators could be co-accommodated with the transported substrate. In the case of the mouse MDR1, there is crystallographic evidence that the binding cavity can accommodate two substrate molecules.15,16 Other binding sites may be postulated, which would allosterically affect affinity and/or capacity in the central binding cavity.
Figure 1.
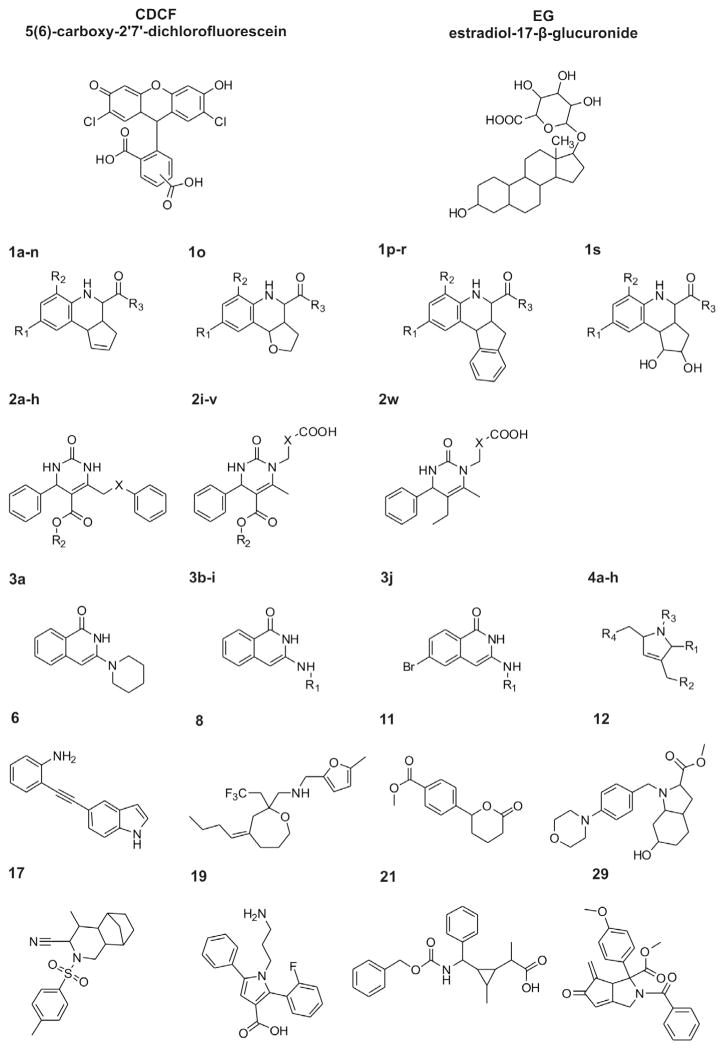
2D structures of CDCF and EG; Markush structures of the compounds presented in Tables 2a–e.
In a comprehensive study of ABCC2 modulators, Pedersen and coworkers tested the effect of 191 chemically diverse drugs and drug-like compounds on the ABCC2-mediated transport of radiolabeled β-estradiol 17-β-D-glucuronide (EG).12 Intriguingly, presence of a net negative charge at physiological pH was not found to be a fundamental characteristic of inhibitors (n = 42 molecules, 37% having a net negative charge), however a net negative charge was seen as preferred in the small sample of stimulators under study (n = 13, 77%; all carrying an anionic center).12 Few reports have otherwise discussed the requirement of modulators to carry an anionic group, and this requirement was never discussed in the context of series of analogues.
The main objective of this study is to generate novel information about the chemical profile of ABCC2 inhibitors, especially information about the SARs in series of analogues, which could be later translated to early stages of drug development.28,29 EG is not the only probe available for vesicular transport assays and, as an alternative, the International Transporter Consortium has recently suggested the use of 5(6)-carboxy-2′,7′-dichlorofluorescein (CDCF).13 CDCF is suitable for high-throughput setups due to its detection by fluorescence and lower costs, but has never been previously used for larger screening purposes.27 By comparing the effects of compounds on CDCF and EG, we have recently introduced the concept of probe-selective compounds, that is, compounds that exert different modulatory effect on EG and CDCF.30 Comparison with a larger dataset has however not yet been reported. Effects of multiple probes would thus be interesting to compare on a larger dataset.
Here, using a vesicular transport assay conducted with both EG and CDCF we screened for ABCC2 modulation a library of 432 small molecules composed around series of analogues. This screening data was used to build classification models, and the most predictive SVM models used to highlight the descriptors important for discriminating inhibitors from inactive compounds. One of the classification models was validated using a foreign set of compounds that confirmed the predictivity assessed using a test-set. For a smaller set of 86 molecules, dose–response experiments were carried out using both substrates probes, EG and CDCF. 64 of these compounds belong to four series of analogues and the SARs relationships of those are discussed in the light of the presence or absence of anionic functional groups.
2. Materials and methods
2.1. Material
Cloned human ABCC2, pGEM3-ABCC2 (U49248), was a kind gift from Dr. Piet Borst (The Netherlands Cancer Institute). HyQ®SFX-Insect MP medium was obtained from Hyclone (Logan, UT, USA). [3H]-EG (1.0 mCi/ml) was purchased from Perkin–Elmer (Boston, MA, USA). EG, CDCF and ATP were purchased from Sigma–Aldrich (St. Louis, MO, USA). The chemical library of 432 compounds used in this study is a subset of the University of Pittsburgh Center for Chemical Methodologies and Library Development (UPCMLD) library (http://ccc.chem.pitt.edu/UPCMLD/index.html; purity data is supplied in Supporting information S2). The UPCMLD library was obtained by diversity-oriented synthesis strategies in a Center Core facility and guided by innovative organic synthesis applied to multicomponent reactions, organometallic imine additions, and natural product inspired heterocyclic chemistry (for lead references on the preparation of library compounds, see Refs. 19–2131–33). The compounds were first diluted to 100 mM with DMSO and stored at −80 °C. Before screening, the compounds were further diluted in DMSO to appropriate concentrations and stored at −20 °C. 2D structures of the 432 tested compounds are provided as (Appendix A).
2.2. Vesicle preparation and vesicular transport assay
Human ABCC2 was expressed in Spodoptera frugiperda (Sf9) insect cells using the Bac-to-Bac® expression system (Invitrogen Life Technologies, Carlsbad, CA, USA). The inside-out membrane vesicles containing ABCC2 were prepared and vesicular transport assay was performed as previously described.30 The base-line of ATP-dependent transport of probes (EG or CDCF) was set as 100%. The final DMSO concentration in the reaction was 1.5%, which does not interfere with the transport of the probes (data not shown). The modulation effect was then calculated as the ratio between the ATP-dependent probe transport with and without the tested compound. For primary screening all library compounds were tested as single point at 80 μM. The compounds selected for dose–response experiments were evaluated with three to seven concentrations in triplicates.
2.3. IC50 calculation and curve fitting
The IC50 values were estimated using one-way ANOVA with Dunnett’s post test dynamic curve fitting four parameter logistic (GraphPad Prism version 6.0e, GraphPad Software, San Diego California USA, www.graphpad.com) Eq. 1.
| (1) |
We constrained Bottom to null as a negative value is only an artefact in the methods of detecting relative transport. No constraints were put on Top as we were expecting some points to give values higher than 100% (stimulation).
2.4. Classification models
The three-dimensional structures of the 432 molecules were constructed by Sybyl-X 1.2 software (Tripos, USA) and their molecular descriptors calculated using PaDEL,34 Discovery Studio35 and Volsurf+.36 Empty descriptors were then removed, and for correlated descriptors (pairwise correlation coefficient above 0.9) a single representative kept. SVM calculations were performed with the LIBSVM software using radial basis function kernels. Descriptors were selected to the SVM model based on F-scores,37 which measures the relative importance of each descriptor in classifying the molecules in classes (higher F-scores means better discrimination between the classes). A toolbox containing a collection of MATLAB modules for developing Kohonen Maps and Counter propagation Artificial Neural networks was also used.38,39 The descriptors computed by Volsurf+ were used to investigate the chemical space using Self-Organizing Maps (Java SOMToolbox).
2.5. External validation
A virtual collection of 52,563 compounds was obtained from the UPCMLD. To work in the same chemical space only compounds with a Tanimoto score higher than 0.7 (compared to compounds of the training set) were filtered and 44 compounds were selected, and their activities tested with the vesicular transport assay.
3. Results and discussion
3.1. Primary screening and dose–response analysis using two probes
3.1.1. Vesicular transport assay
In this work, we assessed 432 compounds for their modulatory effect on the ATP-dependent and ABCC2-mediated transport of two substrates used as chemical probes, EG and CDCF. The assay has been presented previously30 and is conducted in isolated ‘inside-out’ membrane vesicles from Spodoptera frugiperda Sf9 cells overexpressing ABCC2 (Fig. 1, chemical structures of EG and CDCF). Probe transport is detected by measuring radioactivity counts for EG and fluorescence for CDCF and the rate of probe transport without ATP is used to set the baseline. The vesicular transport assay does not provide a direct measurement about the transport of the tested compounds, but uncovers modulators, that is, molecules that inhibit or stimulate transport of the probe(s). In this context, some of the compounds investigated may be passively transported through partitioning to the membrane (or possibly actively transported by endogenous transporters present in insect cells40 without interfering with the results of the transport assay (see Ref. 12).
Each of the EG and CDCF transport assays have previously been individually parameterized to offer an optimal signal-to-noise ratio for each of the two probes.12,27 Under our experimental conditions, the IC50 of CDCF for the inhibition of EG transport is 36 μM (95% confidence interval 1–71 μM); vice-versa, the IC50 of EG is 83 μM (73–93 μM) for the inhibition of CDCF transport.
3.1.2. Primary screening
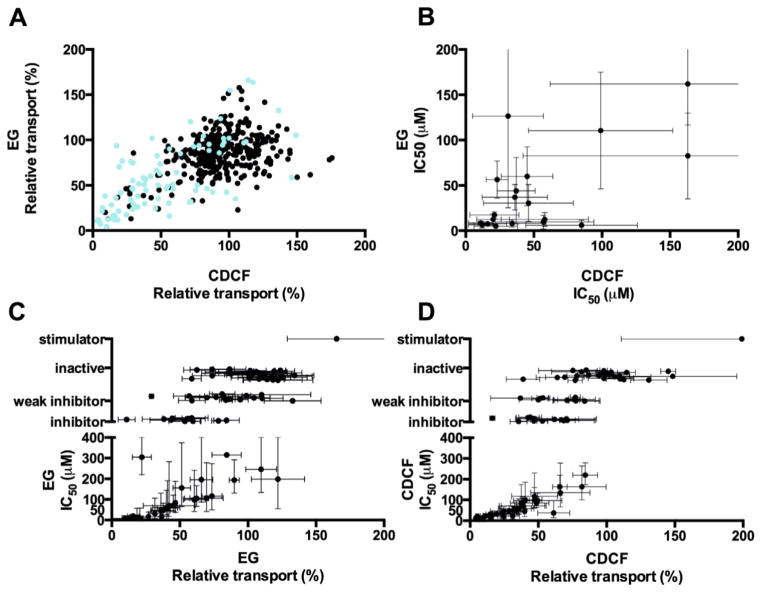
Overall, ABCC2 appears to be relatively readily amenable to modulation (Fig. 2A, Table 1, 2D structures in Appendix A). As a result of the primary screening, 96 compounds (22% of the library) were found to have a modulatory effect (transport activity 0–50% or >150%) on either EG or CDCF transport at the tested concentration, 80 μM. These compounds are divided into 84 potential inhibitors (64 for EG and 65 for CDCF) and 12 stimulators (6 EG stimulators and 6 CDCF stimulators). When milder boundaries (transport activity of 0–80% or >120%) are considered, 268 compounds (62% of the library) had an effect on either EG or CDCF transport. These high numbers suggest a larger than previously thought implication of ABCC2 in pharmacokinetics and drug–drug interactions, since they mean that a previously unexpected number of small molecules are potentially able to interfere with the ABCC2-mediated transport.
Figure 2.
Comparisons of the modulatory effects inferred from CDCF and EG transport inhibition. (A) 432 compounds tested in primary screening, including the 86 compounds selected for dose–response measurements (cyan); (B–D) 86 compounds further analyzed with dose–response measurements. In (B) plot of IC50s of compounds that are both CDCF and EG inhibitors; in (C and D) the average of three measurements at 80 μM with standard deviations and estimated IC50s with 95% confidence intervals.
Table 1.
Modulatory effects of 432 compounds on CDCF and EG transport (primary screen)
| EG transport
|
|||||||
|---|---|---|---|---|---|---|---|
| Inhibitora | Borderline inhibitorb | Inactivec | Borderline stimulatord | Stimulatore | Total | ||
| CDCF transport | Inhibitora | 45 | 16 | 4 | 0 | 0 | 65 |
| Borderline inhibitorb | 11 | 24 | 34 | 1 | 0 | 70 | |
| Inactivec | 7 | 61 | 163 | 12 | 6 | 249 | |
| Borderline stimulatord | 1 | 11 | 28 | 2 | 0 | 42 | |
| Stimulatore | 0 | 3 | 2 | 1 | 0 | 6 | |
| Total | 64 | 115 | 231 | 16 | 6 | 432 | |
% I, relative probe transport at 80 μM.
% I < 50%.
50% ≤ % I < 80%.
80% ≤ % I < 120%.
120% ≤ % I < 150%.
% I ≥150%.
Tables 2a–e.
Substituents and modulatory effects and/or estimated IC50s for 86 compounds selected from dose–response measurements. Unless mentioned, 5–7 different concentrations in triplicates were measured
| Table 2a
| |||||||
|---|---|---|---|---|---|---|---|
| # | –R1 | –R2 | –R3 | CDCF
|
EG
|
||
| IC50 (95% CI), μM | Hill slope (95% CI) | IC50 (95% CI), μM | Hill slope (95% CI) | ||||
| 1a | –H | –H | –OH | 16 (14–19) | −1.6 (−1.9, −1.2) | 9 (6–14) | −1.7 (−2.6, −0.76) |
| 1b | –H | –H | –OCH2CH3 | Inhibitora | 31 (9–106) | −0.9 (−1.8, −0.0) | |
| 1c | –H | –H |
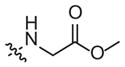
|
58 (32–105) | −0.7 (−1.0, −0.4) | 18 (7–45) | −0.7 (−1.1, −0.3) |
| 1d | –H | –H |
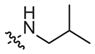
|
99 (53–185) | −0.7 (−1.1, −0.4) | 156 (65–376) | −1.4 (−2.9, −0.1) |
| 1e | –CH3 | –H | –OH | 22 (16–31) | −1.4 (−1.9, −0.8) | 7 (3–18) | −1.2 (−2.3, −0.1) |
| 1f | –CH3 | –H | –OCH2CH3 | 52 (29–95) | −0.6 (−0.9, −0.4) | Inhibitorb | |
| 1g | –CH3 | –F | –OCH2CH3 | 11 (9–13) | −0.9 (−1.0, −0.8) | 10 (7–14) | −2.3 (−3.7, −0.9) |
| 1h | –CH3 | –H |
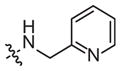
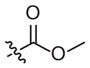
|
85 (41–175) | −0.8 (−1.3, −0.3) | 10 (2–58) | −0.7 (−1.4, −0.0) |
| 1i | –OCH3 | –H | –OCH2CH3 | 20 (12–33) | −0.8 (−1.1, −0.5) | 17 (8–39) | −0.9 (−1.5, −0.3) |
| 1j | –CF3 | –H | –OCH2CH3 | 23 (18–30) | −1.2 (−1.5, −0.9) | Inhibitora | |
| 1k | –OCF3 | –H | –OCH2CH3 | 23 (8–70) | −0.9 (−1.5, −0.2) | 71 (42–122) | −1.1 (−1.7, −0.5) |
| 1l | –OCF3 | –H | –OH | 12 (10–15) | −1.4 (−1.8, −1.0) | 9 (4–22) | −0.9 (−1.5, −0.3) |
| 1m | –OCF3 | –H |
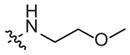
|
Weak inhibitor | Inactivea | ||
| 1n |
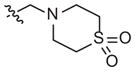
|
–H | –OCH2CH3 | Inhibitorb | 11 (5–24) | −0.8 (−1.3, −0.4) | |
| 1o* | –CF3 | –H | –OH | Inactive | Inactivea | ||
| 1p# | –H | –H | –OCH2CH3 | Inhibitorb | Weak inhibitora | ||
| 1q# | –CH3 | –F | –OH | 21 (18–25) | −2.2 (−3.1, −1.2) | 20 (15–29) | −1.4 (−1.9, − 0.8) |
| 1r# | –CF3 | –H | –OH | 48 (34–68) | −1.2 (−1.6, −0.8) | Weak inhibitora | |
| 1s* | –H | –H | –OCH2CH3 | Weak inhibitora | 105 (66–167) | n.d.c | |
| Table 2b
| |||||||
|---|---|---|---|---|---|---|---|
| # | –X– | –R2 | –R3 | CDCF
|
EG
|
||
| IC50 (95% CI), μM | Hill slope (95% CI) | IC50 (95% CI), μM | Hill slope (95% CI) | ||||
| 2a | –O– | –CH2CH3 | 4-Br | Inactive | Inactivea | ||
| 2b | –CH2– | –CH3 | 3-NO2 | Inactive | Inactivea | ||
| 2c | –CH2– | –CH3 | 4-Br | Inactivea | Inactivea | ||
| 2d | –CH2– | –CH3 | 4-Cl | 117 (60–230) | −0.6 (−1.0, −0.3) | Inhibitora | |
| 2e | –CH2– | –CH3 | 4-NO2 | Inactivea | Inactivea | ||
| 2f | –CH2– | –CH2CH3 | 4-CN | Weak inhibitora | Inactivea | ||
| 2g | –CH2– | –CH2CH3 | 4-Cl | Inactive | Weak inhibitora | ||
| 2h | –CH2– | –CH2CH3 | 3-NO2 | Inactivea | Inactive | ||
| 2i* | –(CH2)2– | –CH2CH3 | –H | 85 (63–114) | −1.4 (−1.9, −0.8) | Weak inhibitora | |
| 2j* | –(CH2)2– | –CH2CH3 |
 2,3 |
71 (47–107) | n.d.b | Weak inhibitora | |
| 2k* | –(CH2)2– | –CH3 |
 2,3 |
Weak inhibitora | Inactivea | ||
| 2l* | –(CH2)2– | –CH3 | 4-Cl | Inactivea | Inactive | ||
| 2m* | –(CH2)2– | –CH3 |
 4 |
Weak inhibitora | 246 (133–454) | n.d.b | |
| 2n* | –(CH2)2– | –CH2CH3 |
|
31 (26–37) | −1.9 (−2.3, −1.5) | 198 (55–718) | n.d.b |
| 2o* | –(CH2)4– | –CH2CH3 | 2-Cl; 3-Cl | 46 (33–66) | −1.8 (−2.7, −1.0) | 45 (16–130) | −1.1 (−2.2, −0.0) |
| 2p* | –(CH2)4– | –CH2CH3 | 4-Cl | Weak inhibitora | Weak inhibitora | ||
| 2q* |
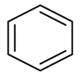
|
–CH2CH3 | 4-NO2 | 34 (25–46) | −1.3 (−1.7, −0.9) | 11 (5–25) | −1.2 (−2.1, −0.3) |
| 2r* | –(CH2)2– |
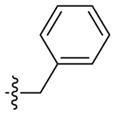
|
4-Cl | 47 (31–72) | −1.6 (−2.4, −0.8) | Weak inhibitora | |
| 2s* | –(CH2)2– |
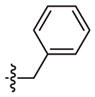
|
2-Cl | 58 (43–79) | −1.6 (−2.4, −0.9) | Weak inhibitora | |
| 2t* | –(CH2)2– |
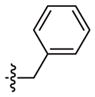
|
4-OCH3 | 135 (66–278) | n.d.b | Weak inhibitora | |
| 2u* | –(CH2)2– | –CH3 |
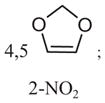
|
57 (37–87) | −1.4 (−2.2, −0.7) | 15 (4–57) | −1.3 (−3.3, −0.7) |
| 2v* | –(CH2)2– |
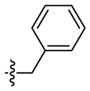
|
3-OCH3 | 97 (72–132) | −1.7 (−2.7, −0.7) | Inhibitora | |
| 2w# | –(CH2)2– | — | 2-Cl, 3-Cl | Weak inhibitor a | Weak inhibitora | ||
| Table 2c
| |||||
|---|---|---|---|---|---|
| # | –R1 | CDCF
|
EG
|
||
| IC50 (95% CI), μM | Hill slope (95% CI) | IC50 (95% CI), μM | Hill slope (95% CI) | ||
| 3a* | Inactivea | Inactivea | |||
| 3b | –(CH2)6OH | 36 (24–55) | −1.5 (−2.1, −0.8) | 47 (27–82) | −1.0 (−1.5, −0.5) |
| 3c |
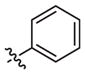
|
163 (101–264) | −1.7 (−2.6, −0.7) | 194 (130–292) | −2.2 (−3.3, −1.1) |
| 3d |
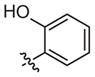
|
Inhibitorb | 61 (26–144) | −1.3 (−2.7, −0.1) | |
| 3e |
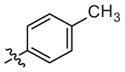
|
72 (46–114) | −1.3 (−2.0, −0.6) | Weak inhibitora | |
| 3f |
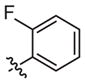
|
Inhibitora | Weak inhibitora | ||
| 3g |
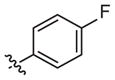
|
100 (63–160) | −1.3 (−2.1, −0.5) | Weak inhibitora | |
| 3h |
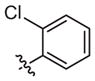
|
Inactive | Weak inhibitorb | ||
| 3i |
|
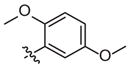
Weak inhibitora | Inhibitora | ||
| 3j# |
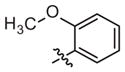
|
6 (2–23) | −0.9 (−1.5, −0.2) | 305 (220–423) | −1.1 (−1.7, −0.4) |
| Table 2d
| ||||||||
|---|---|---|---|---|---|---|---|---|
| # | –R1 | –R2 | –R3 | –R4 | CDCF
|
EG
|
||
| IC50 (95% CI), μM | Hill slope (95% CI) | IC50 (95% CI), μM | Hill slope (95% CI) | |||||
| 4a |
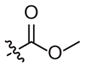
|
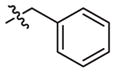
|
–CH3 | –CH3 | Weak inhibitor | Weak inhibitor | ||
| 4b |
|
|
|
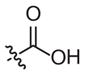
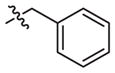
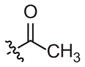
–CH3 | 163 (121–219) | −1.2 (−1.6, −0.8) | 116 (49–273) | −0.9 (−1.6, −0.2) |
| 4c |
|
|
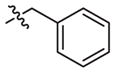
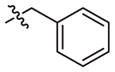
|
–CH3 | Inactivea | Inactivea | ||
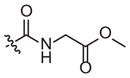
| 4d |
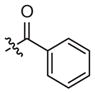
|
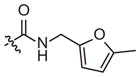
|
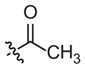
|
–CH3 | Inhibitora | Inactivea | ||
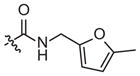
| 4e |
|
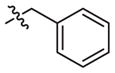
|
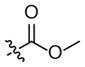
|
–CH3 | 37 (14–97) | −0.6 (−0.9, −0.3) | Weak inhibitora | |
| 4f |
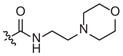
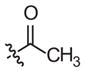
|
–CH3 |
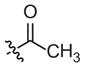
|
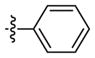
|
Inactivea | 70 (18–283) | −1.0 (−2.1, −0.1) | |
| 4g |
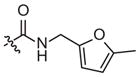
|
–CH3 |
|
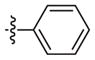
|
Weak inhibitora | 59 (18–191) | −0.8 (−1.3, −0.2) | |
| 4h |
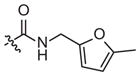
|
–CH3 |
|
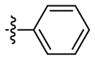
|
Weak inhibitora | 99 (40 240) | n.d.c | |
| Table 2e
| ||||
|---|---|---|---|---|
| # | CDCF
|
EG
|
||
| IC50 (95% CI), μM | Hill slope (95% CI) | IC50 (95% CI), μM | Hill slope (95% CI) | |
| 5 | Inactivea | Inactivea | ||
| 6 | 23 (14–38) | −1.5 (−2.4, −0.5) | Weak inhibitora | |
| 7 | Inactivea | Inactivea | ||
| 8 | 32 (26–39) | −1.5 (−1.8, −1.1) | Inhibitorb | |
| 9 | Weak inhibitora | Weak inhibitora | ||
| 10 | Inhibitora | Weak inhibitora | ||
| 11 | 45 (19–105) | −0.8 (−1.3, −0.3) | 83 (37–188) | −0.8 (−1.4, −0.2) |
| 12 | Inactivea | 196 (87–439) | −0.7 (−1.3, −0.2) | |
| 13 | Weak inhibitora | Weak inhibitora | ||
| 14 | Inactivea | Weak inhibitora | ||
| 15 | Inactivea | Inactivea | ||
| 16 | Inactivea | Inactivea | ||
| 17 | Stimulator | Inactivea | ||
| 18 | Inactivea | Inactive | ||
| 19 | Weak inhibitora | 106 (41–278) | −1.0 (−1.9, −0.0) | |
| 20 | Inactivea | Inactivea | ||
| 21 | 220 (173–279) | −1.6 (−2.1, −1.1) | Stimulatora | |
| 22 | Inactivea | Inactivea | ||
| 23 | Inactivea | Inactivea | ||
| 24 | Inactivea | Inactivea | ||
| 25 | Inactivea | Inactivea | ||
| 26 | Inactive | Inactivea | ||
| 27 | Inactivea | Inactivea | ||
| 28 | Weak inhibitora | Weak inhibitora | ||
| 29 | 7 (5–11) | −1.1 (−1.4, −0.7) | Inhibitorb | |
| 30 | Inactivea | Inactive | ||
Modifications of the cyclopentene ring.
Benzene ring fused to cyclopentene (Fig. 1).
Modulatory effect estimated from three concentrations only.
4-Parameter logistic model curve could not be fitted.
Hill coefficient could not be calculated with significant accuracy (within three units).
The scaffold is subdivided into 2a–2h,
2i–v and
2w (Fig. 1).
Modulatory effect estimated from three concentrations only.
Hill coefficient could not be calculated with significant accuracy (within three units).
Piperidine group.
Modifications of the phenyl ring (Fig. 1).
Modulatory effect estimated from three concentrations only.
4-Parameter logistic model curve could not be fitted.
Modulatory effect estimated from three concentrations only.
4-Parameter logistic model curve could not be fitted.
Hill coefficient could not be calculated with significant accuracy (within three units).
Modulatory effect estimated from three concentrations only.
4-Parameter logistic model curve could not be fitted.
Arguably, caution should be taken with the primary screening data since only a single measurement at one concentration was made. When the primary screening data is completed with more concentration points, as is done for 86 compounds selected for further analysis, 62% (53 out of 86) of the compounds for EG and 79% (68 out of 86) for CDCF remain in the inhibitor/stimulator/inactive classes predicted by the primary screen. Most of the compounds that change classes shift into the nearest class, for example, inhibitor to weak inhibitor.
Comparing the functional assignment into inhibitory, stimulatory and inactive classes obtained using either the EG or the CDCF probes in primary screening, the agreement is very high, 91% (Table 1) (borderline groups, which may be ‘shifting’ classes easily, not being considered). Furthermore, a weak linear relationship (correlation coefficient of 0.36) is seen when the numerical values of relative transport rate inhibition obtained for EG are compared to those obtained for CDCF (Fig. 2A). Relative transport rate inhibition values depend directly on the assay set-up; here we have used in both cases concentrations close to half-times Km (50 μM EG, and 5 μM CDCF); other conditions such as incubation times (10 and 30 min) are different.
3.1.3. Dose–response curve analysis and IC50 estimation
86 compounds were selected for further analysis (Fig. 2B–D; 2D structures and IC50 curves in Appendices B and C.1). The selection process aimed to conciliate simultaneously three objectives: (1) populate certain scaffolds with priority to obtain ‘instant SARs’; (2) investigate molecules with large variation of effects between the two probes; (3) investigate the strongest inhibitors. For 50 (CDCF) and 38 (EG) of these compounds, five to seven concentration points were measured in triplicates and, for inhibitors, IC50s and Hill coefficients estimated from the dose–response curves using 4-parameter logistic regression (see Section 2). The remaining compounds from the set, weaker inhibitors and inactive compounds (included on purpose to the selection), were tested with three concentrations points only.
As a result, 25 EG and 38 CDCF inhibitors (23 common to both) were identified. The remaining compounds were weak inhibitors and inactive compounds. Overall, at almost all concentrations, the standard deviation of the CDCF measurements is lower than the EG measurements, and therefore the CDCF assay appears more robust (Appendix Table C.2). For the majority of the inhibitors, IC50 values could be estimated without any difficulties from the data collected. A few compounds with a very weak slope in the dose response curve, for example, 1f, 3h (EG) and 1n (CDCF), or a very steep slope, for example, 29 (EG) and 1p and 3d (CDCF), did not fit the 4-parameter logistic regression; for those, IC50s are not provided. Many compounds, such as 1c, 2h, 2n, 2m, 2q, 3d, 8, 18, 19 (EG) and 1k, 2t, 3j, 6 (CDCF), showed a weak, often non-significant stimulation effect towards the left part of their dose–response curve (at low concentrations), which made the curve-fitting and estimation of IC50 values difficult. This behaviour has been previously reported.25 Furthermore, the low-concentration stimulatory effect probably introduced uncertainties in the estimates of Hill coefficients, and for four compounds too large deviations were obtained. Last, while for most compounds the measurement at the highest concentration (400 μM) is not raising any alarm, for five compounds, 2g, 3i (EG and CDCF), 2f (EG) and 1n, 12 (CDCF), the curves do not reach complete inhibition and stay at the level of the 80 μM datapoint. This behaviour may reflect experimental uncertainties, or possibly be an artefact resulting from lack of solubility leading to aggregation or from leakage of vesicles caused by partitioning of compounds into the membrane. 30 Intuitively, aggregation and leakage should affect both EG and CDCF similarly, and there are only two compounds (2g, 3i) fitting this profile. Partial inhibition, which may be the result of allosteric effects, may also be invoked as a hypothesis. To the best of our knowledge, partial inhibition has never been suggested nor demonstrated for any ABC transporter.
3.2. Identification of novel ABCC2 modulators
3.2.1. Chemical scaffolds
The library is constituted of series of analogues (annotated and grouped molecular structures are provided in Appendix A). There are many ways to classify compounds into scaffold groups,41,42 and here we used a variant of the Schuffenhauer rules.42 Under this classification scheme, 349 molecules (81% of library) can be grouped together into 32 series of analogues having three or more representatives. Roughly half of the compounds can be assigned to the 10 most populated series of analogues. The set of 86 compounds selected for testing using dose–response curves contain 62 molecules that can be categorized in four series of analogues (Fig. 1, Tables 2a–e): cyclopentahydroquinoline (referred to later as Scaffold 1, 19 molecules selected out of 25 analogues present in primary screen), 4-phenyl-3,4-tetrahydropyrimindin-2-one (Scaffold 2, 24 out of 59), 3-(phenylamino)-1,2-dihydroisoquinolin- 1-one (Scaffold 3, 10 out of 19), 2-benzyl-2,5-dihydro-1H-pyrrole (Scaffold 4, 8 out of 24). Scaffold 1 contains in proportion the largest percentage of inhibitors of at least one probe, 68% in the primary screen. This finding is in agreement with the known tendency of tetrahydroquinolines to be promiscuous binders in bioassays (see, for example Ref. 43,44).
3.2.2. Most potent and probe-selective modulators
The dose–response analysis revealed 16 compounds with IC50s lower than 30 μM on either of the two probes (Tables 2a–e): these are compounds 1a, 1c, 1e, 1g–l, 1n, 1q; compounds 2q, 2u; compound 3j; and compounds 6 and 29. Even when taken outside of the context of their analogue series, by themselves, these compounds provide us with information about the chemical structures of potent ABCC2 inhibitors, which could help to predict ABCC2-mediated drug–drug interactions. Furthermore, these compounds are potentially interesting as biochemical probes for studying MRP function; nonetheless, to be used for that purpose their modulatory effect should be tested on other ABC transporters as well.
Another interesting aspect sought here is the compound’s ability to affect differently the transport of different probes, since probe-selective compounds could become useful chemical tools in deciphering the complex transport mechanism of ABCC2. Probe selectivity can be demonstrated from a large difference in IC50s, or from different modulatory effects (stimulator, inactive, inhibitor) observed within the window of concentrations considered. In itself, that some compounds exert different modulatory effects on CDCF and EG is not surprising since these probes have themselves different transport characteristics: While CDCF shows a classical ‘Michaelis’ type of transport kinetics,27 EG shows a sigmoidal curve characteristic of self-stimulation.24,45
Here, the most striking trend is that most of EG inhibitors, 24 out of 25 (96%), also inhibit CDCF transport (23 inhibitors, 1 weak inhibitor); and reciprocally most of the CDCF inhibitors, 37 out of 38 (97%) inhibit EG transport (23 inhibitors, 14 weak inhibitors) (Table 3). For example, the strongest inhibitors 1a, 1g, 1i, 1l, and 1q have IC50s within the same order of magnitude for EG and CDCF. This suggests that the strongest inhibitors detected using the vesicular transport assay interfere significantly with the ABCC2 transport mechanism, in such a way that both probes are affected. Overall, very few inhibitors appear to be probe-selective in terms of IC50. By far the most interesting is 3j that displays true probe-selectivity, being 50-fold selective for CDCF with an IC50 of 6 μM, as compared to an IC50 of 305 μM for EG. Among the most probe selective agents are also compounds 1h and 2n that are, respectively, about 8-fold selective towards EG and 6-fold selective towards CDCF, but this should be taken with caution for 1h since the 95% confidence intervals overlaps.
Table 3.
Modulatory effects of 86 selected compounds on CDCF and EG transport (dose–response curves)
| EG transport
|
||||||
|---|---|---|---|---|---|---|
| Inhibitor | Weak inhibitora | Inactive | Stimulatorb | Total | ||
| CDCF transport | Inhibitor | 23 | 14 | 1d | 0 | 38 |
| Weak inhibitora | 1 | 14 | 4 | 1f | 20 | |
| Inactive | 1c | 4 | 22 | 0 | 27 | |
| Stimulatorb | 0 | 0 | 1e | 0 | 1 | |
| Total | 25 | 32 | 28 | 1 | 86 | |
Dose–response curve shifted to the right, IC50 ≫100 μM.
% I more than 150% at 80 μM concentration.
Compound 4f.
Compound 4d.
Compound 17.
Compound 21.
Different functional effects were seen at high concentrations for 4d, 4f, 17 and 21. The clearest difference is seen for 21 that display an inhibitory effect on CDCF transport and no apparent effect on EG transport; and for 17 that has a clear stimulatory effect on the transport of CDCF and no apparent effect on EG transport. In Kidron et al., the three CDCF inhibitors (indomethacin, diclofenac, estrone-3-sulfate) had inhibitory activity in the 70–170 μM range, while being stimulators of EG, and the two inhibitors of EG (budes-onide, thioridazine) had vice-versa IC50s in the 40–60 μM range, while stimulating the transport of CDCF.30 Caution should always be taken when discussing differences in functional effects since it is usually not possible to measure the rightmost part of the curve (high concentrations) due to limits in compounds solubility and to artefacts that arise when high concentrations of compounds are used. For the weakest inhibitors, detection of probe selectivity is thus not attainable.
3.3. The importance of anionic functional groups for ABCC2 modulators
3.3.1. Preference for anionic modulators
The primary screening data additionally serves the purpose of revealing trends in the chemical profile of ABCC2 modulators. The first step of our analysis led us to scrutinize easily interpretable molecular descriptors for trends in their ability to discriminate modulators from inactive compounds: molecular weight, logP, polar surface area, and Number of Charged Centers predicted at pH 7.0 (NCC7), 3.0 (NCC3) and 11.0 (NCC11). NCC3 and NCC11 provide estimates about the amount of respectively basic and anionic functional groups. As a result, we do not find any of these descriptors to be preferred for inhibitors in primary screening (Appendix Table C.3). Furthermore, in secondary screening, an anionic character neither appears to be a necessary feature of ABCC2 inhibitors since only half (8/16) of the most potent molecules bear an anionic functional group. For inhibitors, this result is fully in agreement with previous studies.12 Concerning stimulators and substrates, we are not able to compare our results with the Pedersen study since we did not find enough stimulators for a meaningful assessment and since our assay do not provide information as to whether compounds are actually transported.
3.3.2. SAR in Scaffold 1
The dose–response analysis confirmed the high activity seen in the primary screening for Scaffold 1, (Table 2a, 1aFig. 1). Molecules sharing this chemical scaffold can further be categorized based on the modifications present in the cyclopentene ring: –1n, the majority of the compounds, are cyclopentahydroquinolines; in 1o, the only inactive compound, a tetrahydrofuran replaces the cyclopentene; 1p–1r have an additional benzene ring fused to the cyclopentene ring and 1s has a 2,3-cyclopentanediol in place of the cyclopentene. Out of 19 compounds tested, only one is inactive on both probes and six compounds exhibit inhibition with IC50 lower than 30 μM on both CDCF and EG. Compounds with a carboxylic acid at the tetrahydroquinoline 2-position (R3 = OH) have the lowest IC50 values (compounds 1a, 1e, and 1l have 10–20 μM IC50’s). Comparing 1a to 1b, as well as 1e to 1f, a switch from an ester to a carboxylic acid at the 2-position is accompanied by lowering IC50s by 2–3 fold, an effect probably associated with the anionic character of the carboxylate. Neutral compounds such as 1g, 1h, 1i, 1j, and 1k however appear to be also good modulators (IC50 < 30 μM) of both EG and CDCF.
3.3.3. SAR in Scaffold 2
Another promising scaffold for SAR analysis is Scaffold 2 (Table 2b). Molecules with this scaffold are divided into two groups. In the first group, molecules containing a phenyl ring connected by a one-carbon linker (or ether bond) (2a–h) that have low or no inhibitory activity. Molecules 2i–w of the second group feature at equivalent position a carboxylic acid group connected by two or four carbon atoms, and are mostly active. Differences in modulatory activity between 2a–h and 2i–w are thus likely associated with the presence of this carboxylic acid. Among other types of substitutions found within the 2i–w series, the presence of an additional aromatic group at R2 or R3 generally leads to an increase of inhibitory activity; for instance, adding a phenyl ring at position R2 leads to compound 2r with an inhibitory activity not found for 2l. Among 2i–w, many compounds are able to modulate CDCF transport much better than EG transport, making 2i–w a promising scaffold for further development of probe-selective compounds.
3.3.4. Scaffolds 3, 4 and other compounds
Scaffold 3 contains only neutral molecules (Table 2c). The most active members of this series, with estimated IC50 values in the 15–70 μM range for CDCF transport, have a hexyl alcohol (3b), or methylated phenyl ring (3e) as R group, while the analogues with fluoro- or chloroarene-groups are inactive or only weakly active. Compound 3j is the most active CDCF inhibitor in this series and bears a bromine atom at the isoquinolinone (Fig. 1). Scaffold 4 has only one member that contains a carboxylic acid, 4b, which has weak activity on both probes (Table 2d). Among the eight compounds tested for this scaffold, 4b ranks second for EG and fifth for CDCF. The dihydropyrrole 4e is the most active modulator on CDCF transport and is only weakly active on EG. Furthermore, this scaffold features two probe-selective molecules: 4d is EG selective, and 4f is CDCF selective. The remaining most active compounds, 6, 8, 11 and 29, have IC50 values in the 7–45 μM range, none of which bears a carboxylic acid group.
3.4. Molecular profile of inactive compounds and modulators based on molecular descriptors
3.4.1. Descriptor analysis using Self-Organizing maps
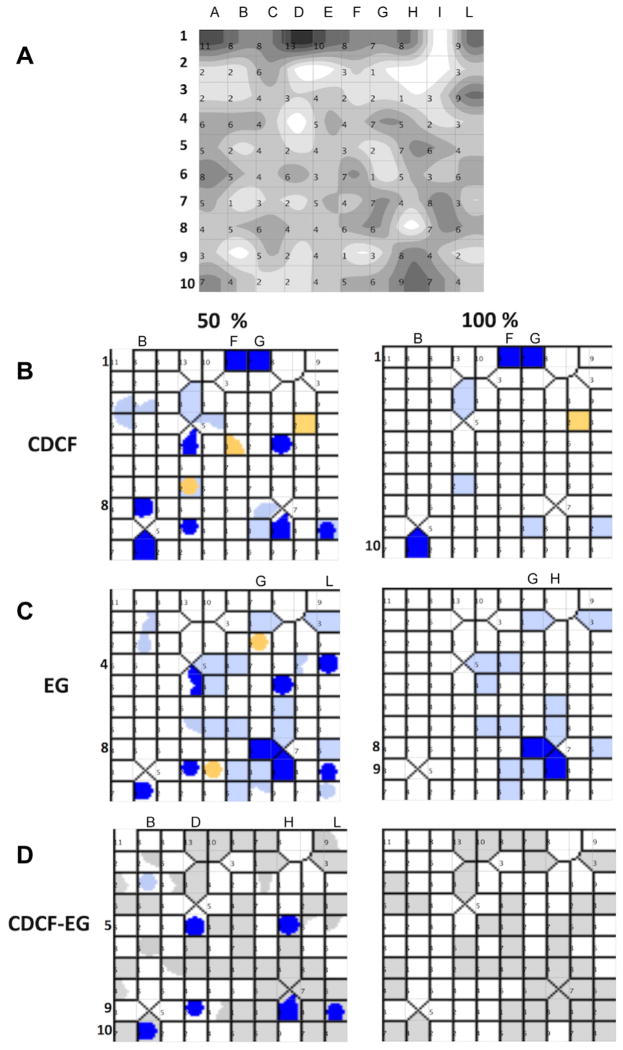
In the last stage of this study, we used automated computational methods to analyze the compounds through numerical descriptors, looking for trends that can be revealed across analogue series and would have been missed by the manual analysis. The chemical space was first visualized using Self-Organizing Maps calculated using the Volsurf descriptors (Fig. 3).
Figure 3.
Modulatory properties of the 432 compounds tested in primary screening mapped on the chemical space using self-organizing maps and Volsurf+ descriptors. (A) Number of compounds present at each node, the darker the node the more molecules are present in the node. 2D maps of CDCF (B) and EG (C) inhibitors and (D) inhibitors of both probes. Nodes contain: inhibitors (dark blue), borderline inhibitors (light blue), inactive (white), borderline stimulators (yellow). Nodes with at least 50% of molecules in the node in the same class (left panels) and nodes with 100% of molecules in the same class (right panels).
The most striking result is that modulators do not appear clustered in a single region of the 2D space: in six separate regions, at least 50% of the compounds are both EG and CDCF inhibitors (Fig. 3D left; B10, D5, D9, H5, H9 and L9). Compounds located inside each of these regions have similar descriptors and therefore similar chemical structures. These regions are also well separated in the 2D space, which shows that diverse chemical structures can lead to modulation. Four regions additionally contain either only CDCF inhibitors (two regions, 15 molecules in total; regions B8 and F1–G1) or only EG inhibitors (two regions, 4 molecules in total; regions G8 and L4). Furthermore, in 12 other regions more than half of the total compounds in that region are inhibitors (EG, 6 regions; CDCF, 6 regions; Fig. 3B–D right panels). The chemical scaffolds 1–4 presented in this article are distributed across several regions, showing that the information given by the Self-Organizing Maps and by the scaffold-based classification is different (Appendix Table C.2).
3.4.2. SVM modelling to find discriminative descriptors
We then used SVM modelling to identify the descriptors able to best discriminate modulatory classes. Chronologically, this was done in two phases. Firstly, a single classification model was built using the SVM method and Volsurf+ descriptors computed for mixed EG-CDCF inhibitors (details about test and training set are provided in Appendix Table C.4). Accuracy on test set of 66% and a Matthews correlation coefficient (MCC) of 0.44 were obtained for that model (67% in cross-validation on the training set). These are characteristic of a moderately predictive model. We interrogated this model with a fully external set of 44 compounds that was obtained for the only purpose of validation. These compounds were retrieved out of a library of ~52,000 compounds and physically tested in the same assay conditions (see Section 2).
As a result of the foreign set testing, the SVM/Volsurf+ model was able to correctly predict 16 out of 34 compounds as inhibitors of both CDCF or EG, and to correctly predict all 10 inactive compounds (confusion matrix as Appendix Table C.4B). Several metrics also suggests reasonable accuracy as well as usefulness for pre-screening of molecules: enrichment compared to random screening is about 3.6-folds; the area under curve in receiver-operator analysis, which can be interpreted as the probability that a randomly chosen active compound is ranked higher than a randomly chosen decoy, is 0.67. These results correspond to the accuracy anticipated from the test set. From the point of view of developing a model for practical use, it is very interesting that the sensitivity (recall, true positive rate) is of 100%, that is, no false positive have been generated.
Next, we explored four sets of descriptors (three unique sets, and one merging them) together with the SVM modelling for their ability to classify inhibitors, divided in three datasets (CDCF, EG, and both CDCF and EG). In total, 12 QSAR models of ABCC2 inhibition were thus constructed (Table 4). The three sets of descriptors are: 48 Volsurf+ descriptors (selected from an initial set of 116 descriptors), PaDEL, 129 molecular descriptors (856), and Discovery Studio, 66 (324) molecular descriptors. As a result, the accuracy of the models was generally within the same range as the previously SVM/Volsurf+ model, although some performed better. Of all the 12 SVM models built, the best one in terms of testing accuracy (SVM/PaDEL descriptors/EG and CDCF inhibitors) gave 74% accuracy on the test set, 70% cross-validation accuracy on the training set, and an MCC of 0.54. The models built combining all descriptors performed slightly better than those built using single descriptor sets. Considering only inhibitors of both CDCF and EG in the training dataset did not lead to models with higher predictivity. We also explored another set of 12 models constructed using XY-fused Kohonen Networks, but this did not lead to any improvement in model accuracy (Appendix Fig. C.4C). These numbers are in line with other classification models of ABCC2 published in the literature, in particular the performance of Pedersen’s classification models: 86% in training set, 71–72% in test set for classifying inhibitors and non-inhibitors, respectively.12
Table 4.
Outcome of 12 SVM classification models constructed with four descriptor sets and three overlapping datasets
| CDCF inhibitors dataset
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DS descriptors
|
PADEL descriptors
|
Volsurf+descriptors
|
All descriptors combined
|
||||||||
| Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC |
|
|
|
|
|
||||||||
| 62 | 53 | 0.44 | 80 | 66 | 0.58 | 68 | 62 | 0.48 | 80 | 60 | 0.63 |
| Most important feature and (f-score) | |||||||||||
| Num_Ringsa (30) | nT10Ringb (0.26) | LgS3c (0.13) | Num_Rings (0.30) | ||||||||
| ESP_point_count__4_Propgen_VAMPd (0.13) | nwHBae (0.25) | DRDRACf (0.13) | nT10Ring (0.26) | ||||||||
| Rg (0.10) | nwHBa (0.25) | ||||||||||
| EG inhibitors dataset
| |||||||||||
| Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC |
|
|
|
|
|
||||||||
| 74 | 52 | 0.55 | 75 | 60 | 0.52 | 66 | 45 | 0.41 | 75 | 60 | 0.59 |
| Most important feature and (f-score) | |||||||||||
| Num_Rings (0.30) | nT10Ring (0.34) | HL1h (0.13) | nT10Ring (0.34) | ||||||||
| Alog Pi (0.16) | WTPT-2j (0.21) | IT3k (0.13) | Num_Rings (0.30) | ||||||||
| R (0.10) | WTPT-2 (0.21) | ||||||||||
| Inhibitors of both CDCF and EG dataset
| |||||||||||
| Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC | Cross-validation accuracy (%) | Testing accuracy (%) | MCC |
|
|
|
|
|
||||||||
| Most important feature and (f-score) | |||||||||||
| 76 | 63 | 0.51 | 74 | 70 | 0.54 | 67 | 66 | 0.44 | 76 | 60 | 0.59 |
| Num_Rings (0.28) | nT10Ring (0.32) | DRDRAC (0.09) | nT10Ring (34) | ||||||||
| JYl (0.10) | WTPT-2 (0.23) | R (0.07) | Num_Rings (30) | ||||||||
| HL2 (0.02) | WTPT-2 (0.23) | ||||||||||
Num_Rings, number of rings in smallest set of smallest rings.
nT10Ring, number of ‘10-membered’ rings.
LgS3, logarithm of solubilities at pH 3.
ESP_point_count__4_Propgen_VAMP, semi empirical QM properties.
nwHBa, count of E-States for weak H-Bond acceptors.
DRDRAC, 3D pharmacophoric descriptor (Dry–Dry-Acceptor triplet).
R, rugosity.
HL1, hydrophilic–lipophilic balance.
AlogP, Log (octanol–water partition coefficient).
WTPT-2 Weighted Path Descriptor.
IT3—INTEraction enerGY moments.
JY, topological descriptors (Balaban Indices).
Based on these models, the descriptors that are most useful to discriminate between inhibitors and inactive molecules are related to the number and size of rings, solubility and hydrophobicity (Table 4, Appendix Table C.4): number of rings in smallest set of rings (Num_Rings), number of ‘10 membered’ rings (nT10Ring), weighted path descriptor (WTPT-2), logarithm of solubility at pH 3 (lgS3). Notably, the importance of an anionic character for inhibition was not identified in this analysis either. Similarly to our analysis, in Pedersen et al.,12 the discriminant analysis showed that for a pool of 191 molecules and 669 molecular descriptors, size (molecular weight), lipophilicity (octanol–water partition coefficient), hydrophobicity (surface area of non-polar atoms), and aromaticity (surface area of unsaturated non polar atoms) were the most important descriptors for discerning inhibitors from non-inhibitors.
4. Conclusions
In this manuscript, we extended the work of Pedersen et al.,12 using an entirely novel dataset of 432 compounds densely populated around analogues series, and tested by two chemical probes. We reveal four series of analogues compounds, potent modulators of ABCC2 activity, screened from a library containing 32 groups of at least three analogues. 16 compounds 1a, 1c, 1e, 1g, 1h, 1i, 1j, 1k, 1l, 1n, 1q, 2q, 2u, 3j, 6, 29 were found with IC50 lower than 30 μM. Many of these compounds belong to Scaffold 1, tetrahydro- quinolines, which is known to be promiscuous. Nonetheless, the landscape of their SARs is varied. These molecules could be later tested for their selectivity profile towards other transporters in order to develop them into biochemical tool compounds useful to study in vivo the specific role of ABCC2.
The screening approach in this study using both EG and CDCF shows that the two probes generally bring similar information, even though some exceptions are found: compound 3j appears to be both very potent at CDCF and highly (50-fold) selective compared to EG. It is thus a good candidate for development as a tool compound to dissect the ABCC2 transportmechanism. Overall, the vesicular assay using CDCF as a probe is more robust, in addition to being more affordable, making it a more attractive probe for high-throughput screening than the radioactively labeled EG.
The screening data can be used to develop computational models of ABCC2 interactions, and by extension could be used to predict better drug–drug interactions mediated by ABCC2. The 24 classification models that were built show reasonable accuracies, with ~70% for the best models. Descriptors related to the number of rings were consistently found to distinguish inhibitors from inactive compounds. Furthermore, we identified that lipophilicity and solubility are important descriptors for ABCC2 inhibitors. Predictivity and important descriptors are similar compared to the Pedersen et al., study, which confirms them to be important for ABCC2 inhibition since our model is built on a fully independent dataset.
The present study brings new knowledge about the SARs of ABCC2 inhibitors (Tables 2a–e, f–h). The analysis of analogue series shows that there is no requirement for a negatively charged carboxylic acid substituent in modulators of ABCC2-mediated transport of EG or CDCF. Nonetheless, inside the analogue series, compounds with an anionic character are often more potent compared to the neutral compounds of the same scaffold. Furthermore, there is a trend for associating changes to carboxylic acid with a gain in potency, as seen for Scaffold 1 and Scaffold 2. In order to generalize this trend, we would need at hand larger series of analogues that have been designed for this purpose, which is beyond the scope of this study.
Supplementary Material
Acknowledgments
Funding sources
This study was supported by the Centre for Drug Research (Faculty of Pharmacy, University of Helsinki), the Academy of Finland, the Orion-Farmos Foundation, Orion Corporation Ltd and the Drug Discovery and Chemical Biology (DDCB), Biocenter Finland network. P.W. thanks the National Institutes of Health P50 CMLD Program (GM067082) for support. G.W. would like to thank the Magnus Ehrnrooth Foundation, the Oskar Öflund Foundation, and the Finnish Cultural Foundation for financial support. The Center for International Mobility (CIMO) supported both G.W. and P.K.
The doctoral program in Informational and Structural Biology is acknowledged for organizing and providing support to PhD studies. The Drug Discovery and Chemical Biology consortium is thanked for IT maintenance in HX’s group. We thank Leena Pietilä, Staffan Berg and Nenad Manevski for technical assistance. CSC-IT Center for Science Ltd is thanked for providing computational resources.
Abbreviations
- ABC
ATP-binding cassette
- CDCF
5(6)-carboxy-2′,7′-dichlorofluorescein
- EG
β-estradiol 17-(β-D-glucuronide)
- MRP2
Multidrug Resistance associated Protein 2
- SARs
structure activity relationships
- SVM
support vector machine
Footnotes
5. Author contributions
All authors have contributed to this manuscript. The experimental assays were designed and conducted by G.W., P.K. and H.K. The QSAR modelling was designed and conducted by L.G. and H.X. P.K. conducted the scaffold decomposition analysis. The manuscript draft was written for a large part by H.X. in close collaboration with G.W. and H.K. All authors participated in analysis of the experiments and finalizing the manuscript. All authors have given approval to the final version of the manuscript. G.W., P.K. have contributed equally.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2015.04.029. These data include MOL files and InChiKeys of the most important compounds described in this article.
References and notes
- 1.Taniguchi K, Wada M, Kohno K, Nakamura T, Kawabe T, Kawakami M, et al. Cancer Res. 1996;56:4124. [PubMed] [Google Scholar]
- 2.Yamasaki M, Makino T, Masuzawa T, Kurokawa Y, Miyata H, Takiguchi S, et al. Br J Cancer. 2011;104:707. doi: 10.1038/sj.bjc.6606071. http://dx.doi.org/10.1038/sj.bjc.6606071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evers R, de Haas M, Sparidans R, Beijnen J, Wielinga PR, Lankelma J, et al. Br J Cancer. 2000;83:375. doi: 10.1054/bjoc.2000.1262. http://dx.doi.org/10.1054/bjoc.2000.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y, König J, Buchholz JK, Spring H, Leier I, Keppler D. Mol Pharmacol. 1999;55:929. [PubMed] [Google Scholar]
- 5.Smitherman PK, Townsend AJ, Kute TE, Morrow CS. J Pharmacol Exp Ther. 2004;308:260. doi: 10.1124/jpet.103.057729. http://dx.doi.org/10.1124/jpet.103.057729.either. [DOI] [PubMed] [Google Scholar]
- 6.Paulusma C. Biochem J. 1999;401:393. [PMC free article] [PubMed] [Google Scholar]
- 7.Kamisako T, Leier I, Cui Y, König J, Buchholz U, Hummel-Eisenbeiss J, et al. Hepatology. 1999;30:485. doi: 10.1002/hep.510300220. http://dx.doi.org/10.1002/hep.510300220. [DOI] [PubMed] [Google Scholar]
- 8.Leier I, Hummel-Eisenbeiss J, Cui Y, Keppler D. Kidney Int. 2000;57:1636. doi: 10.1046/j.1523-1755.2000.00007.x. http://dx.doi.org/10.1046/j.1523-1755.2000.00007.x. [DOI] [PubMed] [Google Scholar]
- 9.Chu XY, Kato Y, Niinuma K, Sudo KI, Hakusui H, Sugiyama Y. J Pharmacol Exp Ther. 1997;281:304. [PubMed] [Google Scholar]
- 10.Hooijberg JH, Broxterman HJ, Kool M, Assaraf YG, Peters GJ, Noordhuis P, et al. Cancer Res. 1999;59:2532. [PubMed] [Google Scholar]
- 11.Sasaki M, Suzuki H, Ito K, Abe T, Sugiyama Y. J Biol Chem. 2002;277:6497. doi: 10.1074/jbc.M109081200. http://dx.doi.org/10.1074/jbc.M109081200. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen JM, Matsson P. J Med Chem. 2008;51:3275. doi: 10.1021/jm7015683. http://dx.doi.org/10.1021/jm7015683. [DOI] [PubMed] [Google Scholar]
- 13.Hillgren KM, Keppler D, Zur AA, Giacomini KM, Stieger B, Cass CE, et al. Clin Pharmacol Ther. 2013;94:52. doi: 10.1038/clpt.2013.74. http://dx.doi.org/10.1038/clpt.2013.74. [DOI] [PubMed] [Google Scholar]
- 14.Izzedine H, Launay-Vacher V, Isnard-Bagnis C, Deray G. Am J Kidney Dis. 2003;41:292. doi: 10.1053/ajkd.2003.50037. http://dx.doi.org/10.1053/ajkd.2003.50037. [DOI] [PubMed] [Google Scholar]
- 15.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, et al. Science. 2009;323:1718. doi: 10.1126/science.1168750. http://dx.doi.org/10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Jaimes KF, Aller SG. Protein Sci. 2014;23:34. doi: 10.1002/pro.2387. http://dx.doi.org/10.1002/pro.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shintre Ca, Pike ACW, Li Q, Kim J-I, Barr AJ, Goubin S, et al. Proc Natl Acad Sci USA. 2013;110:9710. doi: 10.1073/pnas.1217042110. http://dx.doi.org/10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson RJP, Locher KP. Nature. 2006;443:180. doi: 10.1038/nature05155. http://dx.doi.org/10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 19.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Proc Natl Acad Sci USA. 2007;104:19005. doi: 10.1073/pnas.0709388104. http://dx.doi.org/10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jardetzky O. Nature. 1966;211:969. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 21.Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, et al. Proc Natl Acad Sci USA. 2008;105:10338. doi: 10.1073/pnas.0804659105. http://dx.doi.org/10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez C, Koshy C, Yildiz O, Ziegler C. Nature. 2012;490:126. doi: 10.1038/nature11403. http://dx.doi.org/10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 23.Radestock S, Forrest LR. J Mol Biol. 2011;407:698. doi: 10.1016/j.jmb.2011.02.008. http://dx.doi.org/10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Zelcer N, Huisman MT, Reid G, Wielinga P, Breedveld P, Kuil A, et al. J Biol Chem. 2003;278:23538. doi: 10.1074/jbc.M303504200. http://dx.doi.org/10.1074/jbc.M303504200. [DOI] [PubMed] [Google Scholar]
- 25.Bodo A, Bakos E, Szeri F, Varadi A, Sarkadi B. J Biol Chem. 2003;278:23529. doi: 10.1074/jbc.M303515200. http://dx.doi.org/10.1074/jbc.M303515200. [DOI] [PubMed] [Google Scholar]
- 26.Madon J, Eckhardt U, Gerloff T, Stieger B, Meier PJ. FEBS Lett. 1997;406:75. doi: 10.1016/s0014-5793(97)00245-7. [DOI] [PubMed] [Google Scholar]
- 27.Heredi-Szabo K, Kis E, Molnar E, Gyorfi A, Krajcsi P. J Biomol Screen. 2008;13:295. doi: 10.1177/1087057108316702. http://dx.doi.org/10.1177/1087057108316702. [DOI] [PubMed] [Google Scholar]
- 28.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KLR, Chu X, et al. Nat Rev Drug Disc. 2010;9:215. doi: 10.1038/nrd3028. http://dx.doi.org/10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu X, Korzekwa K, Elsby R, Fenner K, Galetin A, Lai Y, et al. Clin Pharmacol Ther. 2013;94:126. doi: 10.1038/clpt.2013.78. http://dx.doi.org/10.1038/clpt.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidron H, Wissel G, Manevski N, Häkli M. Eur J Pharm Sci. 2012;46:100. doi: 10.1016/j.ejps.2012.02.016. http://dx.doi.org/10.1016/j.ejps.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Werner S, Turner D, Lyon M, Huryn D, Wipf P. Synlett. 2006:2334. http://dx.doi.org/10.1055/s-2006-949648.
- 32.Wipf P, Werner S, Woo GHC, Stephenson CRJ, Walczak MAA, Coleman CM, et al. Tetrahedron. 2005;61:11488. http://dx.doi.org/10.1016/j.tet.2005.09.016. [Google Scholar]
- 33.Pierce JG, Kasi D, Fushimi M, Cuzzupe A, Wipf P. J Org Chem. 2008;73:7807. doi: 10.1021/jo801552j. http://dx.doi.org/10.1021/jo801552j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap CW. J Comput Chem. 2011;32:1466. doi: 10.1002/jcc.21707. http://dx.doi.org/10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 35.Accelrys Software Inc. Discovery Studio Modelling Environment, Release 4.0. Accelrys Software; San Diego: 2013. (n.d.) [Google Scholar]
- 36.Cruciani G, Pastor M, Guba W. Eur J Pharm Sci. 2000;11:S29. doi: 10.1016/s0928-0987(00)00162-7. http://dx.doi.org/10.1016/S0928-0987(00)00162-7. [DOI] [PubMed] [Google Scholar]
- 37.Fan R, Chen P, Lin C. J Mach Learn Res. 2005;6:1889. [Google Scholar]
- 38.Ballabio D, Consonni V, Todeschini R. Chemom Intell Lab Syst. 2009;98:115. http://dx.doi.org/10.1016/j.chemolab.2009.05.007. [Google Scholar]
- 39.Ballabio D, Vasighi M. Chemom Intell Lab Syst. 2012;118:24. http://dx.doi.org/10.1016/j.chemolab.2012.07.005. [Google Scholar]
- 40.Szeri F, Iliás A, Pomozi V, Robinow S, Bakos É, Váradi A. Biochim Biophys Acta—Biomembr. 2009;1788:402. doi: 10.1016/j.bbamem.2008.11.007. http://dx.doi.org/10.1016/j.bbamem.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Bemis GW, Murcko MA. J Med Chem. 1996;39:2887. doi: 10.1021/jm9602928. http://dx.doi.org/10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 42.Schuffenhauer A, Ertl P, Roggo S, Wetzel S, Koch MA, Waldmann H. J Chem Inf Model. 2007;47:47. doi: 10.1021/ci600338x. http://dx.doi.org/10.1021/ci600338x. [DOI] [PubMed] [Google Scholar]
- 43.Quan ML, Wong PC, Wang C, Woerner F, Smallheer JM, Barbera FA, et al. J Med Chem. 2014;57:955. doi: 10.1021/jm401670x. http://dx.doi.org/10.1021/jm401670x. [DOI] [PubMed] [Google Scholar]
- 44.Atkinson SJ, Soden PE, Angell DC, Bantscheff M, Chung C, Giblin KA, et al. Medchemcomm. 2014;5:342. http://dx.doi.org/10.1039/c3md00285c. [Google Scholar]
- 45.Bodó A, Bakos É, Szeri F, Váradi A, Sarkadi B. Toxicol Lett. 2003;140–141:133. doi: 10.1016/s0378-4274(02)00497-6. http://dx.doi.org/10.1016/S0378-4274(02)00497-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.