Abstract
Background Aims
Ex vivo expansion and serial passage of human Bone Marrow Stromal Cells (BMSCs, also known as bone marrow-derived mesenchymal stem cells) is required to obtain sufficient quantities for clinical therapy. The BMSC confluence criteria used to determine passage and harvest timing vary widely and the impact of confluence on BMSC properties remains controversial. The effects of confluence on BMSC properties were studied and confluence-associated markers were identified.
Methods
BMSC characteristics were analyzed as they grew from 50% to 100% confluence including viability, population doubling time (PDT), apoptosis, colony formation, immunosuppression, surface marker expression, global gene expression and microRNA expression. In addition, culture supernatant protein, glucose, lactate and pH levels were analyzed
Results
Confluence-dependent changes were detected in the expression of several cell surface markers, 39 culture supernatant proteins, 26 microRNAs and 2078 genes. Many of these surface markers, proteins, microRNAs and genes have been reported to be important in BMSC function. The PEDF/VEGF ratio increased with confluence, but 80% and 100% confluent BMSCs demonstrated a similar level of immunosuppression of mixed lymphocyte reactions. In addition, changes in lactate and glucose levels correlated with BMSC density.
Discussion
BMSC characteristics change as confluence increases. 100% confluent BMSCs may have compromised pro-angiogenesis properties, but may retain their immunomodulatory properties. Supernatant lactate and glucose levels can be used to estimate confluence and ensure consistency in passage and harvest timing. Flow cytometry or microRNA expression can be used to confirm that the BMSCs have been harvested at the appropriate confluence.
Keywords: Bone marrow stromal cells, BMSCs, cellular therapy, gene expression profiling, confluence, microRNA
Introduction
Bone marrow stromal cells (BMSCs), which are also known as mesenchymal stem cells (MSCs), were first isolated from the bone marrow as the progeny of colony-forming-unit fibroblasts (CFU-Fs)1, 2. Since BMSCs include skeletal stem cells, they have been used in clinical trials to treat osteonecrosis of the femoral head3, osteoarthritis/cartilage repair4, 5 and degenerative disc disease6. BMSCs also have anti-inflammatory and immunosuppressive properties in vitro and may hold promise in treating immune diseases, acute Graft-versus-Host-Disease (aGvHD)7, Crohn’s disease8 and in preventing the rejection of transplanted solid organs9 . BMSCs therapy has proven to be safe and capable of improving the clinical performance and the quality of life of patients with ischemic cardiomyopathy (ICM)10. The mechanisms underlying the ability of BMSC to improve heart function could be that MSCs differentiate into cardiomyocytes, vascular smooth muscle and endothelial cells11 ; secrete cell survival factors including VEGF, bFGF, IGF and SDF12 or stimulate the proliferation of endogenous cardiac stem cells (CSCs)13 .
For clinical purposes, BMSCs are isolated from bone marrow aspirates or bone biopsy specimens and are expanded extensively ex vivo to obtain adequate numbers of cells. Most expansion protocols involve adherent culture on plastic surfaces and serial passage. Many variables must be considered when optimizing BMSC expansion, among which cell confluence is a critical factor since the degree of confluence may affect the biological properties of BMSCs. Generally, BMSCs are sub-cultured or harvested when they reach a specified degree of confluence, but there is no universal standard concerning optimal confluence. For example, investigators have used different criteria to describe the conditions when the cells need to be sub-cultured, such as 50–60% confluence14, 15, 70–90% confluence16, sub-confluent (70–80%)17, 80% confluence18–21, 80–90% confluence22, 90% confluence23, 24, near confluence25, approaching confluence26 or confluent27–30. More importantly, the outcomes of BMSC clinical trials have been variable. Since BMSCs used to treat GVHD and other conditions are cultured to variable levels confluence it is important to better understand how confluence at the time of harvest effects the properties of BMSCs 31.
As a Good Manufacturing Practice (GMP)-compliant laboratory, we aim to achieve optimal expansion of BMSCs by establishing the best seeding density and timing of passage and harvest in order to maintain consistent BMSC quality. When sub-culturing or passaging BMSCs in small flasks, the cells are easily observed and confluence assessed under a microscope, but when BMSCs are cultured under large scale GMP conditions they are most often cultured in multi-layer flasks or cell factories and cannot be assessed under microscope. For cell expansion under these circumstances, developing a measure to determine BMSC confluence is in urgent need.
In this study, we grew the BMSCs to 50%, 80% and 100% confluence, which were determined by the proportion of culture area that was covered by BMSC, then investigated the impact of confluences on BMSC phenotype including cellular proliferation, apoptosis, surface marker expression, growth factor and cytokine production, immunosuppressive activity, microRNA expression profile, and gene expression profile. We found multiple changes in BMSCs as their confluence increased from 50% to 100%.
Materials and Methods
BMSC Culture
Bone marrow collection and BMSC culture were performed according to Standard Operating Procedures (SOP) established in our lab32. These studies were approved by the NHLBI Institutional Review Board (IRB) under clinical protocol 10-CC-0053, which was also registered on clinicaltrial.gov under NCT01071577.
For the primary culture, 5 to 10 mL of bone marrow was collected from the posterior iliac crest of healthy donors after obtaining informed consent. The marrow was collected in Bone Marrow Prep Syringes (Pharmacy Department, NIH, Bethesda, MD) and then washed with 2.5x volume of HBSS (Lonza, Walkersville, MD). The cells were then re-suspended with BMSC culture media (BMSC CM) [alpha MEM with 2 mM glutamine (Lonza), supplemented with 20% lot-selected FBS (Hyclone, Thermo Fischer Scientific, Waltham, MA) and 10 µg/ml Gentamicin] and plated at a density of 2 x 105/cm2 in T-75 flasks (Corning Life Sciences, Corning, NY). The cells were incubated at 37°C in 5% CO2 and non-adherent cells were removed after 24 hours. The media was changed every three days thereafter until the colonies reach 80% confluence. The primary BMSCs were washed with 10mL HBSS twice, digested with 5mL TrypLE Express (Invitrogen, Life Technologies, Grand Island, NY) and centrifuged at 406 x g for 10 minutes. The cell number was then counted and viability was assessed using the Trypan Blue exclusion method. The cells harvested at this stage were designated as Passage 1 and serial passage numbers were designated thereafter.
Passage 2 BMSCs from 7 donors were thawed from a working cell bank and plated at a density of 3 x 103/cm2 in T-75 flasks, the donors of these cells that were used for each experiment in this study were listed in supplemental file 1. The BMSCs were cultured to 80% confluence and harvested; the cells at this stage were called passage 3 and were plated to T75 flask for further culture. When passage 3 BMSCs reached 50%, 80% and 100% confluence as determined by microscope observation by 3 trained investigators (the representative figures of 50%, 80% and 100% confluence were shown in supplemental file 2), they were harvested as passage 4 cells. The cell densities were 1.34 x104 ± 5.23 x103, 3.28 x104 ± 8.81 x103 and 4.58 x104 ± 2.40 x104 cells per cm2 under 50%, 80% and 100% confluence respectively (Figure 1A). The BMSCs were then subjected to assays including surface marker expression, apoptosis, cell proliferation, and suppression on the proliferation of mixed lymphocytes. The culture supernatant of BMSCs at different confluences were also collected and used to measure the concentrations of cytokines and growth factors. Total RNA was extracted from an aliquot of the cells for global microRNA and gene expression profiling.
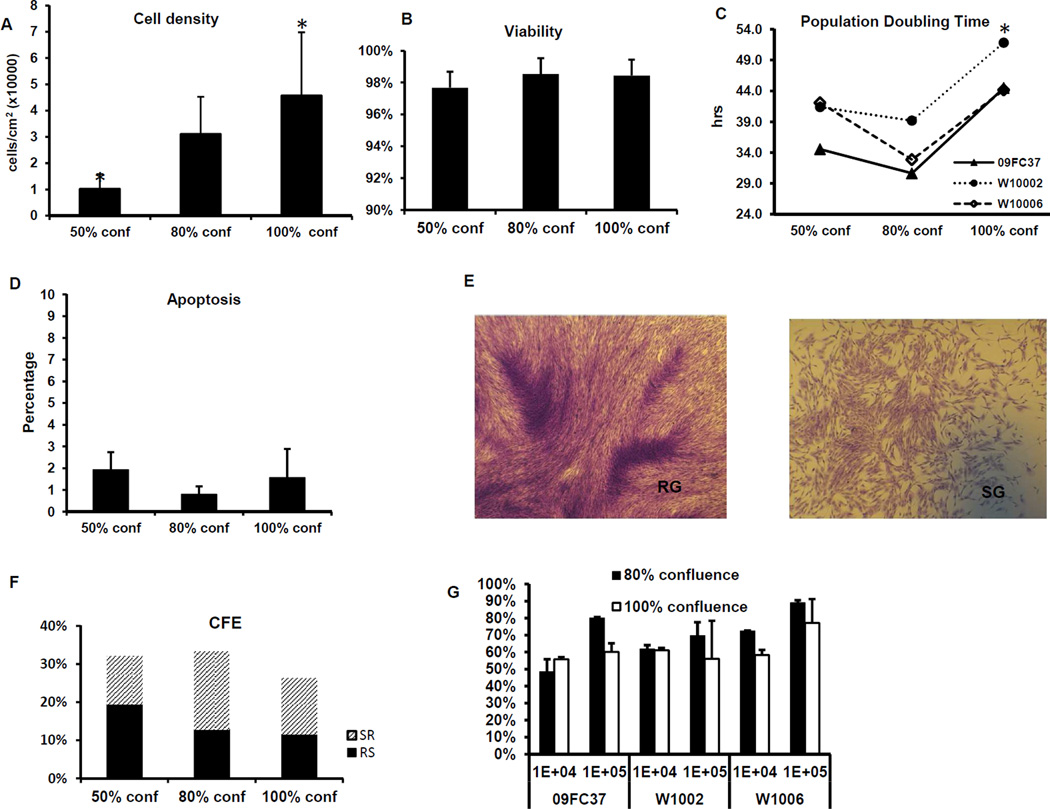
Figure 1. Changes in the phenotypes of 50%, 80% and 100% confluent BMSCs.
The BMSCs were cultured to 50%, 80% and 100% confluences, the cell densities per cm2 culture area were calculated (A) and the viability was determined by Trypan Blue staining (B). In addition, BMSC population doubling time (hours) was calculated (C) and the rate of apoptosis was measured by Annexin V- flow cytometry analysis (D). Representative figures of Rapidly Growing (RG) and Slow Growing (SG) colonies are shown (E). Colony Formation Efficiency (CFE) was counted and the percentage of RG and SG colonies was also shown (F). Immunosuppression of BMSCs of mixed lymphocyte reactions was measured by H3-thymdine incorporation methods; the percentage of suppression was calculated by normalizing the values to the mixed lymphocyte reaction without BMSCs (G). The * indicates p values <0.05 when compared with 80% confluence and the error bars indicate one standard deviation.
Cell Proliferation, Viability and Colony Formation Efficiency (CFE) Assays
When passage 3 BMSCs reached 50%, 80% and 100% confluence, they were harvested (passage 4), the cell number was counted and viability was measured by the Trypan Blue exclusion method. For proliferation analyses, passage 4 BMSCs from the 3 confluences were further plated in T75 flasks at a density of 3000/cm2 and cultured for 5 days and harvested. The proliferation of BMSC was determined by cell counting, Population doubling (PD) and population doubling time (PDT) was calculated by the method described previously33. For CFE assays, passage 4 BMSCs from 50%, 80% and 100% confluences were plated at 1.67/cm2 in T25 flasks in duplicate and cultured for 13 days without changing culture medium. The colonies were then fixed with methanol for 30 minutes and stained with saturated methyl-violet water solution for 20 minutes. Colonies (>50 cells) were then counted under low magnitude light microscope field (25 X).
Apoptosis Analysis by Flow Cytometry
The rate of apoptosis of 50%, 80% and 100% confluent BMSCs was measured with Annexin V-Apoptosis kit (BD Bioscience, San Jose, CA). Briefly, cells were washed twice with cold PBS, re-suspended in binding buffer and then incubated with FITC-labeled Annexin V for 15 minutes at room temperature in the dark. Afterwards, the cells were washed and stained with Propidium Iodide (PI); the cell counts were acquired on an Accuri C6 flow cytometer (Accuri, Ann Arbor, MI) and data analyzed by CFlow software (Accuri), Annexin V-positive and PI-negative cells were enumerated as apoptotic cells.
Flow Cytometry Analysis of Surface Marker Expression
Cell surface markers were measured on 50%, 80% and 100% confluent BMSCs by Flow Cytometry assay; the markers were selected according to either global gene expression profiling results or from the literature34–36. The following antibodies were used, PODXL-FITC (MBL International Inc, Nagoya, Japan), CD49f-PE, CD54-APC, TLR1-PE, CD106-PE, LAMP1 (CD107a)-APC, CD146-PE, CCR7-PE, CD55-FITC, TLR4-PE (BD Biosciences), CD200-APC, CD10-APC (Biolegend, San Diego, CA, USA), and FZD4-APC (R&D, Minneapolis, MN, USA), isotype controls were IgG1-FITC, IgG1-PE, IgG2a-FITC (BD Bioscience), IgG1-APC (eBioscience, San Diego, CA, USA). In brief, BMSCs were incubated with antibody cocktail for 30 minutes in the dark at room temperature and washed with PBS containing 1% BSA; the cells were then suspended in 0.3 mL PBS with 1% BSA, counterstained with 7-AAD; twenty-thousand events were acquired on an Accuri C6 flow cytometer. The data were analyzed with FlowJo software (TreeStar, Ashland, OR). Positive cells were identified as those whose intensities were greater than 99 percentile of the isotype control.
Measurement of Glucose and Lactate, Growth Factors and Cytokines in Culture Supernatants
The concentrations of glucose and lactate in the culture supernatant of BMSC were measured daily by using an i-STAT Analyzer (Abbott, Princeton, NJ). The raw data were imported into Excel spreadsheets; glucose consumption rate (GCR) and lactate generation rate (LGR) were calculated by dividing the time when the consecutive measurements were completed. The concentrations of cytokine and growth factors in BMSC supernatants were measured with SearchLight Protein Array (Aushon Biosystems, Billerica, MA). Briefly, supernatants were collected daily and centrifuged for 10 min at 1400 rpm, aliquoted and stored at −80 °C . The 64 proteins that were measured included cytokines, growth factors, chemokines, and enzymes; the list is shown in Supplemental Table 1.
Suppression of Mixed Lymphocyte Reaction (MLR)
The immunosuppressive properties of 80% and 100% confluent BMSCs were compared using MLR assay (SAIC-Frederic, Frederic, MD). Briefly, Ficoll-separated peripheral blood mononuclear cells (PBMC) were plated in 96-well plates at 1x105 responders per well. Responders were co-cultured with stimulator PBMCs at the same concentration. BMSCs grown to 80% and 100% confluence were added at the following concentrations, 104 and 105 cells /well. After 6 days of culture in a humidified 5% CO2 incubator at 37°C, 0.5 µCi of 3H-thymidine (3H-TdR) was added to each well for 4 hours, and the lymphocyte proliferation was measured using a liquid scintillation counter. The effect of BMSCs on MLR was calculated as the percentage of the suppression compared with the proliferative response of the positive control without BMSC, where the positive control was set to 0% suppression. The experiments were performed in triplicate.
Gene and microRNA Expression Profiling
Total RNA was extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany) and assessed using the Nano Drop 2000 (Thermo Scientific, Wilmington, DE). Total RNA (0.5 µg) was labeled with Cy5-CTP using Quick Amp Labeling kit (Agilent, Santa Clara, CA). An equal amount of human reference RNA (Stratagen, La Jolla, CA) was labeled with Cy3-CTP. 825 ng of labeled cRNA was pooled, fragmented and then hybridized on 4 x 44K microarrays (Agilent) for 17 hours at 65°C. The microarrays were then washed and scanned using an Agilent Scanner and data were acquired using Agilent Feature Extraction Software. The data were submitted to Gene Expression Omnibus (GEO) under accession number GSE60525.
MicroRNA profiling was performed by a protocol established in our lab37. Briefly, 4 µg total RNA was labeled with miRCURY™ LNA Array Power Labeling Kit (Exiqon, Woburn, MA). The total RNA from Epstein-Barr virus (EBV)-transformed lymphoblastoid cell line was used as the reference. The test sample was labeled with Hy5 and the reference with Hy3. After labeling, the test sample and the reference were co-hybridized to the custom-made microRNA array at room temperature overnight in the present of blocking reagents; the slides were then washed and scanned by GenePix scanner Pro 4.0 (Axon, Sunnyvale, CA, USA). The data were submitted to Gene Expression Omnibus (GEO) under accession number GSE60449.
Data Processing and Statistical Analyses
Gene and microRNA expression analysis was performed by procedures as reported previously37, 38. Raw data were uploaded into mAdb database (http://madb.nci.nih.gov/) and then analyzed by BRB-ArrayTools39 (http://linus.nci.nih.gov/BRB-ArrayTools.html) and Partek Genomics Suite (Partek Inc, Saint Louis, MO). Tests for differences between different degrees of confluence were conducted for individual genes and microRNAs using two-way ANOVA, considering P values of <0.001 as significant for genes, and <0.05 as significant for microRNAs. Clusters and Heatmaps were generated by MultiExperiment Viewer (MeV4.6.2, http://www.tm4.org/mev/)40. We also did functional annotation on the differentially expressed genes, aiming to identify the gene sets underlying the different degree of confluence. In this way, the genes were first organized into higher-level sets, which can be downloaded from Genomica Repository (http://genomica.weizmann.ac.il/). The confluences in which different sets are “induced” or “repressed” were identified following methods described previously41. The significance of the gene sets and confluences were calculated by using Genomica41.
Results
Effect of Confluence on BMSC Proliferation, Viability, Apoptosis, CFE and Immunosuppression
All the BMSCs used in this study were derived from 7 healthy donors who were less than 44 years old and all the cells were from early passages (passage 4) in order to reduce the influence of passage as we have described previously42. The viability was >95% for all 3 confluences and there was no significant difference in viability among the three confluences (Figure 1B).
To address the influence of contact inhibition on cell growth, the BMSCs were harvested after becoming 50%, 80% and 100% confluent and re-seeded into T75 flasks. After 5 days of culture, the cell number was counted and the population doubling time (PDT) was calculated. For all 3 donors tested, the PDT of 80% confluent BMSCs (34.2 hrs) was slightly shorter than 50% confluent cells (39.3 hrs, p=0.22), but much shorter than 100% confluent cells (46.8 hrs, p=0.02) (Figure 1C).
We also measured apoptosis using an Annexin V Flow Cytometry assay, in which the apoptotic cells are positive for Annexin-V but negative for propidium iodide (PI). All the BMSC samples displayed low apoptosis rate (<3%), and there was no difference among the 3 degrees of confluence (Figure 1D).
Colony Formation Efficiency (CFE), is a good measure of BMSC quality, since only a proportion of the BMSCs can re-adhere to plastic surfaces and proliferate in a density independent fashion. To determine the influence of 100% confluence, we performed CFE assays and documented the quantities of the two types of colonies that grew (Figure 1E); one type was the rapidly growing (RG)colonies, which were composed of fibroblastic cells with extended cell processes that formed dense colonies, the other was the slowly growing (SR) colonies, which were composed of larger cells and formed sparse colonies43. Our analysis revealed similar quantities of total CFE from 80% and 50% confluent BMSCs, but less from 100% confluent BMSCs. When we enumerate the two types of colonies, 50% confluent BMSCs produced the highest percentage of RS colonies, while 80% and 100% confluent cells yielded similar percentages of RS colonies, but the difference was not statistically significant among the three degree of confluence (Figure 1F).
To assess the immunomodulatory effects of the BMSCs their ability to inhibit mixed lymphocyte reactions was compared among 80% and 100% confluent cells from 3 donors. Two doses of BMSCs, 10,000 and 100,000 cells per well, were tested. At the lower dose of 10,000 cells per well (BMSC: PBMC ratio=1:10), the immunosuppression was very similar between 80% and 100% confluent BMSCs. At the higher dose of 100,000 cells per well (BMSC: PBMC ratio=1:1), 80% confluent BMSCs were slightly more suppressive than 100% confluent BMSCs (Figure 1G), but the difference was not statistically significant (p=0.11).
Proteins in BMSC Culture Supernatants
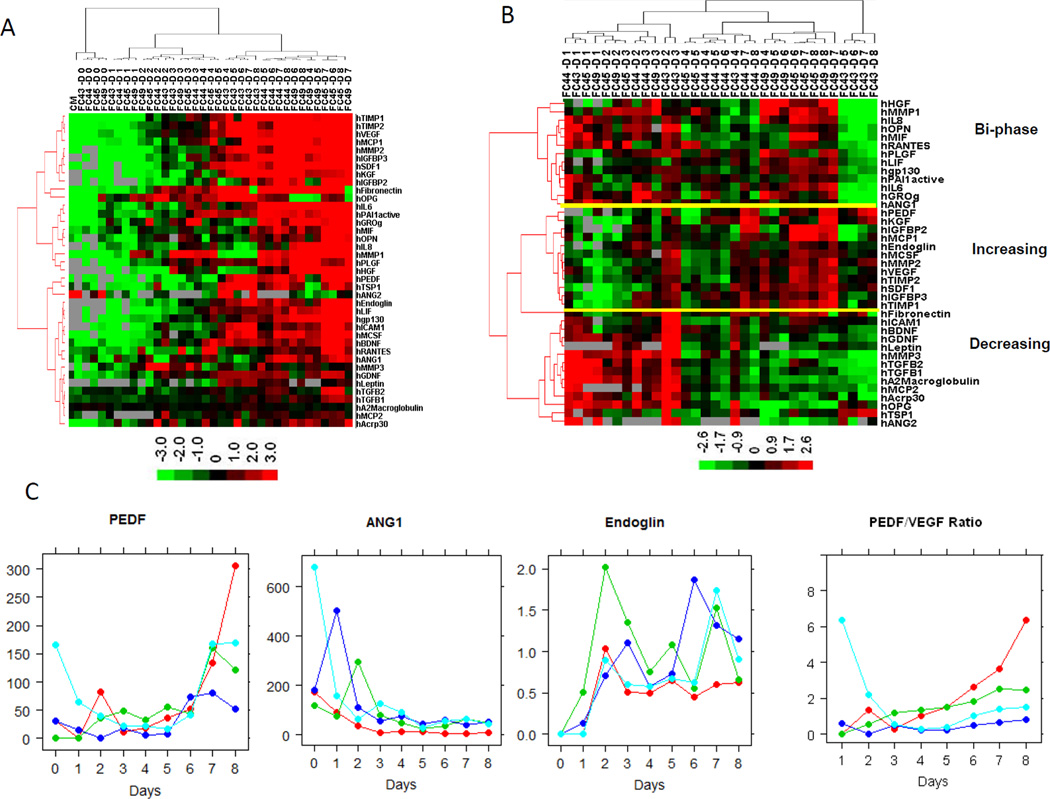
A total of 64 proteins, including cytokines, growth factors, chemokines and enzymes, were measured in the supernatant of 50%, 80% and 100% confluent BMSCs from 4 donors. Among these 64 proteins, 10 were undetectable, 15 were below the limit of detection, and 39 proteins were detected during the culture period (Supplemental File 3). The baseline levels of these 39 proteins in the culture medium (without contact with cells) were undetectable or very low except for TGFB1, PEDF and TSP1. At the initiation of cell culture (Day 0), the concentrations of most proteins were low, except for PEDF, TGFB1, TGFB2, TIMP1 and FN1. Afterwards, the protein concentrations increased daily during the culture (Figure 2A and Supplemental Table 1). To decide whether these increases were caused only by the increasing number of cells or by the increasing number of cells plus increasing protein production per cell, we normalized the factor concentrations by cell number. We found that protein concentrations were not linearly correlated with the cell number, thus suggesting cell-cell contact caused by confluence changed the quantities of proteins secreted. The cell number-normalized protein concentrations displayed distinct patterns; some were either steadily increasing or steadily decreasing, while some were bi-phasic (Figure 2B). For example, PEDF, IGFBP2, IGFBP3 and MMP2 displayed a steadily increasing product pattern (a representative plot of PEDF is shown in Figure 2C). In contrast ANG1, ANG2, TGFB2, GDNF, BDNF, BNGF, OPG, ICAM1 and PLGF showed a steadily decreasing pattern (i.e., ANG1 in Figure 2C). Finally, Endoglin (ENG), IL6, IL8, OPN, FN1 and TSP1 showed a bi-phasic pattern (i.e., Endoglin in Figure 2C)
Figure 2. Concentration of proteins in BMSC culture supernatants.
The culture supernatants of 4 BMSCs were collected daily for 8 days and the concentrations of proteins were measured by SearghLight. A heat-map of the unsupervised hierarchical clustering analysis of the raw concentrations of 39 proteins were shown in panel A and the protein concentrations normalized by the cell number in panel B. Representative plots of daily normalized supernatant concentrations of PEDF, ANG1 and Endoglin and the ratio of PEDF/VEGF are shown in panel C. In the heat-map, each row stands for one protein, and each column stands for one BMSC supernatant. The donor ID and day of culture were indicated for each sample on the top of heatmap. CM stands for culture medium.
Effects of Confluence on BMSC Surface Marker Expression
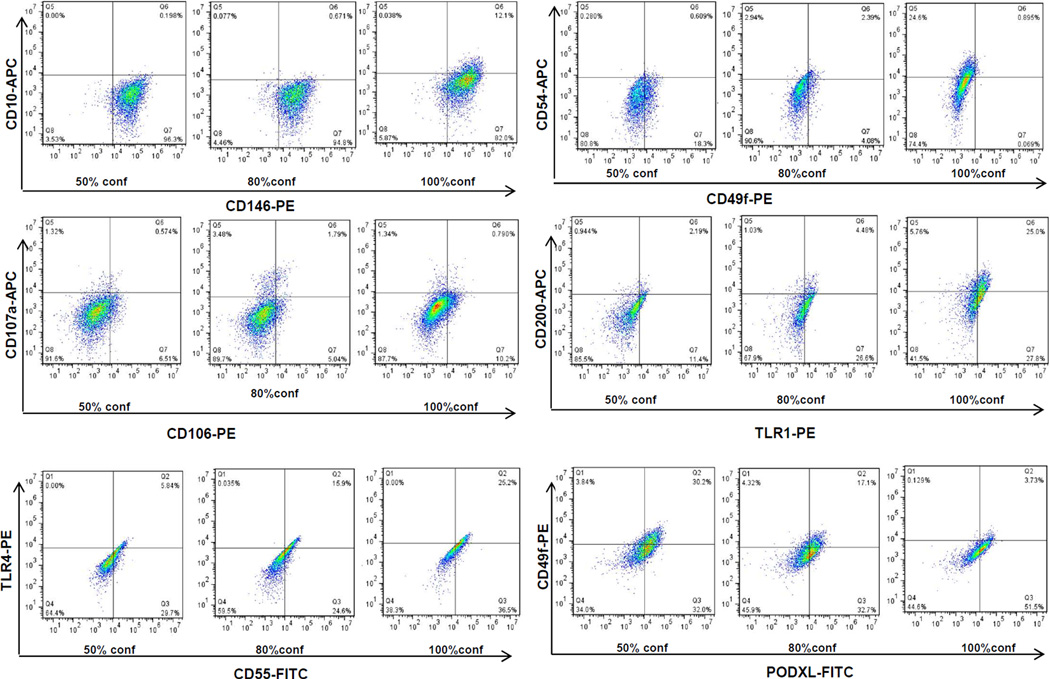
The expression of BMSC surface markers were also affected by confluence. Among these markers, CD10, CD54, CD106, CD200 and TLR4 were up-regulated in 100% confluent BMSCs compared with 50% and 80% confluent cells (Figure 3), while CCR7 and FZD4 were not detected (positive percentage <1%). This suggests that CD10, CD54, CD200 and TLR4 may serve as biomarkers of BMSC confluence. In particular, CD10, which is expressed by BMSCs as well as by mesenchymal cells from other tissues44–46, was barely detectable at 50% and 80% confluence, but a sharp increase in its expression was observed when BMSCs reached 100% confluence (Figure 3). Of note, the expression of CD49f was down-regulated with confluence in both donors; PODXL was slightly down-regulated at 100% confluence in one donor and down-regulated at 80% and 100% confluence in another. These data were in agreement with a previous report34; the only difference being that a comparison was made only between low density cells and those making initial contact cells in the published paper and it is unclear what would occur when the BMSCs reached 100% confluence.
Figure 3. Changes in BMSC surface marker expression at 50%, 80% and 100% confluence.
BMSCs from 2 donors were cultured to 50%, 80% and 100% confluence and the expression of surface markers were measured by flow cytometry. Representative plots from one donor were shown. The markers were indicated by x-axis, and y-axis.
Glucose and Lactate Levels in BMSC Supernatants
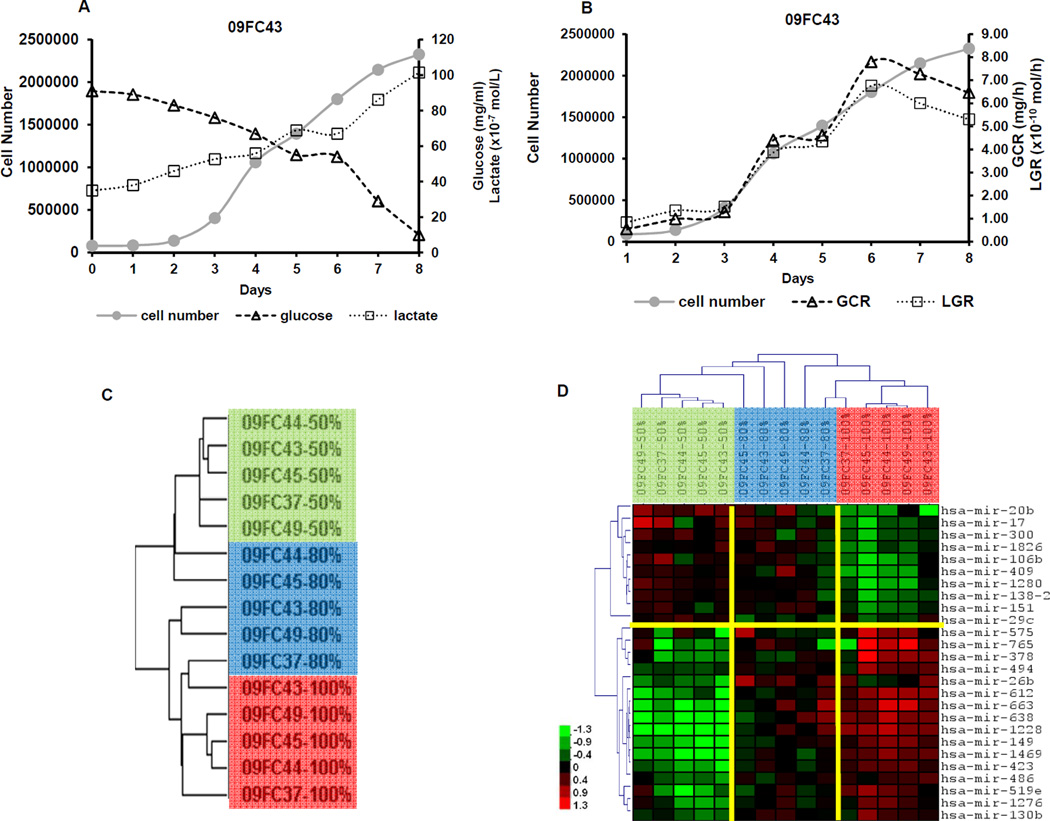
One goal of this study is to discover biomarkers that can be used for monitoring BMSC confluence during the manufacturing process. To identify surrogate markers of BMSC confluence, we measured glucose and lactate levels and pH values of the BMSC culture supernatants. The pH values fluctuated daily, ranging from 7.27 to 7.81 and there was no relationship between pH and BMSC confluence (data not shown). The glucose levels decreased daily despite small fluctuations after medium change on day 3, 5 and day 7 and they were inversely correlated with cell number with correlation coefficients ranging from 0.89 to 0.96, which were significant for BMSCs for all 4 donors (p<0.01) (Table 1 and Figure 4A). In contrast, the lactate values increased daily and also correlated with cell number with correlation coefficients ranging from 0.88 to 0.96 (p<0.01, Figure 4A). We also calculated the rate of the change in glucose and lactate values noticing that both the glucose consumption rate (GCR) and lactate generation rate (LGR) were significantly correlated with cell number (Table 1 and Figure 4B). Based on these data, we concluded that the values of glucose and GCR, or lactate and LGR are good indicators of cell number and can be used to determine the timing of BMSC passage or harvesting.
Table 1.
Correlation between Glucose, Lactate, Glucose Consumption Rate (GCR), Lactate Generation Rate (LGR) and Cell Number
| BMSC ID | Glucose | Lactate | GCR | LGR | ||||
|---|---|---|---|---|---|---|---|---|
| R | p-Value | R | p-Value | R | p-Value | R | p-Value | |
| 09FC43 | −0.96 | 4.77E-05 | 0.95 | 7.74E-05 | 0.96 | 2.11E-04 | 0.94 | 4.96E-04 |
| 09FC44 | −0.89 | 1.35E-03 | 0.88 | 1.55E-03 | 0.97 | 5.60E-05 | 0.98 | 2.79E-05 |
| 09FC45 | −0.95 | 1.14E-04 | 0.93 | 2.81E-04 | 0.97 | 7.21E-05 | 0.89 | 3.26E-03 |
| 09FC49 | −0.9 | 1.06E-03 | 0.88 | 1.65E-03 | 0.76 | 2.84E-02 | 0.8 | 1.65E-02 |
Figure 4. Measuring glucose and lactate in the culture supernatant and microRNA profiling of BMSCs at 50%, 80% and 100% confluence.
The BMSC number and culture supernatant concentrations of glucose and lactate were measured daily from 4 donors; the Glucose Consumption Rate (GCR) and Lactate Generation Rate (LGR) were calculated. Representative plot of raw values (A) and calculated GCR and LGR (B) from one donor, 09FC43, are shown.
Unsupervised hierarchical clustering analysis of 166 informative human microRNA separated samples into clusters of 50% confluence (green bar), 80% confluence (blue bar) and 100% confluence (red bar) (C). Twenty-six differentially expressed microRNAs were identified by Two-way ANOVA (p<0.05). Hierarchical clustering analysis using the 26 microRNA separated the samples into clusters of 50% confluent (green bar), 80% confluent (blue bar) and 100% confluent BMSCs (red bar). The names of the 26 microRNA are shown on the right of each row (D). Scale Bar indicates the expression level.
MicroRNA expression profiling
The microRNA profiling of 50%, 80% and 100% confluent BMSCs was conducted on custom-made microarrays with 1920 probes. After filtering out the array spots which did not meet the criteria of size (between 20 and 300nm), presence (greater than 50%) and intensity (greater than 20), and species (mouse, rat, etc), a total of 166 human microRNAs were considered evaluable. Unsupervised hierarchical clustering using these 166 microRNAs separated the BMSC samples into distinct clusters based on their degrees of confluence: 50% confluent BMSCs were located primarily within one cluster, 100% confluent BMSCs were located within another, while 80% confluent BMSCs were segmented within either 50% confluence (2 of 5 samples) or 100% confluence (3 of 5) (Figure 4C). For further analysis, 26 microRNAs that were differentially expressed among the 3 degrees of confluence were identified by using 2-way ANOVA, considering P<0.05 as significant. Hierarchical clustering analysis using these 26 microRNAs separated the 15 BMSC samples into 2 major clusters; one with all five 50% confluent samples and three 80% confluent samples; another one with all five 100% confluent samples and two samples of 80% confluence. Within one cluster, the 50% confluent BMSC samples formed a tight sub-cluster when compared with the 80% confluent BMSCs, as did the 100% confluent samples (Figure 4D). Among the differentially expressed microRNAs miR-17, miR-106b, miR-138, miR-26b, miR-130b, miR-494, miR-663 and miR-638 have been previously reported to be expressed in BMSCs and may be involved with osteogeneic or chondrogenic differentiation47–49, and miR-17, miR-29c and miR-486 may be associated with aging or senescence of BMSC50, 51 (Table 2).
Table 2.
MicroRNAs Differentially Expressed Among 50%, 80% and 100% BMSCs
| microRNAs | Confluence | P-Value | MSC activities | ||
|---|---|---|---|---|---|
| 50% | 80% | 100% | |||
| hsa-miR-17 | −3.13 | −3.41 | −4.18 | 2.9E-02 | Down-regulated in aging human MSC, targeting CDKN1A63 Accelerated 3T3L1 adipocyte differentiation by targeting Rb2/p13064 promote proliferation of USSC and prevent cell cycle arrest during neuronal lineage by targeting CCND1, E2F1, CDKN1A, PTEN, RB1, RBL1, RBL2, WEE165 |
| hsa-miR-20b | −2.50 | −3.10 | −4.22 | 4.4E-03 | up-regulated in ES-MSCs and targeting EPAS166 expressed in WJ-MSC and down-regulated during WJ-MSC neurogenesis67 |
| hsa-miR-106b | −2.40 | −2.61 | −3.37 | 1.1E-02 | Up-regulated in osteo-differentiated MSCs68 promote proliferation of USSC and prevent cell cycle arrest during neuronal lineage by targeting CCND1, E2F1, CDKN1A, PTEN, RB1, RBL1, RBL2, WEE165 |
| hsa-miR-29c | 0.57 | 0.08 | 0.20 | 1.7E-02 | Up-regulated in replicative senescence69, possibly targeting DNMT3A, DNMT3B69,70 increased during osteoblastic differentiation of murine cell line and involved in Wnt Signaling Pathway by targeting Sparc71 reduced expression of extracellular matrix genes72 |
| hsa-miR-138 | 0.92 | 0.71 | 0.14 | 1.4E-02 | Down-regulated during osteoblast differentiation of hMSCs; inhibited osteoblast differentiation of hMSCs in vitro and reduced ectopic bone formation in vivo by targeting FAK73 Down-regulated in osteo-differentiated MSCs68 Up-regulated in osteo-differentiated MSC49 Down-regulated during adipogenic differentiation of human ASC; inhibited adipogenesis by targeting EID-19 up-regulated during WJ-MSC neurogenesis67 |
| hsa-miR-26b | −0.39 | 0.82 | 0.49 | 4.3E-03 | up-regulated during osteoblast differentiation of hMSCs73 up-regulated during chondrogeneic differentiation of human BMSC, possibly targeting COL4A1, COL2A1, and COL6A174 up-regulated expression in wound tissue at the stage of active granulation formation75 Detected in human ES derived MSC76 |
| hsa-miR-130b | −0.62 | −0.07 | 0.08 | 2.1E-02 | Up-regulated in osteo-differentiated MSCs68 and chondro-differentiated MSCs74 Detected in human ES derived MSC76 up-regulated expression in wound tissue at the stage of active granulation formation75 |
| hsa-miR-494 | −0.23 | 0.20 | 0.59 | 4.9E-03 | Differentially expressed in osteo-differentiated MSC with Donor variance68 |
| hsa-miR-663 | −1.40 | 0.36 | 0.93 | 1.4E-04 | Differentially expressed in osteo-differentiated MSC with Donor variance68 Exclusively expressed in chondroblast derived from human MSC49 2 fold higher in MV than ES-MSC76 |
| hsa-miR-638 | −1.67 | 0.05 | 0.66 | 1.1E-05 | Exclusively expressed in chondroblast derived from human MSC49 down-regulated in osteo-differentiated MSC49 2 fold higher in MV than MSC76 |
| hsa-miR-486 | −0.37 | 0.20 | 0.59 | 2.8E-03 | Up-regulated in aging human ASCs, induces a premature senescence-like phenotype and inhibits proliferation; inhibits adipogenic and osteogenic differentiation by targeting SIRT177 |
USSC: unrestricted somatic stem cells from human cord blood. ES-MSC: human ES-derived MSC. WJ-MSC: Wharton's jelly matrix-derived mesenchymal stem cell. ASC: Adipose-tissue derived stem cell. MV: microvesicles
Gene Expression Profiling of BMSCs
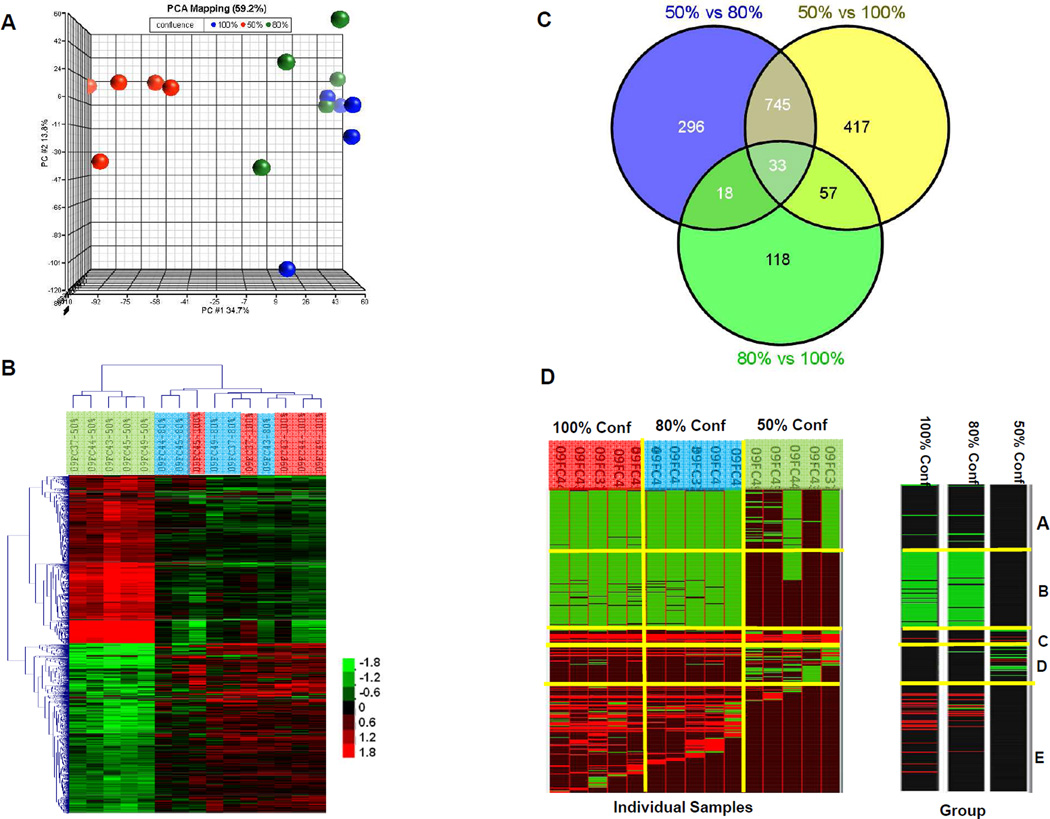
Gene expression profiling was also performed on 50%, 80% and 100% confluent BMSCs. Among all the genes, 11,795 passed the variance criteria. Principle Component Analysis (PCA) using these genes separated the samples into 2 groups, one included 50% confluent BMSCs, while the other included 80% and 100% confluent BMSC (Figure 5A). These results were consistent with published data that BMSCs of high confluence have different gene expression profiles compared to those of low confluence52. We then identified 2708 genes that were differentially expressed among the 3 confluences by using 2-way ANOVA (p-value <0.001). Hierarchical clustering analysis using these genes revealed that 50% confluent BMSCs formed one cluster, while 80% and 100% confluent BMSCs formed another (Figure 5B). Among these differentially expressed genes, most were differentially expressed between 50% confluent and 80% confluent BMSCs (p<0.001, FC≥1.5, n=1092), or between 50% confluent and 100% confluent BMSCs (p<0.001, FC≥1.5, n=1252), while only 226 genes were differentially expressed between 80% and 100% confluent BMSCs (Figure 5C).
Figure 5. Gene expression profiling of BMSCs at 50%, 80% and 100% confluence.
Principal component analysis (PCA) separated the BMSCs into a group with 50% confluent BMSCs (red spheres) and another group with both 80% (green spheres) and 100% confluent BMSCs (blue spheres) (A). Differentially expressed genes (p<0.001) were used for hierarchical clustering analysis which separated the samples into a cluster with 50% confluence (green bar) and another cluster with 80% confluence (blue bar) and 100% confluence (red bar). Scale Bar indicates the expression level. (B). The number of genes whose expression differed significantly among groups (p<0.001, Fold Change >1.5) are displayed using a Venn Diagram (C). The annotation of differentially expressed genes was conducted using Genomica. Each row stands for one gene set, and each column stands for one individual BMSC sample (D, left) or one of the 3 groups of 50%, 80% and 100% confluent BMSCs (D, right). The green grids indicated “repressed” and red grids indicated “activated”.
To obtain an overview of the functions of these differentially expressed genes, we conducted a gene set enrichment analysis. A heat-map was generated based on the significance that was defined by the percentage of hit genes with each gene set using Genomica software with default settings41. The significantly changed (p<0.05, FDR<0.1) gene sets, either “induced” or “repressed”, as defined by Segal41 were grouped into 5 clusters (Figure 5D).
Most of the gene sets in cluster A were related to the metabolic process; at the individual sample level, these gene sets were repressed in 5/5 80% confluent samples, in 5 of 5 100% confluent samples and in 2/5 50% confluent samples. However, as we grouped the BMSCs samples according to their degrees of confluence, these gene sets did not differ significantly among the 50%, 80% and 100% confluent groups (Figure 5D).
The gene sets in cluster B which were repressed by 80% and 100% confluent BMSC samples included Regulation of Mitosis, Cell Cycle Checkpoint and Regulation of Cyclin-Dependent Protein Kinase Activity. These results were in accordance with published data52.
The majority of gene sets in cluster C were induced in all the individual samples but did not differ significantly at the group of confluence level (Figure 5D, right panel). The exception was Growth Factor Activity, which was particularly induced by 100% confluence and the genes contributing to this set were IGF1, GRN, CSF1, KITLG, FGF5, VEGFC, S100A6, CLCF1 and DKK1.
The gene sets in Cluster D were either induced or repressed by 50% confluence but not by 80% or 100% confluence, interestingly, two gene sets associated with microRNAs, hsa-miR-184 and hsa-miR-205 were identified in this cluster.
Gene sets in cluster E were mainly induced by 80% and 100% confluence but not by 50% confluence. These genes included Extracellular Region, Immune Response, Cell Adhesion, Cell Migration and Inflammatory Response.
The Extracellular Region set contained a large number of genes that are functionally important in both stromal and stem cell functions. Among the genes in the Extracellular Region set that are likely involved with stromal cell functions were those encoding extracellular matrix proteins: FBN1, FBLN1, FBLN5, ECM2, LUM, DCN, COLQ, COL16A1 and COL10A1; enzymes and inhibitors: PLAU, SERPINE2, PAPPA, TIMP3, TIMP1, CTSD, ADAMTS10 and ADAMTS1; growth factors: SERPINF1 (PEGF), CSF1, KITLG, TNFSF15, FGF5, CLCF1, ANGPTL2 and IGF1 and chemokines: CCL13, CCL7 and CCL2. The Extracellular Region included stem cell genes that included Wnt signaling transducers: WNT7B, DKK1, DKK2, DKK3 and WISP1; TGF-beta signaling pathway transducers: DCN, LTBP3, BGN and HTRA1; associated with osteoblastic differentiation: TNFRSF11B, SPOCK1, BGN; and associated with chondrogeneic differentiation VCAN, COMP and COL10A1. Evaluation of the expression of the individual Wnt signaling pathway inhibitor genes DKK1, DKK2 and DKK3; the angiogenesis inhibitors SERPINF1, TNFSF15 and ADAMTS1 revealed that they were up-regulated by 100% confluent BMSCs, suggesting that 100% confluence may comprise some of the beneficial properties of BMSCs including their stem cells qualities and support of angiogenesis.
The genes contributing to Immune Response were FTH1, GPX1, CCL2, ITGB5, TNFAIP6, VCAM1, GBP2, DES, MYLK, FAP, CDH13, TIMP3, TIMP1 and CTSB. Among these genes TNFAIP6 is an important regulator of immunosuppression and anti-inflammation response53, 54. Moreover, genes involved in Inflammatory Response were particularly up-regulated by 100% confluence and these genes were PLA2G4C, SERPING1, ANXA1, CCL13, CCL7, CCL2, TNFAIP6, SAA2, CFHR1, CFH, IL1RAP, C1S, C1R and PRKCA.
Discussion
In the field of cellular therapy, the degree of confluence at which to passage or harvest BMSCs remains an important unanswered question. Using multiple different measures, we found that the characteristics of BMSCs change as they grow from 50% to 80% and to 100% confluence, particularly when they reach 100% confluence. Our results suggest that harvesting cells at 80% confluence is optimal since BMSC proliferation slows when they reach 100% confluence and changes in expression of biomarkers in 100% confluent cells suggests that their pro-angiogenesis activities may be compromised. We found that culture supernatant levels of lactate or glucose levels could be used as a surrogate marker of cell number and confluence; flow cytometry or microRNA expression analysis can be used to confirm that the BMSCs have been harvested at the appropriate level of confluence.
Our assessment of BMSC growth showed that confluence affected BMSC proliferation and colony formation, particularly for 100% confluent cells. BMSC PDT fell from 50% to 80% confluence and then increased when they become 100% confluent. The quantity of colonies formed by BMSCs was similar for 50% and 80% confluent cells but was less for 100% confluent cells.
We also found that significant changes in protein expression occurred as confluence increased from 80% to 100% which was manifested by changes in the make-up of BMSC culture supernatant and the expression of surface markers. The supernatant factors that increased steadily included PEDF, which has been reported to be the most potent natural angiogenesis inhibitor and the VEGF/PEDF ratio is closely correlated with the degree of neovascularization55. PEDF has also been reported to impair the beneficial effects of MSCs against myocardial infarction injury56. We observed an increase in the PEDF/VEGF ratio in 3 of the 4 donors (Figure 2C) which suggests that growing BMSCs to greater than 100% confluence may comprise their pro-angiogenesis properties. Interestingly, bFGF (FGF2), another functionally important BMSC growth factor, was not detected in the culture supernatant, but we were able to detect it in the cytoplasm of BMSCs at a very high level (data not shown). This may, therefore, partially explain the in vivo paracrine effects of BMSC, since the trophic factors such as bFGF would be released from the dead cells after infusion53.
The expression of several BMSC surface markers increased as confluence increased; but many did not increase until the cells reached 100% confluence. It is not known if the increase in expression of these markers are important to the function of BMSCs, but the expression of CD54 (ICAM1) and CD106 (VCAM) have been reported to be induced by IFN-γ or TNF-α35, 57, 58 and have been associated MSC-mediated immunosuppression59. We also found that CD55 was up-regulated in 100% confluent BMSCs. CD55 has previously been detected on BMSCs and can suppress complement activation60. Since BMSCs are able to activate the complement system when exposed to human sera61, it would be interesting to investigate the function CD55 on BMSCs.
In contrast to the changes in protein expression and production, transcription analysis revealed that the most gene expression changes occurred when the confluence increased from 50% to 80%. The genes that were up-regulated in 100% confluent BMSCs compared to 80% confluent cells included Wnt signaling pathway inhibitor genes and angiogenesis inhibitors. This also suggests that 100% confluent BMSCs many have reduced pro-angiogenic activity despite the differences between 80% and 100% BMSCs in the expression of genes and proteins involved with the immune response, there was little difference in the suppression of mixed lymphocyte reactions between 80% and 100% confluent cells. This suggests that it is possible that there is little difference in immunomodulatory functions of BMSCs grown to 80% and 100% confluence.
Since an important regenerative medicine application of BMSCs is bone regeneration, we explored changes in the expression of genes involved with bone formation. We previously demonstrated that in vivo bone formation ability of 100% confluent BMSCs was reduced slightly compared with 70% confluent BMSCs, but this difference was donor dependent62. In this study we found that Regulation of Ossification (Gene Ontology term, GO: 0030278) was over-represented by 80% confluent BMSCs (p=4.62E-03). The genes contributing to this GO were ANKH, BMPR1B, CSF1, CTNNBIP1, EGR2, IGF1, JUND, PBX1, PRL, RUNX2 and SKI. Interestingly, the expression of these genes was higher in 80% confluent BMSCs compared to 50% confluent cells, but they did not significantly differ between 80% confluence and 100% confluence. However, since the BMSC skeletal stem cell sub-population is quite rare, and our gene expression profiling was performed on bulk RNA, the real signature of SSC may be masked by the majority of the more committed stromal cells. We are now performing single-cell analysis to further characterize the BMSCs, which may provide insightful data on the effects of confluences on SSC.
From the perspective of a GMP-compliant manufacturing facility, harvesting BMSCs at the same degree of confluence for each production lot is essential to provide consistently high quality BMSCs. The practice in our clinical cell therapy laboratory is to harvest BMSCs when they become 80% confluent. Based on the results of this study we will continue this practice. The results of our study suggest that culturing to 100% confluence is detrimental in some respects to BMSCs. BMSCs grown to 50% confluence also differ from 80% confluent cells, but the impact of these differences is less certain. However, harvesting at 50% confluence reduces yields and, as a result, increases the cost of production.
It would be desirable to identify markers that are associated with cell confluence and which could provide a global evaluation of BMSC confluence. Since BMSCs are grown as adherent cell cultures, the cells cannot be sampled to measure surface marker, microRNA or gene expression to determine if they had reached 80% confluence. Culture supernatant, however, can easily be collected and used to assess biomarkers. While the levels of many soluble proteins change during BMSC culture, only when protein levels are normalized for cell number can they be used to predict BMSC confluence. Therefore, supernatant protein levels are of limited use in assessing confluence of these adherent cells. We did, however, find that supernatant glucose and/or lactate values are good indicators of cell density. The GCR and LGR also proved to be highly correlated with cell densities. These measures are markers of BMSC confluence and could be used as an indicator of the optimal harvest time. However, LGR and GCR will depend on culture conditions and culture vessel characteristics, so if changes in lactate and or glucose levels are used, they will have to be validated for each set of culture conditions.
It is often desirable to test intermediate or final products to document their quality. Our results show that the analysis of the expression of surface makers by flow cytometry, particularly CD10 expression, could be used to ensure that BMSCs have been harvested prior to reaching 100% confluence. It may also be possible to use the expression of a panel of microRNA for a more refined measurement to ensure that BMSCs have been harvested when they were greater than 50% confluent but less than 100% confluent.
Of note, these studies involved the culture of bone marrow-derived MSCs in flasks with 20% FBS and the effects of confluence could possibly be different on MSCs grown under other conditions or on MSCs from other tissues. For example, it is not certain if the effects of confluence would be the same on earlier or later passage BMSCs. Confluence may also have different effects on BMSCs grown in other vessels such as bioreactors, with other culture medium such as serum-free culture medium or on other types of MSCs such as adipose tissue-derived MSCs. In addition, the effects of confluence on the in vivo properties of BMSCs could differ from the properties measured in vitro. Further studies on the effects of confluence on other types of MSCs, MSCs grown under other conditions and the effects on their in vivo properties are warranted.
In conclusion, growing BMSCs to higher confluences increases cell density and yields, but harvesting cells at 100% confluence may not result in cells of the highest quality. The measurement of supernatant lactate and glucose levels can be used to estimate BMSC quantity and confluence and can be used to help ensure consistency in the timing of passage and harvest. Flow cytometry or microRNA expression analysis can be used to confirm that the BMSCs have been harvested at the appropriate confluence.
Supplementary Material
Acknowledgements
The authors thank the staff of the Cell Processing Laboratory, Department of Transfusion Medicine, Clinical Center, NIH for their support of these studies. We also thank Dr. Francesco Marincola and Dr. Ena Wang (Department of Transfusion Medicine, Clinical Center, NIH, now at Sidra Medical and Research Center, Qtar) for useful discussions and comments on the manuscript. This work was funded by NIH Bone Marrow Stromal Cell Transplantation Center (BMSCTC) and the Department of Transfusion Medicine, Clinical Center NIH.
Abbreviations
- BMSC
Bone Marrow Stromal Cell
- PEDF
Pigment Epithelium-Derived Factor
- VEGF
Vascular Endothelial Growth Factor
- PD
Population Doubling
- PDT
Population Doubling Time
- CFE
Colony Formation Efficiency
- aGvHD
acute Graft-versus-Host-Disease
- GMP
Good Manufacturing Practice
- SOP
Standard Operating Procedures
- PI
Propidium Iodide
- GCR
Glucose Consumption Rate
- LGR
Lactate Generation Rate
- MLR
Mixed Lymphocyte Reaction
- PCA
Principle Component Analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclose of Interests
The authors have no commercial, proprietary or financial interest in the products or companies described in this article.
Supplementary Material
S1. List of BMSCs used for each experiment
S2: Representative figures of 50%, 80% and 100% confluence.
S3: Table showing concentrations of proteins in BMSC supernatant.
Contributor Information
Jiaqiang Ren, Email: renj@mail.nih.gov.
Huan Wang, Email: heidi.wang@nih.gov.
Katherine Tran, Email: ktran16@jhu.edu.
Ping Jin, Email: pjin@cc.nih.gov.
Luciano Castiello, Email: castiellol@cc.nih.gov.
Ji Feng, Email: fenj2@cc.nih.gov.
Sergei A Kuznetsov, Email: skuznetsov@dir.nidcr.nih.gov.
Pamela G. Robey, Email: probey@dir.nidcr.nih.gov.
Marianna Sabatino, Email: sabatinom@cc.nih.gov.
David F. Stroncek, Email: dstroncek@cc.nih.gov.
Reference
- 1.Friedenstein AJ, Piatetzky S, II, Petrakova KV. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morphol. 1966;16:381–390. [PubMed] [Google Scholar]
- 2.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhao D, Cui D, Wang B, Tian F, Guo L, Yang L, Liu B, Yu X. Treatment of early stage osteonecrosis of the femoral head with autologous implantation of bone marrow-derived and cultured mesenchymal stem cells. Bone. 50:325–330. doi: 10.1016/j.bone.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Davatchi F, Abdollahi BS, Mohyeddin M, Shahram F, Nikbin B. Mesenchymal stem cell therapy for knee osteoarthritis. Preliminary report of four patients. Int J Rheum Dis. 14:211–215. doi: 10.1111/j.1756-185X.2011.01599.x. [DOI] [PubMed] [Google Scholar]
- 5.Wakitani S, Okabe T, Horibe S, Mitsuoka T, Saito M, Koyama T, Nawata M, Tensho K, Kato H, Uematsu K, Kuroda R, Kurosaka M, Yoshiya S, Hattori K, Ohgushi H. Safety of autologous bone marrow-derived mesenchymal stem cell transplantation for cartilage repair in 41 patients with 45 joints followed for up to 11 years and 5 months. J Tissue Eng Regen Med. 5:146–150. doi: 10.1002/term.299. [DOI] [PubMed] [Google Scholar]
- 6.Orozco L, Soler R, Morera C, Alberca M, Sanchez A, Garcia-Sancho J. Intervertebral disc repair by autologous mesenchymal bone marrow cells: a pilot study. Transplantation. 92:822–828. doi: 10.1097/TP.0b013e3182298a15. [DOI] [PubMed] [Google Scholar]
- 7.Le Blanc K, Rasmusson I, Sundberg B, Gotherstrom C, Hassan M, Uzunel M, Ringden O. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet. 2004;363:1439–1441. doi: 10.1016/S0140-6736(04)16104-7. [DOI] [PubMed] [Google Scholar]
- 8.Ciccocioppo R, Bernardo ME, Sgarella A, Maccario R, Avanzini MA, Ubezio C, Minelli A, Alvisi C, Vanoli A, Calliada F, Dionigi P, Perotti C, Locatelli F, Corazza GR. Autologous bone marrow-derived mesenchymal stromal cells in the treatment of fistulising Crohn's disease. Gut. 60:788–798. doi: 10.1136/gut.2010.214841. [DOI] [PubMed] [Google Scholar]
- 9.Tan J, Wu W, Xu X, Liao L, Zheng F, Messinger S, Sun X, Chen J, Yang S, Cai J, Gao X, Pileggi A, Ricordi C. Induction therapy with autologous mesenchymal stem cells in living-related kidney transplants: a randomized controlled trial. JAMA. 307:1169–1177. doi: 10.1001/jama.2012.316. [DOI] [PubMed] [Google Scholar]
- 10.Hare JM, Fishman JE, Gerstenblith G, DiFede Velazquez DL, Zambrano JP, Suncion VY, Tracy M, Ghersin E, Johnston PV, Brinker JA, Breton E, Davis-Sproul J, Schulman IH, Byrnes J, Mendizabal AM, Lowery MH, Rouy D, Altman P, Wong Po Foo C, Ruiz P, Amador A, Da Silva J, McNiece IK, Heldman AW, George R, Lardo A. Comparison of allogeneic vs autologous bone marrow-derived mesenchymal stem cells delivered by transendocardial injection in patients with ischemic cardiomyopathy: the POSEIDON randomized trial. Jama. 2012;308:2369–2379. doi: 10.1001/jama.2012.25321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quevedo HC, Hatzistergos KE, Oskouei BN, Feigenbaum GS, Rodriguez JE, Valdes D, Pattany PM, Zambrano JP, Hu Q, McNiece I, Heldman AW, Hare JM. Allogeneic mesenchymal stem cells restore cardiac function in chronic ischemic cardiomyopathy via trilineage differentiating capacity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14022–14027. doi: 10.1073/pnas.0903201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uemura R, Xu M, Ahmad N, Ashraf M. Bone marrow stem cells prevent left ventricular remodeling of ischemic heart through paracrine signaling. Circulation research. 2006;98:1414–1421. doi: 10.1161/01.RES.0000225952.61196.39. [DOI] [PubMed] [Google Scholar]
- 13.Hatzistergos KE, Quevedo H, Oskouei BN, Hu Q, Feigenbaum GS, Margitich IS, Mazhari R, Boyle AJ, Zambrano JP, Rodriguez JE, Dulce R, Pattany PM, Valdes D, Revilla C, Heldman AW, McNiece I, Hare JM. Bone marrow mesenchymal stem cells stimulate cardiac stem cell proliferation and differentiation. Circulation research. 2010;107:913–922. doi: 10.1161/CIRCRESAHA.110.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong JA, Ko KM, Bae S, Jeon CJ, Koh GY, Kim H. Genome-wide differential gene expression profiling of human bone marrow stromal cells. Stem Cells. 2007;25:994–1002. doi: 10.1634/stemcells.2006-0604. [DOI] [PubMed] [Google Scholar]
- 15.Bae S, Ahn JH, Park CW, Son HK, Kim KS, Lim NK, Jeon CJ, Kim H. Gene and microRNA expression signatures of human mesenchymal stromal cells in comparison to fibroblasts. Cell Tissue Res. 2009;335:565–573. doi: 10.1007/s00441-008-0729-y. [DOI] [PubMed] [Google Scholar]
- 16.Tormin A, Brune JC, Olsson E, Valcich J, Neuman U, Olofsson T, Jacobsen SE, Scheding S. Characterization of bone marrow-derived mesenchymal stromal cells (MSC) based on gene expression profiling of functionally defined MSC subsets. Cytotherapy. 2009;11:114–128. doi: 10.1080/14653240802716590. [DOI] [PubMed] [Google Scholar]
- 17.Walenda T, Bork S, Horn P, Wein F, Saffrich R, Diehlmann A, Eckstein V, Ho AD, Wagner W. Co-culture with mesenchymal stromal cells increases proliferation and maintenance of haematopoietic progenitor cells. J Cell Mol Med. 14:337–350. doi: 10.1111/j.1582-4934.2009.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones E, English A, Churchman SM, Kouroupis D, Boxall SA, Kinsey S, Giannoudis PG, Emery P, McGonagle D. Large-scale extraction and characterization of CD271+ multipotential stromal cells from trabecular bone in health and osteoarthritis: implications for bone regeneration strategies based on uncultured or minimally cultured multipotential stromal cells. Arthritis Rheum. 62:1944–1954. doi: 10.1002/art.27451. [DOI] [PubMed] [Google Scholar]
- 19.Lee CC, Christensen JE, Yoder MC, Tarantal AF. Clonal analysis and hierarchy of human bone marrow mesenchymal stem and progenitor cells. Exp Hematol. 38:46–54. doi: 10.1016/j.exphem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riekstina U, Cakstina I, Parfejevs V, Hoogduijn M, Jankovskis G, Muiznieks I, Muceniece R, Ancans J. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009;5:378–386. doi: 10.1007/s12015-009-9094-9. [DOI] [PubMed] [Google Scholar]
- 21.Rosova I, Dao M, Capoccia B, Link D, Nolta JA. Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells. 2008;26:2173–2182. doi: 10.1634/stemcells.2007-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grisendi G, Anneren C, Cafarelli L, Sternieri R, Veronesi E, Cervo GL, Luminari S, Maur M, Frassoldati A, Palazzi G, Otsuru S, Bambi F, Paolucci P, Pierfranco C, Horwitz E, Dominici M. GMP-manufactured density gradient media for optimized mesenchymal stromal/stem cell isolation and expansion. Cytotherapy. 12:466–477. doi: 10.3109/14653241003649510. [DOI] [PubMed] [Google Scholar]
- 23.Lazarus HM, Koc ON, Devine SM, Curtin P, Maziarz RT, Holland HK, Shpall EJ, McCarthy P, Atkinson K, Cooper BW, Gerson SL, Laughlin MJ, Loberiza FR, Jr, Moseley AB, Bacigalupo A. Cotransplantation of HLA-identical sibling culture-expanded mesenchymal stem cells and hematopoietic stem cells in hematologic malignancy patients. Biol Blood Marrow Transplant. 2005;11:389–398. doi: 10.1016/j.bbmt.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Corcione A, Benvenuto F, Ferretti E, Giunti D, Cappiello V, Cazzanti F, Risso M, Gualandi F, Mancardi GL, Pistoia V, Uccelli A. Human mesenchymal stem cells modulate B-cell functions. Blood. 2006;107:367–372. doi: 10.1182/blood-2005-07-2657. [DOI] [PubMed] [Google Scholar]
- 25.Samuelsson H, Ringden O, Lonnies H, Le Blanc K. Optimizing in vitro conditions for immunomodulation and expansion of mesenchymal stromal cells. Cytotherapy. 2009;11:129–136. doi: 10.1080/14653240802684194. [DOI] [PubMed] [Google Scholar]
- 26.Mankani MH, Kuznetsov SA, Marshall GW, Robey PG. Creation of new bone by the percutaneous injection of human bone marrow stromal cell and HA/TCP suspensions. Tissue Eng Part A. 2008;14:1949–1958. doi: 10.1089/ten.tea.2007.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koc ON, Gerson SL, Cooper BW, Dyhouse SM, Haynesworth SE, Caplan AI, Lazarus HM. Rapid hematopoietic recovery after coinfusion of autologous-blood stem cells and culture-expanded marrow mesenchymal stem cells in advanced breast cancer patients receiving high-dose chemotherapy. J Clin Oncol. 2000;18:307–316. doi: 10.1200/JCO.2000.18.2.307. [DOI] [PubMed] [Google Scholar]
- 28.Koc ON, Day J, Nieder M, Gerson SL, Lazarus HM, Krivit W. Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH) Bone Marrow Transplant. 2002;30:215–222. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- 29.Gentry T, Foster S, Winstead L, Deibert E, Fiordalisi M, Balber A. Simultaneous isolation of human BM hematopoietic, endothelial and mesenchymal progenitor cells by flow sorting based on aldehyde dehydrogenase activity: implications for cell therapy. Cytotherapy. 2007;9:259–274. doi: 10.1080/14653240701218516. [DOI] [PubMed] [Google Scholar]
- 30.Zhukareva V, Obrocka M, Houle JD, Fischer I, Neuhuber B. Secretion profile of human bone marrow stromal cells: donor variability and response to inflammatory stimuli. Cytokine. 50:317–321. doi: 10.1016/j.cyto.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Wolfe M, Pochampally R, Swaney W, R R. Isolation and Culture of Bone Marrow-Derived Human Multipotent Stromal Cells (hMSCs) Mesenchymal Stem Cells: Methods and Protocols. 2008;449:3–425. doi: 10.1007/978-1-60327-169-1_1. [DOI] [PubMed] [Google Scholar]
- 32.Sabatino M, Ren J, David-Ocampo V, England L, McGann M, Tran M, Kuznetsov SA, Khuu H, Balakumaran A, Klein HG, Robey PG, Stroncek DF. The establishment of a bank of stored clinical bone marrow stromal cell products. J Transl Med. 10:23. doi: 10.1186/1479-5876-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren J, Stroncek DF, Zhao Y, Jin P, Castiello L, Civini S, Wang H, Feng J, Tran K, Kuznetsov SA, Robey PG, Sabatino M. Intra-subject variability in human bone marrow stromal cell (BMSC) replicative senescence: Molecular changes associated with BMSC senescence. Stem cell research. 2013;11:1060–1073. doi: 10.1016/j.scr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee RH, Seo MJ, Pulin AA, Gregory CA, Ylostalo J, Prockop DJ. The CD34-like protein PODXL and alpha6-integrin (CD49f) identify early progenitor MSCs with increased clonogenicity and migration to infarcted heart in mice. Blood. 2009;113:816–826. doi: 10.1182/blood-2007-12-128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren G, Zhao X, Zhang L, Zhang J, L'Huillier A, Ling W, Roberts AI, Le AD, Shi S, Shao C, Shi Y. Inflammatory cytokine-induced intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in mesenchymal stem cells are critical for immunosuppression. Journal of immunology. 2010;184:2321–2328. doi: 10.4049/jimmunol.0902023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang YC, Tanimoto K, Tanne Y, Kamiya T, Kunimatsu R, Michida M, Yoshioka M, Yoshimi Y, Kato Y, Tanne K. Effects of human full-length amelogenin on the proliferation of human mesenchymal stem cells derived from bone marrow. Cell and tissue research. 2010;342:205–212. doi: 10.1007/s00441-010-1064-7. [DOI] [PubMed] [Google Scholar]
- 37.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Jin P, Sabatino M, Ren J, Civini S, Bogin V, Ichim TE, Stroncek DF. Comparison of endometrial regenerative cells and bone marrow stromal cells. Journal of translational medicine. 2012;10:207. doi: 10.1186/1479-5876-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 40.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 41.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nature genetics. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 42.Sabatino M, Ren J, David-Ocampo V, England L, McGann M, Tran M, Kuznetsov SA, Khuu H, Balakumaran A, Klein HG, Robey PG, Stroncek DF. The establishment of a bank of stored clinical bone marrow stromal cell products. Journal of translational medicine. 2012;10:23. doi: 10.1186/1479-5876-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prockop DJ, Sekiya I, Colter DC. Isolation and characterization of rapidly self-renewing stem cells from cultures of human marrow stromal cells. Cytotherapy. 2001;3:393–396. doi: 10.1080/146532401753277229. [DOI] [PubMed] [Google Scholar]
- 44.Farias VA, Linares-Fernandez JL, Penalver JL, Paya Colmenero JA, Ferron GO, Duran EL, Fernandez RM, Olivares EG, O'Valle F, Puertas A, Oliver FJ, Ruiz de Almodovar JM. Human umbilical cord stromal stem cell express CD10 and exert contractile properties. Placenta. 2011;32:86–95. doi: 10.1016/j.placenta.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Conrad C, Zeindl-Eberhart E, Moosmann S, Nelson PJ, Bruns CJ, Huss R. Alkaline phosphatase, glutathione-S-transferase-P, and cofilin-1 distinguish multipotent mesenchymal stromal cell lines derived from the bone marrow versus peripheral blood. Stem cells and development. 2008;17:23–27. doi: 10.1089/scd.2007.0159. [DOI] [PubMed] [Google Scholar]
- 46.Buhring HJ, Treml S, Cerabona F, de Zwart P, Kanz L, Sobiesiak M. Phenotypic characterization of distinct human bone marrow-derived MSC subsets. Annals of the New York Academy of Sciences. 2009;1176:124–134. doi: 10.1111/j.1749-6632.2009.04564.x. [DOI] [PubMed] [Google Scholar]
- 47.Gao J, Yang T, Han J, Yan K, Qiu X, Zhou Y, Fan Q, Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. Journal of cellular biochemistry. 2011;112:1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- 48.Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goff LA, Boucher S, Ricupero CL, Fenstermacher S, Swerdel M, Chase LG, Adams CC, Chesnut J, Lakshmipathy U, Hart RP. Differentiating human multipotent mesenchymal stromal cells regulate microRNAs: prediction of microRNA regulation by PDGF during osteogenesis. Experimental hematology. 2008;36:1354–1369. doi: 10.1016/j.exphem.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giraud-Triboult K, Rochon-Beaucourt C, Nissan X, Champon B, Aubert S, Pietu G. Combined mRNA and microRNA profiling reveals that miR-148a and miR-20b control human mesenchymal stem cell phenotype via EPAS1. Physiological genomics. 2011;43:77–86. doi: 10.1152/physiolgenomics.00077.2010. [DOI] [PubMed] [Google Scholar]
- 51.Zou Z, Zhang Y, Hao L, Wang F, Liu D, Su Y, Sun H. More insight into mesenchymal stem cells and their effects inside the body. Expert opinion on biological therapy. 2010;10:215–230. doi: 10.1517/14712590903456011. [DOI] [PubMed] [Google Scholar]
- 52.Lee MW, Kim DS, Ryu S, Jang IK, Kim HJ, Yang JM, Lee DH, Lee SH, Son MH, Cheuh HW, Jung HL, Yoo KH, Sung KW, Koo HH. Effect of ex vivo culture conditions on immunosuppression by human mesenchymal stem cells. BioMed research international. 2013;2013:154919. doi: 10.1155/2013/154919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee RH, Pulin AA, Seo MJ, Kota DJ, Ylostalo J, Larson BL, Semprun-Prieto L, Delafontaine P, Prockop DJ. Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell stem cell. 2009;5:54–63. doi: 10.1016/j.stem.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi H, Lee RH, Bazhanov N, Oh JY, Prockop DJ. Anti-inflammatory protein TSG-6 secreted by activated MSCs attenuates zymosan-induced mouse peritonitis by decreasing TLR2/NF-kappaB signaling in resident macrophages. Blood. 2011;118:330–338. doi: 10.1182/blood-2010-12-327353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan W, Crawford R, Xiao Y. The ratio of VEGF/PEDF expression in bone marrow mesenchymal stem cells regulates neovascularization. Differentiation; research in biological diversity. 2011;81:181–191. doi: 10.1016/j.diff.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Liang H, Hou H, Yi W, Yang G, Gu C, Lau WB, Gao E, Ma X, Lu Z, Wei X, Pei J, Yi D. Increased expression of pigment epithelium-derived factor in aged mesenchymal stem cells impairs their therapeutic efficacy for attenuating myocardial infarction injury. European heart journal. 2013;34:1681–1690. doi: 10.1093/eurheartj/ehr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Najar M, Raicevic G, Id Boufker H, Stamatopoulos B, De Bruyn C, Meuleman N, Bron D, Toungouz M, Lagneaux L. Modulated expression of adhesion molecules and galectin-1: role during mesenchymal stromal cell immunoregulatory functions. Experimental hematology. 2010;38:922–932. doi: 10.1016/j.exphem.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Ghannam S, Pene J, Torcy-Moquet G, Jorgensen C, Yssel H. Mesenchymal stem cells inhibit human Th17 cell differentiation and function and induce a T regulatory cell phenotype. J Immunol. 2010;185:302–312. doi: 10.4049/jimmunol.0902007. [DOI] [PubMed] [Google Scholar]
- 59.Ren G, Roberts AI, Shi Y. Adhesion molecules: key players in Mesenchymal stem cell-mediated immunosuppression. Cell adhesion & migration. 2011;5:20–22. doi: 10.4161/cam.5.1.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Lin F. Mesenchymal stem cells are injured by complement after their contact with serum. Blood. 2012;120:3436–3443. doi: 10.1182/blood-2012-03-420612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moll G, Jitschin R, von Bahr L, Rasmusson-Duprez I, Sundberg B, Lonnies L, Elgue G, Nilsson-Ekdahl K, Mougiakakos D, Lambris JD, Ringden O, Le Blanc K, Nilsson B. Mesenchymal stromal cells engage complement and complement receptor bearing innate effector cells to modulate immune responses. PloS one. 2011;6:e21703. doi: 10.1371/journal.pone.0021703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuznetsov SA, Mankani MH, Robey PG. Effect of serum on human bone marrow stromal cells: ex vivo expansion and in vivo bone formation. Transplantation. 2000;70:1780–1707. doi: 10.1097/00007890-200012270-00018. [DOI] [PubMed] [Google Scholar]
- 63.Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C, Laschober GT, Lepperdinger G, Sampson N, Berger P, Herndler-Brandstetter D, Wieser M, Kuhnel H, Strasser A, Rinnerthaler M, Breitenbach M, Mildner M, Eckhart L, Tschachler E, Trost A, Bauer JW, Papak C, Trajanoski Z, Scheideler M, Grillari-Voglauer R, Grubeck-Loebenstein B, Jansen-Durr P, Grillari J. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 9:291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Schira J, Muller HW, Wernet P. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One. 6:e16138. doi: 10.1371/journal.pone.0016138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giraud-Triboult K, Rochon-Beaucourt C, Nissan X, Champon B, Aubert S, Pietu G. Combined mRNA and microRNA profiling reveals that miR-148a and miR-20b control human mesenchymal stem cell phenotype via EPAS1. Physiol Genomics. 43:77–86. doi: 10.1152/physiolgenomics.00077.2010. [DOI] [PubMed] [Google Scholar]
- 67.Chang SJ, Weng SL, Hsieh JY, Wang TY, Chang MD, Wang HW. MicroRNA-34a modulates genes involved in cellular motility and oxidative phosphorylation in neural precursors derived from human umbilical cord mesenchymal stem cells. BMC Med Genomics. 4:65. doi: 10.1186/1755-8794-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao J, Yang T, Han J, Yan K, Qiu X, Zhou Y, Fan Q, Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem. 112:1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- 69.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, Ho AD. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem. 2009;108:216–224. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soci UP, Fernandes T, Hashimoto NY, Mota GF, Amadeu MA, Rosa KT, Irigoyen MC, Phillips MI, Oliveira EM. MicroRNAs 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiol Genomics. 43:665–673. doi: 10.1152/physiolgenomics.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. 108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han J, Yang T, Gao J, Wu J, Qiu X, Fan Q, Ma B. Specific microRNA expression during chondrogenesis of human mesenchymal stem cells. Int J Mol Med. 25:377–384. doi: 10.3892/ijmm_00000355. [DOI] [PubMed] [Google Scholar]
- 75.Zou Z, Zhang Y, Hao L, Wang F, Liu D, Su Y, Sun H. More insight into mesenchymal stem cells and their effects inside the body. Expert Opin Biol Ther. 10:215–230. doi: 10.1517/14712590903456011. [DOI] [PubMed] [Google Scholar]
- 76.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 38:215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim YJ, Hwang SH, Lee SY, Shin KK, Cho HH, Bae YC, Jung JS. miR-486-5p Induces Replicative Senescence of Human Adipose Tissue-Derived Mesenchymal Stem Cells and Its Expression Is Controlled by High Glucose. Stem Cells Dev. doi: 10.1089/scd.2011.0429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.