Abstract
Background
The clinical response to inhaled corticosteroids (ICS) is associated with single nucleotide polymorphisms (SNPs) in various genes. This study aimed to relate variations in genes in the steroid pathway and asthma susceptibility genes to exacerbations in children and young adults treated with ICS.
Methods
We performed a meta-analysis of three cohort studies: PACMAN (n=357, age: 4-12 years, the Netherlands), BREATHE (n=820, age: 3-22 years, UK) and PAGES (n=391, age: 2-16 years, UK). Seventeen genes were selected based on a role in the glucocorticoid signaling pathway or a reported association with asthma. Two outcome parameters were used to reflect exacerbations: hospital visits and oral corticosteroid (OCS) use in the previous year. The most significant associations were tested in three independent validation cohorts; the CAMP (clinical trial, n=172, age:5-12 years, USA), GALA II (n=745, age:8-21, USA) and PASS cohorts (n=391, age:5-18, UK) to test the robustness of the findings. Finally, all results were meta-analyzed.
Results
Two SNPs in ST13 (rs138335 and rs138337), but not in the other genes, were associated at a nominal level with an increased risk of exacerbations in asthmatics using ICS in the three cohorts studied. In a meta-analysis of all six studies, ST13 rs138335 remained associated with an increased risk of asthma-related hospital visits and OCS use in the previous year,; OR=1.22 (p=0.013) and OR=1.22 (p=0.0017) respectively.
Conclusion and clinical relevance
A novel susceptibility gene, ST13, coding for a co-chaperone of the glucocorticoid receptor, is associated with exacerbations in asthmatic children and young adults despite their ICS use. Genetic variation in the glucocorticoid signaling pathway may contribute to the interindividual variability in clinical response to ICS treatment in children and young adults.
Keywords: Childhood asthma, corticosteroids, exacerbations, pharmacogenomics, ST13
Introduction
Inhaled corticosteroids (ICS) are considered first line therapy for reducing airway inflammation, improving lung function, and controlling asthma stability in patients with persistent asthma [1,2]. While most asthmatic patients have a beneficial response to inhaled corticosteroid therapy, approximately 10% of the patients suffer from severe symptoms despite regular use of corticosteroids [3], and almost half of the costs of asthma management arises from unscheduled health care visits due to exacerbations [4]. Heterogeneity in treatment response may partly be due to genetic variation [5]. An example of genetic variation in the FCER2 gene contributing to exacerbations despite ICS treatment has been published previously [6,7].
Corticosteroids are thought to exert their anti-inflammatory effects primarily by binding to a ubiquitously expressed glucocorticoid receptor (GR) in the cytoplasm [8]. In the absence of glucocorticoids the receptor is predominantly sequestered in the cytoplasm in a multi-protein chaperone complex. Various chaperones and co-chaperones have been described to be involved in the stabilization and maturation of the receptor [9]. Upon binding of glucocorticoids to receptor, the complex translocates to the nucleus where it can block gene expression of a wide range of pro-inflammatory genes and promote the expression of anti-inflammatory genes. To date, there have been few studies addressing variations in corticosteroid receptor complex genes and steroid treatment response in patients with asthma [10,11].
We hypothesized that susceptibility genes might also be associated with an increased risk of exacerbations despite steroid treatment, due to a potential link with exacerbation-prone asthma phenotypes. In the present study we aim to relate genetic variations in genes in the steroid pathway and asthma susceptibility genes to asthma exacerbations despite ICS treatment.
Methods
Study population
Tag SNPs in 17 candidate genes were studied in three independent North-European cohorts of steroid-treated asthmatic children and adolescents: 1) the Pharmacogenetics of Asthma Medication in Children: Medication with Anti-inflammatory effects (PACMAN) cohort study, 2) the BREATHE study and, 3) the Paediatric Asthma Gene Environment Study (PAGES). For the current analyses we excluded participants of non-Northern European origin.
PACMAN
The PACMAN study is an observational cohort study of children (age: 4-12 years) with a reported (regular) use of asthma medication through community pharmacies in the Netherlands. Details of the study protocol have been described elsewhere [12]. We analyzed the PACMAN data obtained between 2009 and 2012. Data were collected with the help of pharmacists belonging to the Utrecht Pharmacy Practice Network for Education and Research (UPPER), and the work was conducted in compliance with the requirements of the IRB of the Department of Pharmacoepidemiology and Clinical Pharmacology, Utrecht University. A detailed history of the subjects is obtained, including information on asthma symptoms, exacerbations and medication use over the preceding 12 months during a study visit in the community pharmacies. Saliva samples are collected for DNA extraction (Oragene DNA Self Collection kit, DNA Genotek, Inc., Ontaria, Canada). The Medical Ethics Committee of the University Medical Centre Utrecht has approved the PACMAN study.
BREATHE
The BREATHE study includes children and young adults (age: 3-22 years) with physician-diagnosed asthma through primary or secondary clinics in either Tayside or Dumfries (Scotland, United Kingdom) [13,14]. We analyzed the BREATHE data obtained between 2004 and 2006. At the asthma clinic a detailed history was obtained, including information on symptoms, treatment and asthma exacerbations over the preceding 6 months. Mouthwash samples were collected and DNA was isolated using Qiagen DNAeasy 96 kits (Qiagen GmbH, Hilden, Germany. The Tayside Committee on Medical Research Ethics has approved the BREATHE study.
PAGES
The PAGES study recruited children and adolescents (age: 2-16 years) with physician-diagnosed asthma through 15 secondary care asthma clinics across Scotland from 2008 to 2011. Details of the study protocol of the PAGES have been described elsewhere [15]. Briefly, a detailed history was obtained including information on symptoms, treatment and exacerbations over the preceding 6 months. Saliva samples were collected for DNA extraction (Oragene DNA Self Collection kit, DNA Genotek, Inc., Ontaria, Canada). The Plymouth and Cornwall Research Ethics Committee has approved the PAGES study.
Validation cohorts
In order to test the robustness of our findings, we assessed the identified significant associations of the first meta-analysis in three additional independent populations: CAMP, the PASS cohort and GALA II and meta-analyzed the results.
CAMP trial
We studied 172 non-Hispanic white corticosteroid-treated children with asthma included in CAMP (USA). CAMP is a multi-center trial that randomized 1,041 children with mild-to-moderate asthma aged 5 to 12 years to the ICS budesonide, nedocromil, or placebo twice daily. The participants were followed for a mean of 4.3 years and follow-up visits took place at 2 and 4 months after randomization and every 4 months thereafter. The design of the study has been described previously [16]. We restricted our analysis to the non-Hispanic white subjects randomized to budesonide with available genotyping data (n=172).
Pharmacogenetics of Adrenal Suppression (PASS) cohort
PASS is a multicenter study of children with asthma (age 5-18 years), treated with corticosteroids, who required assessment of adrenal function with a Low Dose Short Synacthen Test (LDSST). Participants were recruited from November 2008 to September 2011 from 25 sites in the UK. Eligibility criteria were as follows: treatment with ICS >6 months; diagnosis of asthma; under care of a pediatrician experienced in the treatment of asthma; clinical concern about adrenal suppression sufficient to warrant a LDSST. Study participants were recruited either prospectively (if LDSST not yet undertaken) or retrospectively (if LDSST already undertaken). PASS received full ethical approval from Liverpool Paediatric Research Ethics Committee.
Genes-Environment and Admixture in Latino Americans (GALA II) study
The GALA II study is an ongoing multi-center study of Latino children and young adults with and without asthma, as described elsewhere [17]. Subjects were eligible if they were 8-21 years of age, self-identified all four grandparents as Latino, and had <10 pack-years of smoking history. Asthma was defined based on physician diagnosis and report of symptoms and medication use within the last 2 years. For this study we only analyzed asthmatic children with a reported use of SABA and ICS in the past 12 months. Patients included in this study were recruited from urban study centers across the mainland United States and Puerto Rico from 2008 to 2011. All patients completed a questionnaire with questions regarding their medical, asthma, medication use, allergic, social, environmental, and demographic histories. In addition, all participants provided blood for genetic analysis. Local institutional review boards approved the studies, and all subjects and legal guardians provided written informed assent/consent.
Definition of ICS use
Pharmacological management of asthma was categorized based on the British Thoracic Society (BTS) guidelines [2]: step 0: no use of inhaled albuterol on demand in the past month, step 1: inhaled short-acting beta-2 agonists (SABA) as needed, step 2: step 1 plus regular ICS, step 3: step 2 plus regular long-acting inhaled beta-2 agonists (LABA) and, step 4: step 3 plus oral leukotriene receptor antagonists. For the present study we selected children and young adults on BTS treatment steps 2, 3 and 4.
SNP selection and genotyping
Ten genes were selected based on their involvement in the glucocorticoid (GC) receptor complex (NR3C1, HSPCA, HSPA4, FKBP4, ST13), GC transport (SERPINA6) or GC-mediated signalling (CREBBP, TBP, NCOA3, SMAD3). In addition, seven genes were selected based on a previously reported association with asthma susceptibility, severity or asthma medication response (ARG1, 17q21 locus, IL2RB IL18R1, PDE4D, HLA-DQ, BCL2) [18-20]. We selected 50 tag SNPs. SNPs were included if the MAF > 0.2. Tag SNPs were selected using Tagger (http://www.broadinstitute.org/mpg/tagger/server.html) with a gene coverage threshold of 90%. Previously described SNPs in the genes of interest were also selected. Genotyping was performed using the Sequenom Mass Array platform (Sequenom, San Diego, California, USA). Genotype calls of all DNA samples and SNPs were examined for quality. Samples that consistently failed genotyping (≥ 20% of the SNPs) were excluded for further analyses. Subsequently, SNPs with a call rate < 95% were excluded, as well as SNPs not in Hardy-Weinberg equilibrium. A total of 38 SNPs (78%) in twelve genes passed this quality control (see Supplementary Table 1 for selected genes and SNPs). The following genes did not pass quality control and were excluded from further analyses: HSPCA, HSPA4, IL18R1, HLA-DQ and BCL-2. Illumina Infinium II 550 K SNP Chips and 610 Quad Chip (Illumina, Inc, San Diego, California) were used for genotyping in the CAMP study. SNPs of interest for replication were imputed based on 1000 Genomes. GALA II subjects were genotyped using the Axiom® LAT1 array (World Array 4, Affymetrix, Santa Clara, CA) as described elsewhere [21]. Imputed data was obtained using the genotyped SNPs, first phasing the data using SHAPE-IT [22] followed by imputation using IMPUTE2 [23] considering all populations from the 1000 Genomes Project Phase I v3 as a reference [24]. The 2 SNPs selected SNPs for the current analyses were accurately imputed (info score of 0.96 and 0.99 for rs138335 and rs138337, respectively). In the PASS cohort, DNA samples of the participants were shipped to ARK-Genomics (The Roslin Institute, University of Edinburgh) for genome-wide genotyping on the Illumina Human OmniExpressExome-8 v1.0 chip (951,117 SNPs). After sample and SNP quality control measures, genotype data were phased using the software SHAPEIT v2.r644. Imputation of SNPs was then performed using IMPUTE v2.3.0. Statistical analyses were undertaken using PLINK v1.07 and/or SNPtest v2.4.
Definition of outcome
As indicators for asthma exacerbations we studied: 1) asthma-related hospital visits and, 2) course(s) of oral corticosteroid (OCS) use reported by parent or child. The following outcome definitions as a measure for severe exacerbations were used:
-
asthma-related hospital visits reported by the parent of a child:
-
■
BREATHE, PAGES, PASS: asthma-related hospitalization in the past 6 months
-
■
PACMAN, GALA-II: asthma-related ED visits in the past 12 months
-
■
CAMP: asthma-related ED visits and hospitalizations in the first 12 months of the trial.
-
■
-
burst(s) of OCS reported by the parent or child:
-
■
BREATHE, PAGES, PASS: in the past 6 months
-
■
PACMAN, GALA-II: in the past 12 months
CAMP: in the first 12 months of the trial
-
■
Statistical analysis
Logistic regression analysis was used to study the association between the SNPs and risk of exacerbations (OCS use or asthma-related hospital visits). Odds ratios (OR), 95% confidence intervals (CI) and p-values were calculated per study. The model was adjusted for age, gender and BTS treatment step. An additive genetic model was assumed. ORs were meta-analyzed assuming random effects with the inverse variance weighing method. I2 was used to quantify between-study heterogeneity [25]. The Bonferroni-corrected p-value was set at p: 0.0007 (0.05/76). Statistical analysis was carried out using IBM SPSS 19.0 for Windows (SPSS, Inc, Chicago, Ill, USA) and PLINK [26]. Forest plots were made with R and the ‘meta’ package [27].
Functional annotation of associated SNPs
Functional annotation of associated SNPs was carried out querying the Encyclopedia of DNA Elements (ENCODE) data with the online software HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php)[28]. Additional search for evidence of associated loci being expression quantitative trait loci (eQTLs) was performed with using the Geuvadis Data Browser (http://www.ebi.ac.uk/Tools/geuvadis-das)[29].
Results
Characteristics of the study populations
Data were available for 820 children and young adults of the BREATHE cohort, 391 children and adolescents of PAGES and 357 children of the PACMAN cohort (Table 1). Most patients were on BTS treatment step 2 (as-needed short-acting beta-agonist use combined with regular low dose ICS). Compared to the other studies, the participants in the PACMAN cohort reported the lowest rates of asthma-related hospital visits (6.2%) and OCS usage (6.2%) in the past year.
Table 1.
Baseline characteristics study population in the discovery phase
| BREATHE (n=820) | PAGES (n=391) | PACMAN (n=357) | |
|---|---|---|---|
| Child characteristics | |||
| Age, range (yrs) | 9.8 (2-22) | 9.0 (2-16) | 8.7 (4-13)± |
| Male gender, % | 61.2 | 55.8 | 61.1 |
| Asthma exacerbations in preceding 12 months / 6 months | |||
| Asthma-related ED visit/hospital admission*, % | 19.0 (156/819)§ | 15.5 | 6.2 (22/356)§ |
| Oral steroid use*, % | 31.6 (259/819)§ | 43.2 | 6.2 |
| BTS treatment step | |||
| 2, % | 65.9 | 48.8 | 71.7 |
| 3, % | 18.3 | 42.2 | 23.0 |
| 4, % | 15.9 | 9.0 | 5.3 |
BTS, British Thoracic Society,
PACMAN cohort: preceding 12 months, BREATHE/PAGES: preceding 6 months.
data not available for all individuals; (number of individuals / number of individuals with data available). For BREATHE, the individual with missing hospital data is different from the individual with missing OCS data.
Children within the PACMAN cohort were selected between the age of 4-12. However, the child might have been 13 at the moment of the study visit.
Furthermore, data from three additional studies were available for the replication phase; 172 non-Hispanic white children of the CAMP trial, 391 children of the PASS cohort and 745 Latino children and young adults of the GALA II study (Table 2).
Table 2.
Baseline characteristics study population in the replication phase
| CAMP (n=172) | PASS (n=391) | GALA II (n=745) | |
|---|---|---|---|
| Child characteristics | |||
| Age, range (yrs) | 8.8 (5-13)# | 11.1 (5-18) | 12.1 (8-21) |
| Male gender, % | 55.2 | 55.8 | 56.8 |
| Asthma exacerbations in preceding 12 months / 6 months | |||
| Asthma-related ED visit/hospital admission*, % | 13.4 | 75.4 | 42.4 (313/739) |
| Oral steroid use*, % | 47.1 | 51.9 | 41.6 (310/745) |
| BTS treatment step | |||
| 2, % | ¶ | 7.7 | 41.1 |
| 3, % | - | 33.0 | 43.6 |
| 4, % | - | 58.8 | 15.3 |
CAMP is Randomized Clinical Trial of mild-to moderate asthmatics. All children were on 200 μg of budesonide (ICS) plus SABA as needed.
Prospective trial; children were 5-13 years at the start of the trial.
Discovery phase: Associations with severe exacerbations in BREATHE, PAGES and PACMAN
In a meta-analysis of the three North-European cohorts BREATHE, PAGES and PACMAN, we found two out of the 38 SNPs to be associated with an altered risk of severe exacerbations as defined by asthma-related hospital visits. ST13 SNP rs138335 increased the risk of asthma-related hospital visits (OR=1.35 per G allele; 95%CI: 1.07-1.69, p=0.01) (Figure 1). Rs138337 in the same gene, had a comparable effect on the risk of asthma-related hospital visits (OR: 1.36 per G allele, 95%CI: 1.11-1.66, p=0.003) (Figure 2). In addition, rs138335 was also associated with an increased risk of OCS use (OR: 1.33 per G allele; 95%CI: 1.11-1.60, p=0.002) (Figure 3). Supplementary tables 2 and 3 show the summary effect estimates of all investigated SNPs.
Figure 1. Forest plot for the association between ST13 rs138335 and asthma-related hospital visits.
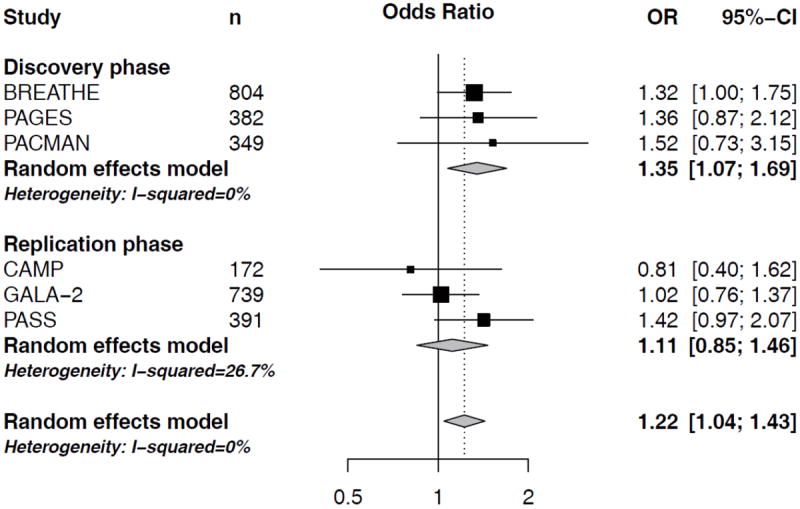
Odds ratios (OR) and corresponding 95%CI per increase in G-allele, controlling for age, sex and treatment step.
Figure 2. Forest plot for the association between ST13 rs138337 and asthma-related hospital visits.
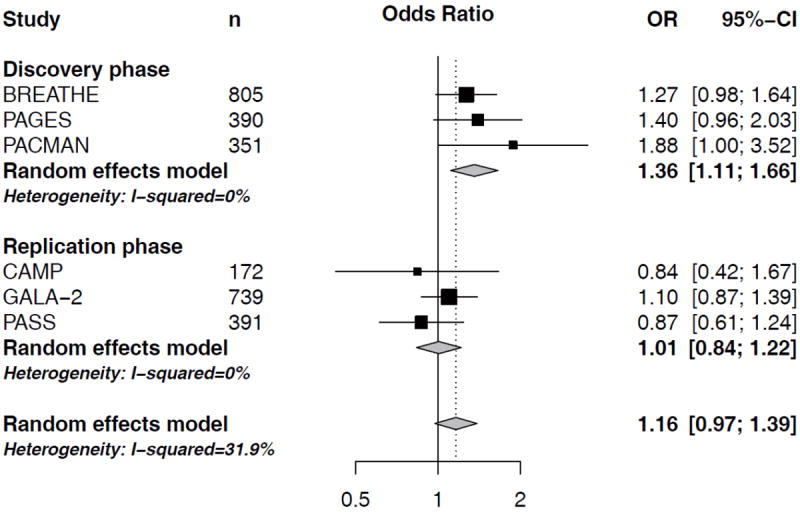
Odds ratios (OR) and corresponding 95%CI per increase in G-allele, controlling for age, sex and treatment step.
Figure 3. Forest plot for the association between ST13 rs138335 and OCS usage.
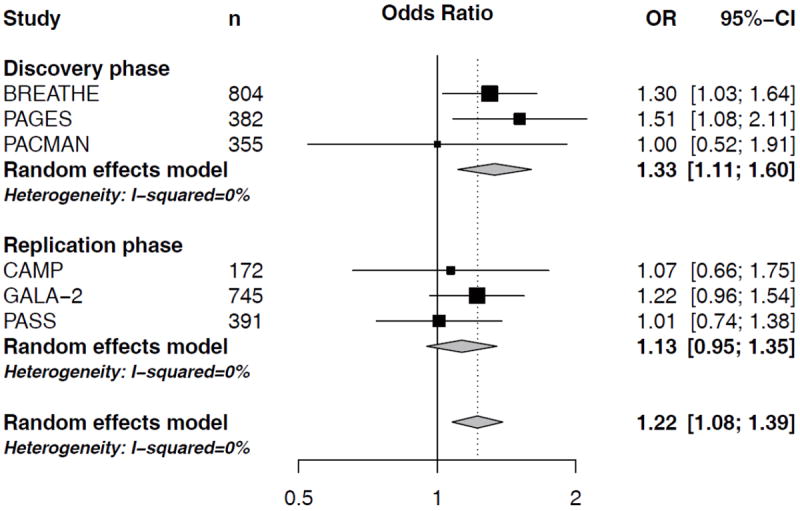
Odds ratios (OR) and corresponding 95%CI per increase in G-allele, controlling for age, sex and treatment step.
Replication phase: ST13 in the CAMP, GALA II and PASS cohorts
In order to assess the robustness of findings we studied rs138337 and rs138335 in three additional independent study populations; the CAMP study (n=172 non-Hispanic white asthmatic children), the PASS cohort (n=391 North-European asthmatic children) and the GALA II study (n=745 Latino asthmatic children). In a meta-analysis of the three cohorts, none of the ST13 SNPs was significantly associated with severe exacerbations (Figure 1-3). When investigating the study populations independently, we observed a trend (p=0.06) in the North-European PASS cohort suggesting that carrying a G-allele at rs138335 increased the risk of asthma-related hospital visits in this study population (OR: 1.42, 95%CI: 0.97-2.07; Figure 1), whereas the other 2 cohorts did not significantly contribute.
Meta-analysis of the six study populations
In the meta-analysis of all six study populations, ST13 rs138335 remains associated with asthma-related hospital-visits (OR per increase in G-allele: 1.22, 95%CI: 1.04-1.43, p=0.013) and OCS usage (OR per increase in G-allele: 1.22, 95%CI: 1.08-1.39, p=0.0017). The effect estimates in the different cohorts largely pointed in the same direction for both outcomes, yet these associations did not pass the Bonferroni-corrected significance threshold of 0.0007.
Functional annotation of associated SNPs
The associated SNPs rs138337 and rs138335 are eQTLs in lymphoblastoid cell lines from Europeans (p=5.8×10-70 and p=1.9×10-36, respectively). In addition, the SNP rs138335 is in strong linkage disequilibrium (LD, r2=0.95) with another SNP (rs138349) that is located in a promoter histone mark, an enhancer histone mark, a DNase I hypersensitive site, and acts as a binding site for an enhancer binding protein and transcription factors. Furthermore, the SNP rs138335 is in high LD (r2=0.86) with the SNP rs2899341, which is located in an enhancer histone mark and in a DNase I hypersensitive site.
Discussion
In a meta-analysis of three independent North-European studies we identified ST13 as a novel risk gene for the occurrence of asthma exacerbations despite inhaled corticosteroid treatment in asthmatic children and young adults. For rs138335 the risk of exacerbations was increased with each substitution of the minor allele for the major allele variant. For rs138337, oppositely, the minor allele variant was found to be associated with an increased risk of exacerbations. The two SNPs were in moderate LD (r2=0.47) in our study. None of the other investigated genes could be linked to an increased risk of severe exacerbations.
SNPs rs138335 and rs138337 both lie in the non-coding intronic regions of the ST13 gene, but our in silico functional evaluation revealed a functional role for these two SNPs as eQTLs and also for SNPs in high LD with them. ST13 encodes a co-chaperone protein (Hsp70 interacting protein; hip) of the steroid-receptor complex and is involved in the functional maturation of the corticosteroid receptor, but the mechanism by which it does so remains to be elucidated [30]. STIP1 (coding for another co-chaperone protein in the GR receptor complex, namely Hsp70/Hsp90-organizing protein: hop) has previously been associated with lung function and lung function improvement in 382 asthmatic patients treated with ICS [10]. At the time of SNP selection, STIP1 was not included in our study. Hip (encoded by ST13) and hop (encoded by STIP1) are thought to function in a cooperative manner in GR maturation [30], building evidence that alterations in the expression or folding of these co-chaperones may influence the binding of corticosteroids to the receptor or downstream signaling and therefore, ICS responsiveness. Functional studies are necessary to support our hypothesis.
A number of limitations need to be noted regarding the present study. Two SNPs in ST13 were associated with both outcomes of exacerbations in the meta-analysis of all six cohorts, but did not pass the Bonferroni-corrected significance threshold. Therefore, we cannot exclude that our findings are false-positives. Even though we were able to analyze a large study population (including 2876 asthmatic children and young adults), a post-hoc power analysis showed we were underpowered to identify a significant association with an OR<1.5 for asthma-related hospital visits and OR<1.4 for OCS use. This underlines the need for large-scale international collaboration in this field [31].
The populations we studied varied in age and severity of asthma symptoms, probably due to the design of the studies. The PACMAN population is recruited in community pharmacies, whereby most participants had well-controlled symptoms [32], while patients in PAGES, BREATHE and CAMP were recruited through primary and secondary care. PASS participants were recruited through secondary care based on clinical concern about adrenal suppression, while participants in GALA II were recruited using a combination of community and clinic-based recruitment. In addition, differences in health system and prescription behavior between the different countries might also play a role [33]. Notwithstanding these differences, statistical heterogeneity (I2) was limited for ST13 in the meta-analysis.
Our study was also limited due to the selection of tagging SNPs with a MAF ≥ 0.20. Due to the sample size, we could not investigate rare variants, which might have had larger effects. Furthermore, the incorporation of common variants with smaller effects in clinical risk models might be valuable for a larger group of the asthma patient population.
In summary, variations in a novel risk gene ST13 seem to be associated with an increased risk of severe exacerbations in children and young adults despite their use of ICS. Although the effect sizes are modest, these results may provide insights into the biological mechanisms that underlie severe exacerbations in asthmatic patients treated with steroids. Heterogeneity in corticosteroid response is probably caused by a complex interaction of genetic and environmental factors. Including ST13 risk status in a multidimensional model with other genetic and non-genetic risk factors (e.g: exposure to tobacco smoke [34] or vitamin D levels [35]) may reveal more precisely interindividual ICS responses.
Supplementary Material
Acknowledgments
The authors would like to thank the children, young adults and parents of the PACMAN cohort study, BREATHE study, PAGES, CAMP study, PASS cohort and GALA II study for their participation.
Sources of funding:
Susanne J.H. Vijverberg and Ellen S. Koster have been paid by an unrestricted grant from GlaxoSmithKline (GSK). Leo Koenderman, Steve Turner, Roger Tavendale, Colin Palmer, Kelan Tantisira, and Sze Man Tse have no financial relationship with a commercial entity that has an interest in the subject of this manuscript. Jan A. M. Raaijmakers is part time professor at the Utrecht University and vice president external scientific collaborations for GSK in Europe, and holds stock in GSK. Anke-Hilse Maitland-van der Zee received an unrestricted grant from GSK. Furthermore, the department of Pharmacoepidemiology and Clinical Pharmacology, Utrecht Institute for Pharmaceutical Sciences, employing authors Susanne J.H. Vijverberg, Ellen S. Koster, Maarten Leusink, Jan A.M. Raaijmakers, and Anke-Hilse Maitland-van der Zee, has received unrestricted research funding from the Netherlands Organisation for Health Research and Development (ZonMW), the Dutch Health Care Insurance Board (CVZ), the Royal Dutch Pharmacists Association (KNMP), the private-public funded Top Institute Pharma (www.tipharma.nl, includes co-funding from universities, government, and industry), the EU Innovative Medicines Initiative (IMI), EU 7th Framework Program (FP7), the Dutch Medicines Evaluation Board, the Dutch Ministry of Health and industry (including GlaxoSmithKline (GSK), Pfizer, and others). This study was part of a short-term research fellowship (Ellen S. Koster) funded by a grant from the Dutch Asthma Fund (Astma Fonds) and a short-term research fellowship (Susanne J.H. Vijverberg) funded by a grant from the Dutch Ter Meulen Foundation. Gerard H. Koppelman was supported by a grant from the Dutch Ter Meulen Foundation and reports to have received grants from the Stichting Astma Bestrijding and the Netherlands Asthma Foundation. AstraZeneca, Boehringer Ingelheim, Chiesi, GSK, Nycomed and Teva have paid money to the University of Groningen for consultancies of Dirkje S. Postma, the University of Groningen also received funding of TIPharma (Dutch ministry of health and sciences) in conjunction with GSK and Nycomed, and unrestricted grants from AstraZeneca and Chiesi, as well as from EU 7th Framework Program (FP7), and the Royal academy of Arts and sciences in The Netherlands for research performed by Dirkje S. Postma. Somnath Mukhopadhyay received an unrestricted grant from MSD (UK) and has received consultative fees from Thermofisher (UK). Kelan Tantisira is supported by NIH grants R01 HL092917, R01 NR013391, and U01 HL065899. The Chief Scientist Officer for Scotland funded the PAGES study. The BREATHE study of asthma in children is funded by Scottish Enterprise Tayside, the Gannochy Trust, the Perth and Kinross City Council and Brighton and Sussex Medical School. PASS was funded by the UK department of health through the “NHS Chair of Pharmacogenomics” (awarded to Professor M Pirmohamed) The contribution from GALA II to this work was supported by grants from National Institutes of Health to E.G.B.: the National Heart, Lung and Blood Institute (HL088133, HL078885, HL004464, HL104608 and HL117004); the National Institute of Environmental Health Sciences (ES015794); the National Institute on Minority Health and Health Disparities (MD006902); the National Institute of General Medical Sciences (GM007546). E.G.B was also funded by the American Asthma Foundation, the RWJF Amos Medical Faculty Development Award, the Sandler Foundation and the Flight Attendant Medical Research Institute. MPY was supported by a postdoctoral fellowship from Fundación Ramón Areces (www.fundacionareces.es).
Abbreviations used
- BTS
British Thoracic Society
- CAMP
Childhood Asthma Management Program
- ED
Emergency Department
- GALA II
Genes-environments & Admixture in Latino Americans
- GC
glucocorticoid
- GR
glucocorticoid receptor
- ICS
Inhaled Corticosteroids
- LABA
long-acting beta-2 agonist
- LD
linkage disequilibrium
- LTRA
leukotriene receptor antagonist
- OCS
Oral Corticosteroids
- PACMAN
Pharmacogenetics of Asthma Medication in Children: Medication with Anti-inflammatory effects
- PAGES
Paediatric Asthma Gene Environment Study
- SABA
short-acting beta agonist
- SNP
Single Nucleotide Polymorphism
- UPPER
Utrecht Pharmacy Practice Network for Education and Research
References
- 1.Global Initiative for Asthma (GINA) Global strategy for asthma management and prevention. Bethesa/Maryland: Updated december 2012. www.ginasthma.org. [Google Scholar]
- 2.Levy ML, Thomas M, Small I, Pearce L, Pinnock H, Stephenson P. Summary of the 2008 BTS/SIGN British Guideline on the management of asthma. Prim Care Respir J. 2009;18(Suppl 1):S1–16. doi: 10.3132/pcrj.2008.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung KF, Godard P, Adelroth E, Ayres J, Barnes N, Barnes P, Bel E, Burney P, Chanez P, Connett G, Corrigan C, de Blic J, Fabbri L, Holgate ST, Ind P, Joos G, Kerstjens H, Leuenberger P, Lofdahl CG, McKenzie S, Magnussen H, Postma D, Saetta M, Salmeron S, Sterk P. Difficult/therapy-resistant asthma: the need for an integrated approach to define clinical phenotypes, evaluate risk factors, understand pathophysiology and find novel therapies. ERS Task Force on Difficult/Therapy-Resistant Asthma European Respiratory Society. Eur Respir J. 1999;13:1198–208. doi: 10.1034/j.1399-3003.1999.13e43.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams AE, Lloyd AC, Watson L, Rabe KF. Cost of scheduled and unscheduled asthma management in seven European Union countries. Eur Respir Rev. 2006;15:4–9. [Google Scholar]
- 5.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–70. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 6.Tantisira KG, Silverman ES, Mariani TJ, Xu J, Richter BG, Klanderman BJ, Litonjua AA, Lazarus R, Rosenwasser LJ, Fuhlbrigge AL, Weiss ST. FCER2: A pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007;120:1285–91. doi: 10.1016/j.jaci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Koster ES, Maitland-van der Zee AH, Tavendale R, Mukhopadhyay S, Vijverberg SJH, Raaijmakers JAM, Palmer CNA. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy. 2011;66:1546–52. doi: 10.1111/j.1398-9995.2011.02701.x. [DOI] [PubMed] [Google Scholar]
- 8.Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006;117:522–43. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Vandevyver S, Dejager L, Libert C. On the Trail of the Glucocorticoid Receptor: Into the Nucleus and Back. Traffic. 2012;13:364–74. doi: 10.1111/j.1600-0854.2011.01288.x. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins GA, Lazarus R, Smith RS, Tantisira KG, Meyers DA, Peters SP, Weiss ST, Bleecker ER. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123:1376–1383.e7. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczepankiewicz A, Breborowicz A, Sobkowiak P, Popiel A. No association of glucocorticoid receptor polymorphisms with asthma and response to glucocorticoids. Adv Med Sci. 2008;53:245–50. doi: 10.2478/v10039-008-0042-8. [DOI] [PubMed] [Google Scholar]
- 12.Koster ES, Raaijmakers JAM, Koppelman GH, Postma DS, van der Ent CK, Koenderman L, Bracke M, Maitland-van der Zee AH. Pharmacogenetics of anti-inflammatory treatment in children with asthma: rationale and design of the PACMAN cohort. Pharmacogenomics. 2009;10:1351–61. doi: 10.2217/pgs.09.79. [DOI] [PubMed] [Google Scholar]
- 13.Palmer CNA, Lipworth BJ, Lee S, Ismail T, Macgregor DF, Mukhopadhyay S. Arginine-16 {beta}2 adrenoceptor genotype predisposes to exacerbations in young asthmatics taking regular salmeterol. Thorax. 2006;61:940–4. doi: 10.1136/thx.2006.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavendale R, Macgregor DF, Mukhopadhyay S, Palmer CNA. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–3. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Turner SW, Ayres JG, Macfarlane TV, Mehta A, Mehta G, Palmer CN, Cunningham S, Adams T, Aniruddhan K, Bell C, Corrigan D, Cunningham J, Duncan A, Hunt G, Leece R, MacFadyen U, McCormick J, McLeish S, Mitra A, Miller D, Waxman E, Webb A, Wojcik S, Mukhopadhyay S, Macgregor D. A methodology to establish a database to study gene environment interactions for childhood asthma. BMC Med Res Methodol. 2010;10:107. doi: 10.1186/1471-2288-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Childhood Asthma Management Program Study Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 17.Nishimura KK, Galanter JM, Roth LA, Oh SS, Thakur N, Nguyen EA, et al. Early-Life Air Pollution and Asthma Risk in Minority Children. The GALA II and SAGE II Studies. Am J Respir Crit Care Med. 2013;188:309–18. doi: 10.1164/rccm.201302-0264OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan YI, Shrine NRG, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, Bush A, Chung KF, Cookson WOCM, Strachan DP, Heaney L, Al-Momani BAH, Mansur AH, Manney S, Thomson NC, Chaudhuri R, Brightling CE, Bafadhel M, Singapuri A, Niven R, Simpson A, Holloway JW, Howarth PH, Hui J, Musk AW, James AL, Brown MA, Baltic S, Ferreira MAR, Thompson PJ, Tobin MD, Sayers I, Hall IP the Australian Asthma Genetics Consortium. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–8. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 19.Litonjua AA, Lasky-Su J, Schneiter K, Tantisira KG, Lazarus R, Klanderman B, Lima JJ, Irvin CG, Peters SP, Hanrahan JP, Liggett SB, Hawkins GA, Meyers DA, Bleecker ER, Lange C, Weiss ST. ARG1 Is a Novel Bronchodilator Response Gene. Am J Respir Crit Care Med. 2008;178:688–94. doi: 10.1164/rccm.200709-1363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SA, Klanderman B, Lasky-Su J, Lazarus R, Murphy AJ, Soto-Quiros ME, Avila L, Beaty T, Mathias RA, Ruczinski I, Barnes KC, Celedon JC, Cookson WO, Gauderman WJ, Gilliland FD, Hakonarson H, Lange C, Moffatt MF, O’Connor GT, Raby BA, Silverman EK, Weiss ST. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galanter JM, Gignoux CR, Torgerson DG, Roth LA, Eng C, Oh SS, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: the Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol. 2014;134:295–305. doi: 10.1016/j.jaci.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A linear complexity phasing method for thousands of genomes. Delaneau O, Marchini J, Zagury J-F. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 23.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. URL http://www.R-project.org. [Google Scholar]
- 28.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson GM, Prapapanich V, Carrigan PE, Roberts PJ, Riggs DL, Smith DF. The Heat Shock Protein 70 Cochaperone Hip Enhances Functional Maturation of Glucocorticoid Receptor. Molecular Endocrinology. 2004;18:1620–30. doi: 10.1210/me.2004-0054. [DOI] [PubMed] [Google Scholar]
- 31.Vijverberg SJ, Raaijmakers JA, Maitland-van der Zee AH. ADRB2 Arg16 and the need for collaboration in childhood asthma pharmacogenomics. Pharmacogenomics. 2013;14:1937–1939. doi: 10.2217/pgs.13.195. [DOI] [PubMed] [Google Scholar]
- 32.Koster ES, Raaijmakers JA, Vijverberg SJ, Koenderman L, Postma DS, Koppelman GH, van der Ent CK, Maitland-van der Zee AH. Limited agreement between current and long-term asthma control in children: the PACMAN cohort study. Pediatr Allergy Immunol. 2011;22:776–83. doi: 10.1111/j.1399-3038.2011.01188.x. [DOI] [PubMed] [Google Scholar]
- 33.Westert GP, Lagoe RJ, Keskimäki I, Leyland A, Murphy M. An international study of hospital readmissions and related utilization in Europe and the USA. Health Policy. 2002;61:269–78. doi: 10.1016/s0168-8510(01)00236-6. [DOI] [PubMed] [Google Scholar]
- 34.Stapleton M, Howard-Thompson A, George C, Hoover RM, Self TH. Smoking and Asthma. J Am Board Fam Med. 2011;24:313–22. doi: 10.3122/jabfm.2011.03.100180. [DOI] [PubMed] [Google Scholar]
- 35.Gupta A, Bush A, Hawrylowicz C, Saglani S. Vitamin D and Asthma in Children. Paediatric Respir Rev. 2012;13:236–43. doi: 10.1016/j.prrv.2011.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


