Correction
In follow-up studies to this work [1], we have identified an error in a single line of code responsible for parsing BLASTZ [2] alignments that affects our previously published results for this alignment tool. This error resulted in a reduction in overall alignment coverage, with a concomitant underestimation of alignment sensitivity and overestimation of alignment specificity. As BLASTZ is an important and widely used alignment tool, we present here the revised results of our performance evaluations for BLASTZ together with previously reported results for the other alignment tools studied, which have been subsequently verified (Figures 1, 2, 3, 4). The general conclusions presented in [1] remain unchanged, although the following sections concerning BLASTZ performance must be modified in light of our recent findings.
Figure 1.
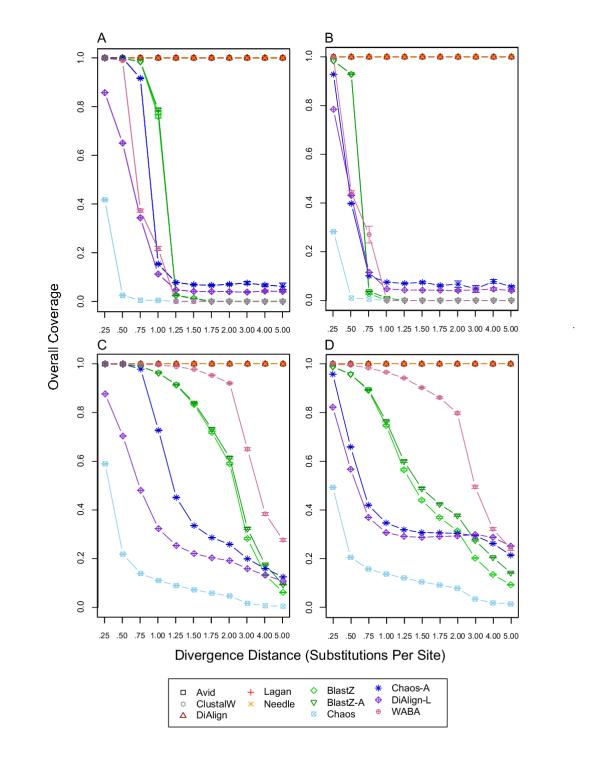
Overall alignment coverage. For each divergence distance and each tool, 1,000 replicates were used to calculate the mean and standard error of overall alignment coverage, which was defined as the fraction of ungapped, orthologous pairs of sites in the simulated alignment that were included in an alignment produced by a tool (for details see Methods in [1]). A) overall coverage without constrained blocks and without insertion/deletion evolution; B) overall coverage without constrained blocks and with insertion/deletion evolution; C) overall coverage with constrained blocks and without insertion/deletion evolution; D) overall coverage with constrained blocks and with insertion/deletion evolution.
Figure 2.
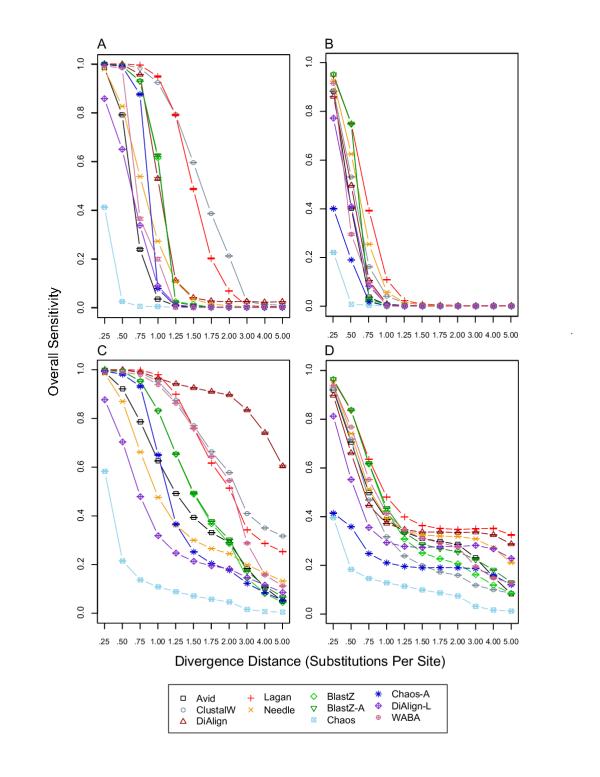
Overall alignment sensitivity. For each divergence distance and each tool, 1,000 replicates were used to calculate the mean and standard error of overall alignment sensitivity, which was defined as the fraction of ungapped, orthologous pairs of sites in the simulated alignment that were aligned correctly in an alignment produced by a tool (for details see Methods in [1]). A) overall sensitivity without constrained blocks and without insertion/deletion evolution; B) overall sensitivity without constrained blocks and with insertion/deletion evolution; C) overall sensitivity with constrained blocks and without insertion/deletion evolution; D) overall sensitivity with constrained blocks and with insertion/deletion evolution.
Figure 3.
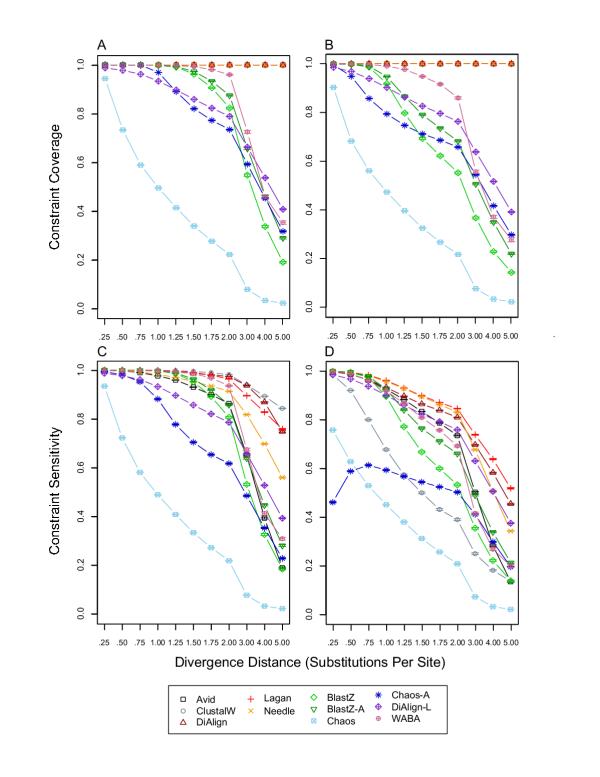
Constraint coverage and sensitivity. For each divergence distance and each tool, 1,000 replicates were used to calculate the mean and standard error of constraint coverage and constraint sensitivity, which were defined as the coverage and sensitivity within interspersed constrained blocks (for details see Methods in [1]). A) constraint coverage without insertion/deletion evolution; B) constraint coverage with insertion/deletion evolution; C) constraint sensitivity without insertion/deletion evolution; D) constraint sensitivity with insertion/deletion evolution.
Figure 4.
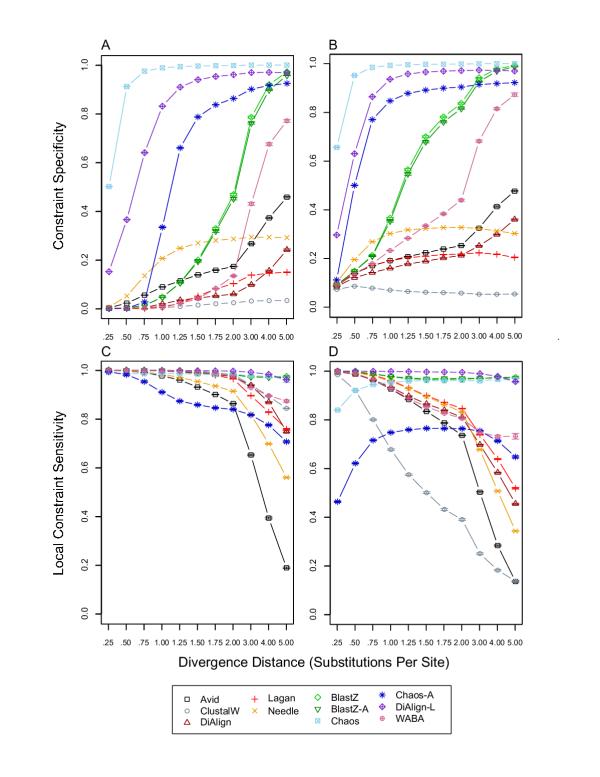
Constraint specificity and local constraint sensitivity. For each divergence distance and each tool, 1,000 replicates were used to calculate a mean and standard error of constraint specificity and local constraint sensitivity. Constraint specificity was defined as the fraction of unconstrained sites in the simulated alignment that were unaligned or gapped in an alignment produced by a tool. Local constraint sensitivity was defined as the constraint sensitivity for just the sites contained in an alignment produced by a tool (for details see Methods in [1]). A) constraint specificity without insertion/deletion evolution; B) constraint specificity with insertion/deletion evolution; C) local constraint sensitivity without insertion/deletion evolution; D) local constraint sensitivity with insertion/deletion evolution.
The true overall alignment coverage for BLASTZ with and without insertion/deletion evolution and with and without blocks of constraint is shown in Figure 1, and reveals increased overall coverage in the presence of constrained blocks for intermediate to high divergence distances (Figures 1C & 1D) relative to previous results ([1] Figure 3C & 3D). As a consequence, the true overall sensitivity for BLASTZ is increased for intermediate to high divergence distances, especially in the presence of insertion/deletion evolution and constrained blocks (Figure 2D) relative to previous results ([1] Figure 4D).
The most important revisions to [1] concern BLASTZ performance in interspersed blocks of constrained sequences (Figures 3, 4). Figure 3 shows that the true constraint coverage, and therefore constraint sensitivity, of BLASTZ is much improved relative to previous results for intermediate to high divergence distances ([1] Figure 5). Thus BLASTZ has increased constraint coverage relative to overall coverage (cp. Figures 1C &1D with 3A &3B), indicating that BLASTZ local alignments preferentially occur in constrained sequences for intermediate to high divergence distances, overturning claims on page 6 of [1] to the contrary. Likewise, the claim that BLASTZ has a "dramatic decrease in constraint sensitivity in the presence of indel evolution" on page 10 of [1] is incorrect. The increase in overall coverage, however, decreases the constraint specificity of BLASTZ for intermediate to high divergence distances (Figure 4A &4B) relative to previous results ([1] Figure 6A & 6B). This decrease in constraint specificity requires reconsideration of the use of BLASTZ local alignments as specific detectors of constrained noncoding sequences discussed page 10 of [1].
Revised performance statistics for BLASTZ are posted along with previous results at [3]. We apologize for any misconception or inconvenience this error may have caused.
Contributor Information
DA Pollard, Email: dpollard@socrates.berkeley.edu.
CM Bergman, Email: cbergman@gen.cam.ac.uk.
J Stoye, Email: stoye@techfak.uni-bielefeld.de.
SE Celniker, Email: celniker@fruitfly.org.
MB Eisen, Email: mbeisen@lbl.gov.
References
- Pollard DA, Bergman CM, Stoye J, Celniker SE, Eisen MB. Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics. 2004;5:6. doi: 10.1186/1471-2105-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–7. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlignmentBenchmarking. http://rana.lbl.gov/AlignmentBenchmarking