Summary
From an shRNA screen, we identified ClpP as a member of the mitochondrial proteome whose knockdown reduced the viability of K562 leukemic cells. Expression of this mitochondrial protease that has structural similarity to the cytoplasmic proteosome is increased in the leukemic cells from approximately half of patients with AML. Genetic or chemical inhibition of ClpP killed cells from both human AML cell lines and primary samples in which the cells showed elevated ClpP expression, but did not affect their normal counterparts. Importantly, Clpp knockout mice were viable with normal hematopoiesis. Mechanistically, we found ClpP interacts with mitochondrial respiratory chain proteins and metabolic enzymes, and knockdown of ClpP in leukemic cells inhibited oxidative phosphorylation and mitochondrial metabolism.
Introduction
The cytoplasmic/nuclear proteasome complex is a well-recognized therapeutic target and proteasome inhibitors have been approved for the treatment of hematologic malignancies (Fisher et al., 2006; O'Connor et al., 2009; Richardson et al., 2005; Vij et al., 2012). Mitochondria also possess a serine protease complex, ClpP, that is structurally similar to the cytoplasmic/nuclear proteasome (Goard and Schimmer, 2013), but little is known about the expression of this complex or the effects of its inhibition in malignant cells.
ClpP is encoded by a nuclear gene, translated in the cytoplasm and imported into the mitochondrial matrix. There, it is assembled into a tetradecamer consisting of 7 repeated symmetric rings arranged in a stable double ringed structure (Corydon et al., 1998; de Sagarra et al., 1999), each end of which is capped by an AAA+ ATPase chaperone, ClpX (Kang et al., 2005). Based on studies of its homologues in bacteria and its structural similarities to the cytoplasmic proteasome, the ClpP complex is thought to degrade damaged or misfolded proteins inside mitochondria.
Inhibition of ClpP in normal mice (Gispert et al., 2013), humans (Jenkinson et al., 2013), and C. elegans (Haynes et al., 2007; Haynes et al., 2010) has not been found to elicit much loss of viability. For example, Clpp−/− mice are viable, but display infertility, slight growth retardation, and acquired hearing loss (Gispert et al., 2013). Homozygous CLPP mutations have also been described in individuals from three human families (Jenkinson et al., 2013). Similar to the mouse phenotype, these individuals have congenital hearing loss and premature ovarian failure. In contrast, deregulating ClpP activity in certain bacteria, either through inhibition (Zeiler et al., 2012) or increased activation (Conlon et al., 2013), is cytotoxic even when they are dormant or have acquired antibiotic resistance (Conlon et al., 2013).
Recently, we (Skrtic et al., 2011) and others (Lagadinou et al., 2013) demonstrated that the leukemic cells including those with stem and progenitor activity from patients with acute myeloid leukemia (AML) have an increased mitochondrial mass and reliance on oxidative phosphorylation. We therefore initiated a study to identify members of the mitochondrial proteome whose inhibition might reduce the viability of AML cells.
Results
A genetic screen identifies ClpP as essential for the viability of leukemia cells
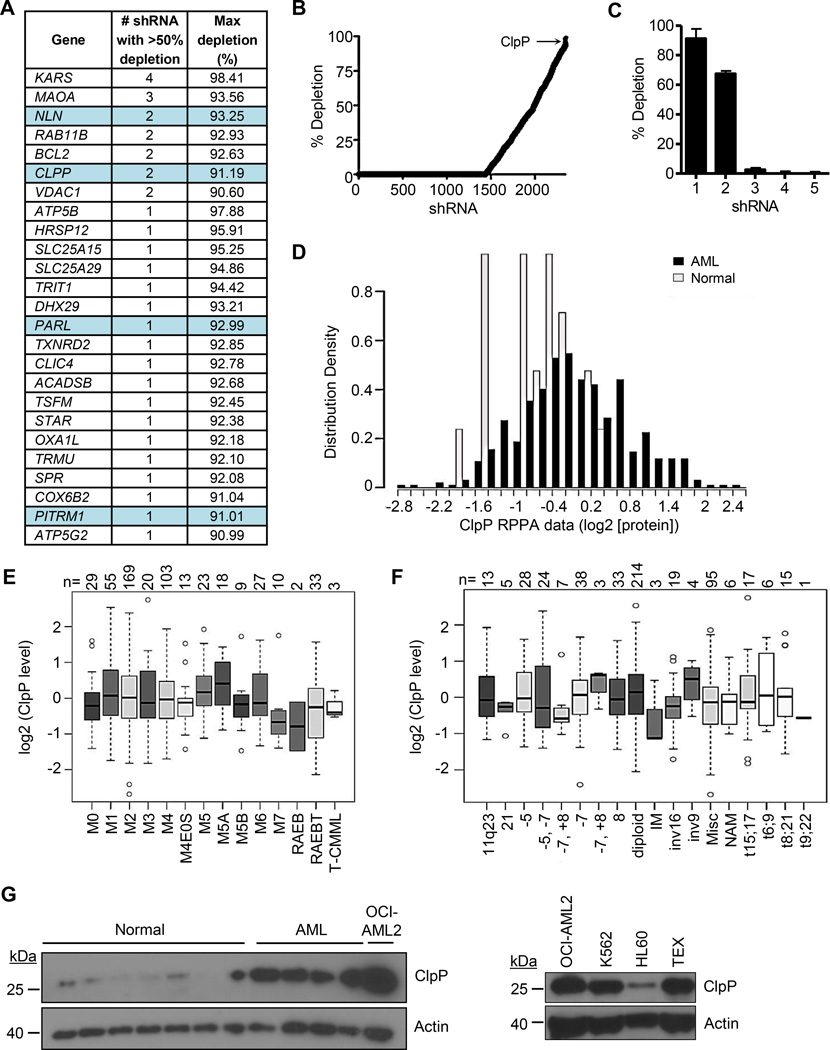
We first sought to determine whether shRNA-mediated knockdown of any of the 1300 members of the mitochondrial proteome would identify candidates that could reduce the viability of human leukemic cells using K562 cells as targets. Accordingly, we transduced K562 cells with a library of 54,021 shRNAs in bar-coded lentiviral vectors targeting 11,255 nuclear-encoded genes. Twenty one days after transduction, cells were harvested, genomic DNA isolated, and the relative abundance of shRNA sequences present in the surviving cells was determined by array analysis of the barcodes. shRNAs able to reduce the viability or growth of K562 cells were inferred to be those not represented in the final cell population (Figure S1A). shRNAs targeting BCR and ABL1 were top hits (Figure S1A–S1C), thus validating the robustness of the screen, as K562 cells are dependent on the BCR-ABL1 fusion oncoprotein for survival (Dan et al., 1998). In this screen, 2422 shRNAs targeted 496 members of the mitochondrial proteome and of the top 25 targets, 4 were mitochondrial proteases (NLN, ClpP, PARL, and PITRM1) with 2 targeting ClpP ranking in the top 1% of all hits in the screen (Figure 1A–C). In contrast, the mitochondrial matrix protease Lon did not appear as a hit in our shRNA screen and repetition of this experiment using shRNAs that produced high levels of target knockdown (both protein and mRNA) confirmed that Lon was not required for the growth and viability of TEX, OCI-AML2, or K562 leukemia cells (Figure S1D and data not shown). Thus some, but not all, mitochondrial proteases are necessary for the viability of AML cells.
Figure 1. Genetic screen identifies ClpP as essential for the viability of leukemia cells.
(A) The list of top 25 shRNAs targeting the mitochondrial proteome.
(B) The percent depletion in rank order of the 2422 shRNA targeting 496 members of the mitochondrial proteome is shown.
(C) The percent depletion of individual clones in the library targeting ClpP.
(D) Expression of ClpP measured in total cell lysates from 511 primary AML cases and 21 samples of normal CD34+ hematopoietic cells using a reverse phase protein array.
(E, F) Expression of ClpP in primary AML cases classified based on their morphologic FAB subtype (E) or cytogenetic abnormalities (F).
(G) Expression of ClpP protein in total cell lysates prepared from normal hematopoietic cells, primary AML samples, and indicated cell lines by immunoblotting.
Error bars represent standard deviation. The box represents the first and third quartiles, whiskers represent the range, the center line represents the median, and the circles outliers.
See also Figure S1 and Table S1.
ClpP is over-expressed in a subset of AML cells and patient samples
To investigate ClpP further as a therapeutic target in human AML, we measured ClpP protein expression in 511 primary AML samples and CD34+ cells from 21 healthy individuals using reverse phase protein arrays. Compared to normal CD34+ hematopoietic progenitor cells, ClpP was over-expressed in 45% of the primary AML samples (Figure 1D). Elevated ClpP was not correlated with CD34 expression based on flow cytometry, remission attainment, overall or event-free survival, or remission duration (Figure S1E–S1G and data not shown). Increased expression of ClpP also showed no association with any morphologic subtype, cytogenetic risk group, or mutational profile (Figure 1E–1F, Figure S1H–M).
Increased expression of ClpP in a subset of AML cell lines and patient samples was confirmed by immunoblotting (Figure 1G, Figure S1N). In addition, we demonstrated that CLPP mRNA was increased in a subset of AML cells compared to normal hematopoietic cells and expressed similarly in the bulk and progenitor (CD34+CD38−) populations of primary AML samples (Figure S1O). Gene expression analysis of previously characterized AML patient samples demonstrated that CLPP mRNA was similarly expressed in fractions enriched or depleted of leukemic stem cells (LSCs) defined by functional assays (p=0.45) (Figure S1P). Analysis of TCGA data indicated that CLPP expression did not correlate with known AML mutations (Table S1).
In primary AML patient samples, higher CLPP expression was associated with an increased expression of genes related to the mitochondrial unfolded protein response (mtUPR). This positive correlation was observed in both the LSC+ and LSC− fractions of primary AML cells (Figure S1Q–S1R). Thus, increased ClpP may be a marker of increased mitochondrial stress in a subset of AML patients that is downstream of multiple genetic pathways.
Analysis of the Cancer Cell Line Encyclopedia indicated that CLPP was highly expressed in AML cells, but also increased in subsets of other hematologic malignancies including multiple myeloma, various lymphomas, and CML. In addition, CLPP was highly expressed in solid tumors, such as prostate cancer and sarcomas (Figure S1S). Therefore, ClpP may also be a relevant target for malignancies beyond AML.
Genetic knockdown of ClpP reduces the growth and viability of AML cells with high ClpP expression
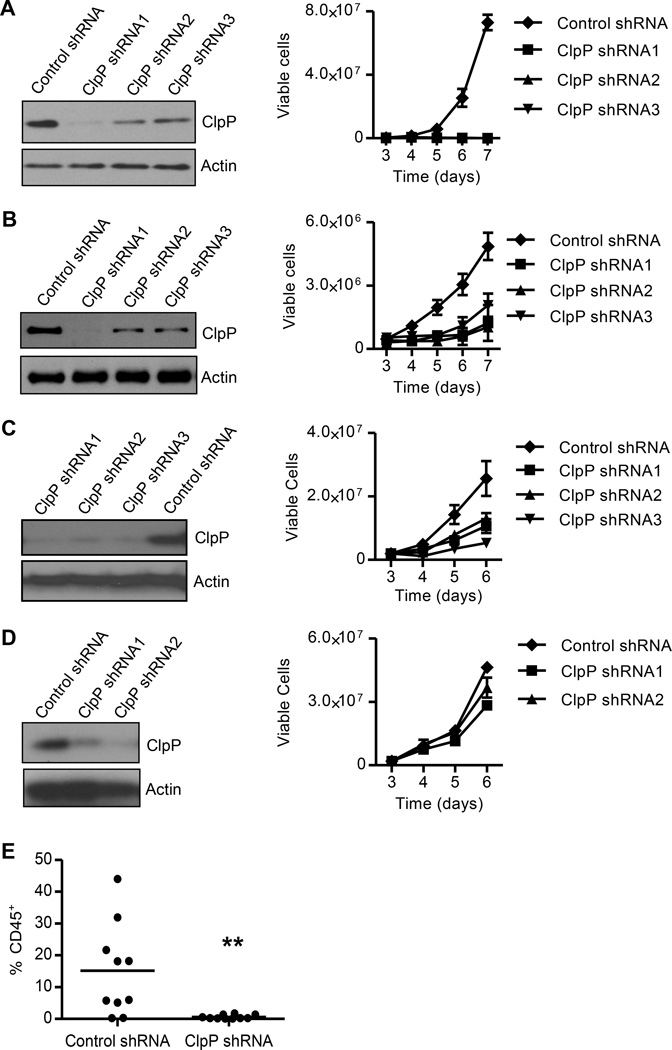
We next tested the effects of ClpP knockdown on the growth and viability of AML cells. K562, TEX, OCI-AML2, and HL60 cells transduced with lentiviral vectors expressing a control sequence or three independent shRNA sequences targeting CLPP. The shRNA sequences targeting CLPP showed robust target knockdown by both quantitative reverse transcriptase-PCR and immunoblotting (Figure 2A–2D and data not shown). Knockdown of ClpP reduced the growth and viability of the K562, TEX, and OCI-AML2 cells that have high ClpP expression (Figure 2A–2C), but not HL60 cells that express low basal levels of ClpP (Figure 2D). In addition, knockdown of ClpP in TEX cells reduced engraftment into the marrow of NOD/SCID-GF mice (Figure 2E).
Figure 2. Knockdown of ClpP reduces the growth and viability of AML cells.
(A – D) The expression of ClpP and Actin in TEX (A), OCI-AML2 (B), K562 (C), and HL60 (D) cells four days after transduced with lentiviral vectors expressing control shRNA or one of the three shRNAs targeting ClpP were detected by immunoblotting.
Viable cell counts of cells seed four days after transduction are shown as mean ± SD. One of three representative experiments is shown.
(E) The percentage of human CD45+ cells in the non-injected femur of sublethally irradiated NOD/SCID-GF mice determined by flow cytometry (n = 10 per group). ** p = 0.005, by t-test. Bar represents mean engraftment.
Clpp knockout mice are viable with normal hematopoiesis
To assess the effects of ClpP inhibition on normal hematopoiesis in vivo, we created constitutive ClpP-deficient mice. In WT mice, ClpP protein was expressed at low levels in bone marrow, lung, kidney, and stomach, but was more abundant in the pancreas, colon, testis, brain, and liver (Figure S2A). ClpP was absent in all tested tissues from Clpp−/− mice (Figure S2B). Clpp−/− mice are viable, although slightly smaller than their WT counterparts (Figure S2C–S2D). Thus, our mice are similar to those obtained by Gispert S., et al., (Gispert et al., 2013), who reported that 80% of their Clpp−/− mice survived over 400 days, but were infertile and had impaired hearing.
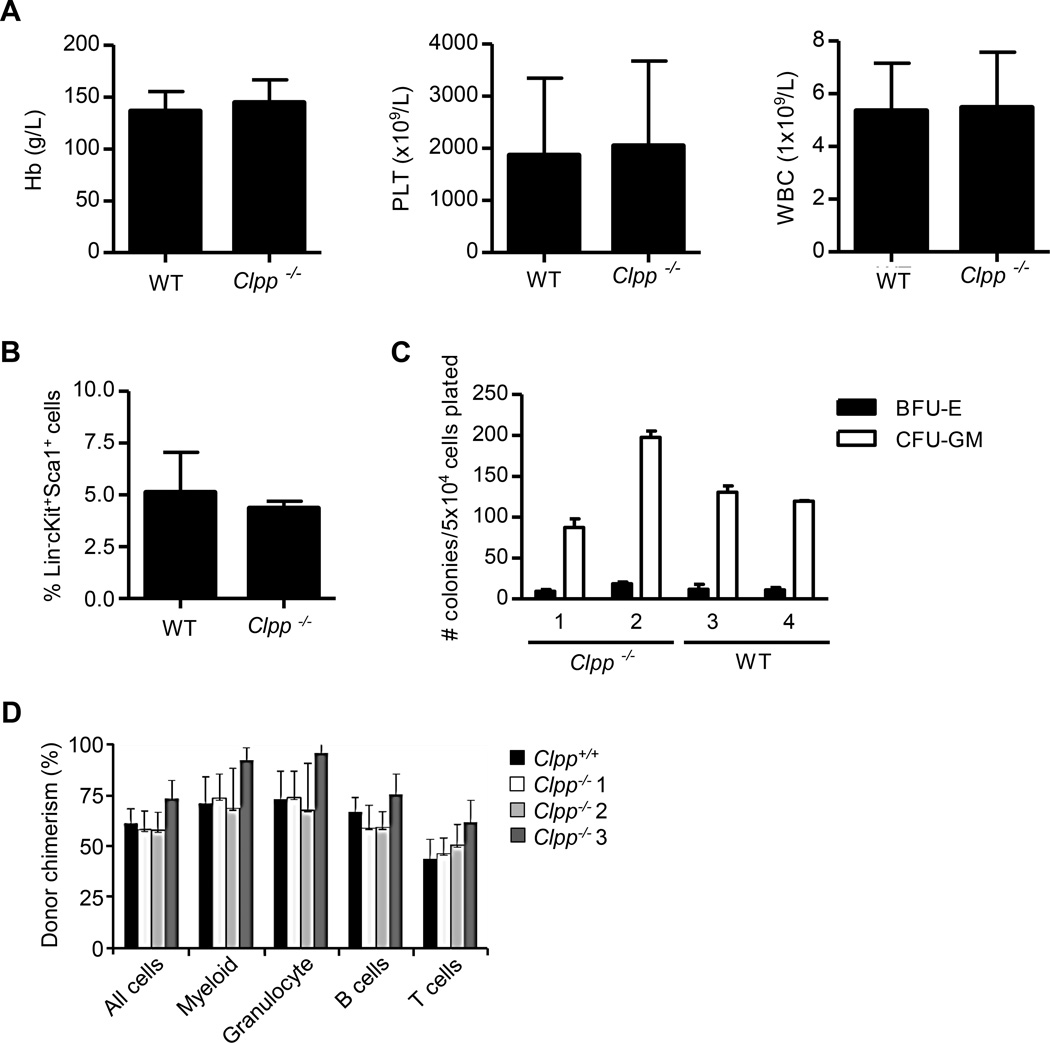
The study by Gispert et al., did not report the effects of Clpp deficiency on hematopoiesis, we therefore studied hematopoiesis in our Clpp−/− mice. Clpp−/− mice had normal numbers of all mature blood cell types in the circulation (Figure 3A) and normal numbers of phenotypically defined (Lin−cKit+Sca1+) progenitor cells in the bone marrow (Figure 3B). In vitro colony assays confirmed the presence of equal numbers of progenitors in the bone marrow of WT and Clpp−/− mice (Figure 3C). Likewise, cells from the marrow of Clpp−/− and WT mice had a similar ability to repopulate the marrow of sublethally irradiated congenic W41/W41 recipients when assessed in a competitive repopulation assay (Figure 3D). T and B cell function and the number of NK cells were also similar between Clpp−/− and WT mice (Figure S2E–S2G). p16 expression in T cells from Clpp−/− and WT mice was also similar (Figure S2H). This lack of any detectable effect of Clpp absence in murine hematopoiesis supports the potential feasibility of using ClpP inhibitors for the treatment of AML.
Figure 3. ClpP knockout mice are viable with normal hematopoiesis.
(A) Mean ± SD hemoglobin (Hb), platelet (PLT), and white blood cell (WBC) counts from peripheral blood samples of WT and Clpp−/− mice (n = 4 per group).
(B) The frequency of Lin-cKit+Sca1+ hematopoietic cells in the bone marrow of WT and Clpp−/− mice. Mean ± SD percentages were determined by flow cytometry.
(C) Erythroid and myeloid colony formation from BFU-E and CFU-GM of hematopoietic cells from WT and Clpp−/− mice (n = 3 per group) plated in vitro under standard conditions. The mean ± SD colony counts are shown.
(D) The mean ± SD proportions of CD45.2 cells from WT or Clpp−/− mice transplanted in sublethally irradiated congenic W41/W41 CD45.1 mice measured by flow cytometry.
See also Figure S2.
ClpP interacts with respiratory chain proteins and mitochondrial metabolism enzymes
The function and substrates of mammalian ClpP are largely unknown. To gain insight into ClpP function, we used the BioID-Mass Spectrometry (BioID-MS) method to identify ClpP interacting partners (Roux et al., 2012). To identify interacting proteins using BioID-MS, a protein of interest is fused in-frame with a mutant E. coli biotin conjugating enzyme, BirA R118G (or BirA*). BirA* efficiently activates biotin, but exhibits a reduced affinity for the activated molecule. Biotinoyl-AMP thus simply diffuses away from BirA* and reacts with nearby amine groups - including those present on lysine residues in neighbouring polypeptides. Following a stringent cell lysis, biotinylated proteins can be affinity purified and identified using mass spectrometry.
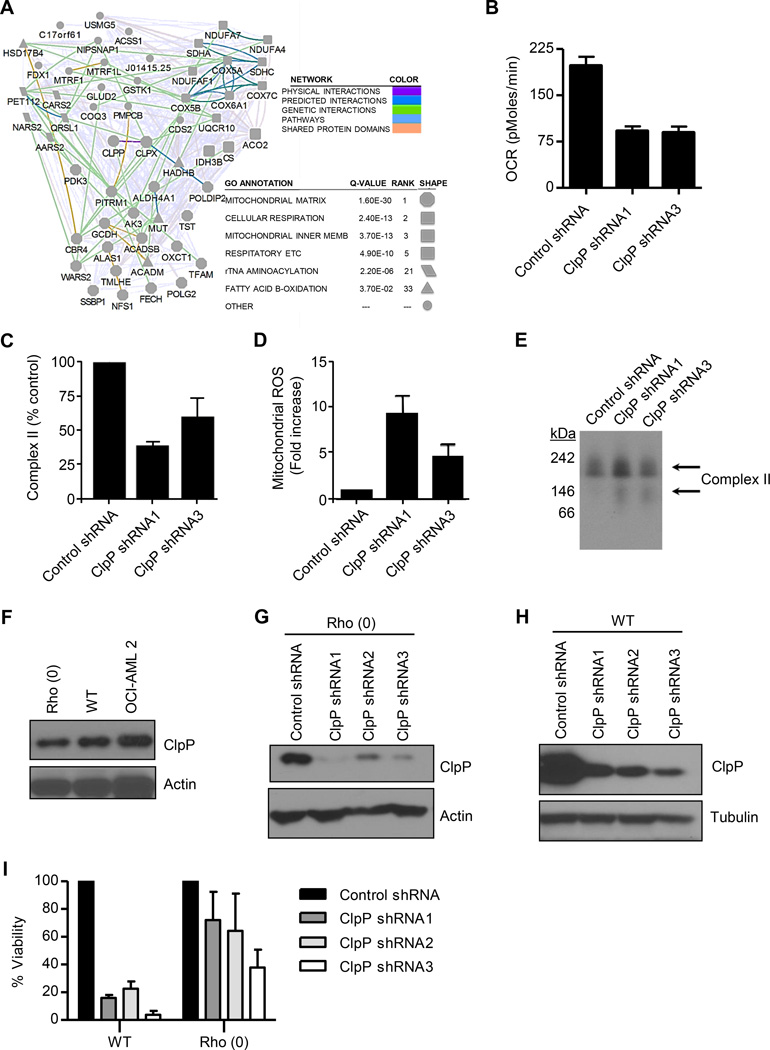
From this protein interaction screen, we identified 191 mitochondrial ClpP-interacting proteins, 49 of which preferentially interacted with WT or mutant ClpP over the unrelated mitochondrial matrix enzyme ornithine transcarbamoylase (OTC) (>3-fold spectral counts for BirA*-ClpP vs BirA*-OTC; Figure 4A and Table S2). The proteins interacting with WT and mutant ClpP were similar. ClpX, the known regulatory subunit of ClpP, was the top hit in our interactome map. Most of the 48 other preferential ClpP interacting proteins were components of the respiratory chain or enzymes involved in mitochondrial metabolism. Thus, these data suggested that ClpP function could be important to maintain the integrity of mitochondrial metabolism.
Figure 4. ClpP regulates mitochondrial metabolism.
(A) Network of proteins that interacted with WT or catalytically inactive ClpP.
(B) Effects of ClpP knockdown on basal oxygen consumption (OCR) in OCI-AML2 cells 4 days after transduction with ClpP shRNA or control sequences. Mean ± SD cell counts from one of three representative experiments are shown.
(C) Effects of ClpP knockdown on complex II activity in OCI-AML2 cells 4 days after transduction with ClpP shRNA or control sequences. Percent mean ± SD enzymatic activity from one of 3 representative experiments is shown.
(D) Effects of ClpP knockdown on mitochondrial ROS production in OCI-AML2 cells 4 days after transduction with ClpP shRNA or control sequences. Mean ± SD mitochondrial ROS positive cells from one of three representative experiments are shown.
(E) Effect of ClpP knockdown in OCI-AML2 cells four days after infection with virus expressing shRNA targeting ClpP or control sequences on SDHA expression and migration on a native gel determined by immunoblotting.
(F) Expression of ClpP in total cell lysates prepared from WT and Rho (0) 143B cells and OCI-AML2 cells.
(G, H) The expression of ClpP and β-actin were detected by immunoblotting in WT (G) and Rho (0) 143B (H) cells transduced with individual shRNA targeting ClpP or control sequences.
(I) The number of viable WT and Rho (0) 143B cells was measured by trypan blue staining 12 days after transduction as described in panel F and G. One of 3 representative experiments showing the percent mean ± SD viable cells relative to cells infected with control shRNA is presented.
See also Figure S3 and Table S2.
Inhibition of ClpP impairs oxidative phosphorylation
Given the interactions of ClpP with respiratory chain proteins and metabolic enzymes, we evaluated the effects of ClpP inhibition on oxidative phosphorylation. We found knockdown of ClpP reduced basal oxygen consumption in OCI-AML2 cells (Figure 4B).
Succinate dehydrogenase subunit A (SDHA), a subunit of respiratory chain complex II, was a top hit in our interaction mapping experiment. Therefore, we also measured the effects of ClpP inhibition on the activity of respiratory chain complex II. Knockdown of ClpP reduced the enzymatic activity of complex II in OCI-AML2 cells (Figure 4C). Consistent with impaired complex II activity, knockdown of ClpP also increased mitochondrial ROS production (Figure 4D). Using non-denaturing gels and immunoblotting, we also observed faster migrating bands of SDHA after ClpP knockdown, potentially representing the accumulation of non-functional misfolded or degraded SDHA (Figure 4E).
To determine whether the changes in oxidative phosphorylation were functionally important for cell death after ClpP inhibition, we examined the impact of ClpP inhibition in Rho (0) cells that lack mitochondrial DNA and oxidative phosphorylation. We knocked down ClpP in WT and Rho (0) 143B cells that expressed similar levels of ClpP protein (Figure 4F–4H). ClpP knockdown reduced the growth and viability of WT 143B cells, but had less effect on the growth and viability of Rho (0) cells (Figure 4I). In addition, we evaluated the impact of knocking down SDHA in OCI-AML2 cells. Knockdown of SDHA impaired complex II activity and reduced the growth and viability in these cells (Figure S3A–C). Thus, these results support SDHA as a functionally important substrate for ClpP and a role for ClpP in maintaining oxidative phosphorylation in a subset of AML cells.
Loss of cell viability was not associated with changes in mtUPR markers Hsp70, or CPN60, or changes in mitochondrial mass or structure in AML cell lines (Figure S3D–S3H).
A small molecule ClpP inhibitor is cytotoxic to AML cells
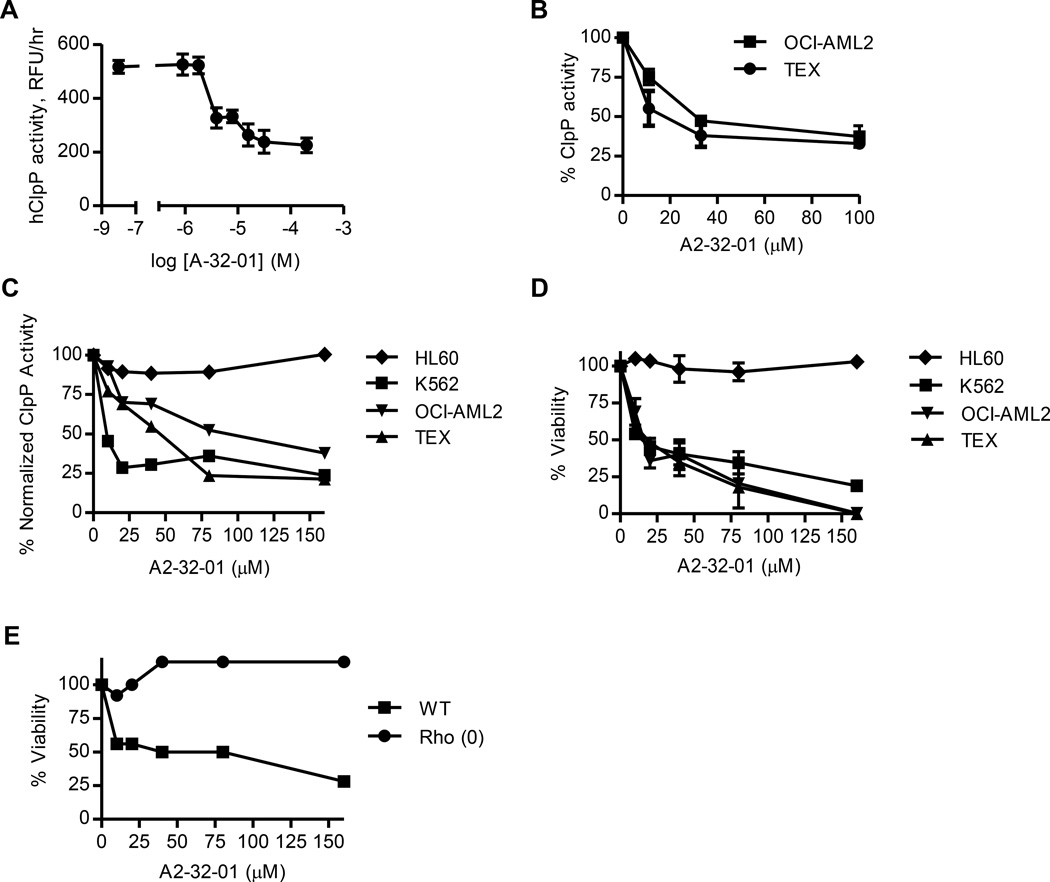
Recently, beta-lactone inhibitors of the bacterial ClpP were developed and shown to be toxic to several bacterial species (Zeiler et al., 2012). As a chemical approach to evaluate the effects of ClpP inhibition on AML and normal cells, we synthesized the reported bacterial ClpP inhibitor (3RS,4RS)-3-(non-8-en-1-yl)-4-(2-(pyridin-3-yl)ethyl)oxetan-2-one (A2-32-01) (Figure S4A). A2-32-01 inhibited the enzymatic activity of recombinant bacterial ClpP similar to the activity of the beta-lactone inhibitors in the initial study (Zeiler et al., 2012) (Figure S4B) as well as the enzymatic activity of recombinant human mitochondrial ClpP (Figure 5A). In addition, A2-32-01 inhibited cleavage of ClpP fluorogenic peptide substrates when added to lysates of mitochondria isolated from AML cells or intact AML cells (Figure 5B). A2-32-01 did not inhibit cytoplasmic chymotrypsin, trypsin, or caspase-like protease enzymatic activity when added to lysates of red blood cells that contain proteasome complexes but lack mitochondria, thus demonstrating its specificity for mitochondrial proteases (Figure S4C).
Figure 5. A2-32-01 inhibits ClpP and is cytotoxic to AML cells.
(A) Effect of A2-32-01 on the ability of recombinant human ClpP and ClpX to cleave the fluorogenic substrate casein-FITC. Mean ± SD.
(B) Effect of A2-32-01 on ClpP activity measured in mitochondrial lysates from OCI-AML2 and TEX cells after 2 hr based on cleavage of the fluorogenic substrate Suc-LY-AMC. Percent mean ± SD.
(C) Effect of 48 hr A2-32-01 treatment on the activity of ClpP in HL60, K562, OCI-AML2, and TEX cells based on the cleavage of the fluorogenic substrate Suc-LY-AMC. Normalized data are shown.
(D) Effect of 48 hr A2-32-01 treatment on the viability of HL60, K562, OCI-AML2, and TEX cells. Percent mean ± SD of viable cells was measured by trypan blue staining.
(E) Effect of 48 hr A2-32-01 treatment on the viability of WT and Rho (0) 143B cells. Percent of viable cells was measured by trypan blue staining.
See also Figure S4.
A2-32-01 induced cell death in TEX, OCI-AML2, and K562 leukemia cells at concentrations that matched its ability to inhibit ClpP activity, but was not toxic to HL60 leukemia cells which express much lower levels of ClpP (Figure 5C–5D). However, HL60 cells were more sensitive to the proteasome inhibitor bortezomib (Figure S4D). Similar to ClpP shRNA, A2-32-01 reduced the growth and viability of WT 143B cells, but had little effect on the growth and viability of their Rho (0) counterparts (Figure 5E). Moreover, addition of A2-32-01 to OCI-AML2 cells produced no additional loss of growth and viability after knockdown of ClpP (Figure S4E). We also evaluated analogues of A2-32-01. The related compound A2-58-06 had activity similar to A2-32- 01, while A2-54-01 did not inhibit ClpP and did not reduce the growth and viability of TEX and OCI-AML2 cells (Figure S4F–S4G).
While A2-32-01 was stable in DMSO, it was rapidly hydrolyzed when dissolved in cell culture medium with >90% of the compound degraded within 1 hour (Figure S4H). We confirmed that degradation products were inactive (data not shown). This rapid degradation likely explains the high IC50 of the compound.
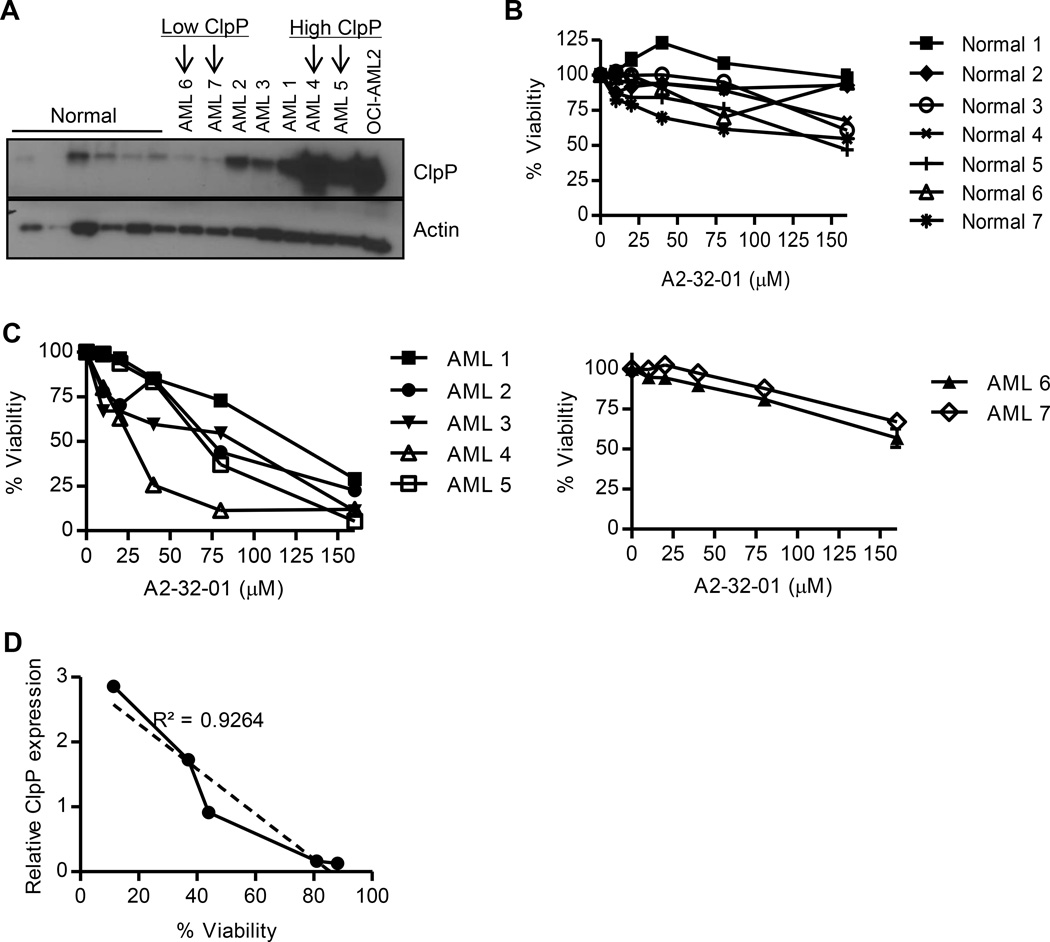
Next, we assessed the impact of A2-32-01 on primary AML and normal hematopoietic cells. A2-32-01 showed significant dose-dependent killing of primary AML patient cells that expressed elevated levels of ClpP. In contrast, the same treatment had little effect on either normal hematopoietic cells or primary AML cells that expressed lower levels of ClpP (Figure 6A–C). In fact, primary AML cells showed a strong positive correlation between ClpP expression and sensitivity to A2-32-01 (Figure 6D). Thus, ClpP expression might serve as a biomarker for patients that would respond to ClpP inhibitors.
Figure 6. ClpP expression correlates with sensitivity to A2-32-01 in primary AML cells.
(A) Expression of ClpP and Actin in primary normal hematopoietic cells, primary AML patient samples, and OCI-AML2 cells was determined by immunoblotting.
(B, C) Effect of A2-32-01 on the viability of primary normal hematopoietic cells (B) and primary AML cells (C) assessed after a 48 hour period of exposure. Mean ± SD percent of viable cells was measured by Annexin V/PI staining and flow cytometry.
(D) Correlation analysis of ClpP expression and cell viability of primary AML cells. Cell viability was measured by Annexin-V/PI staining 48 hours after treatment with 80 µM of A2-32-01.
A2-32-01 shows anti-AML activity in xenograft models of human leukemia
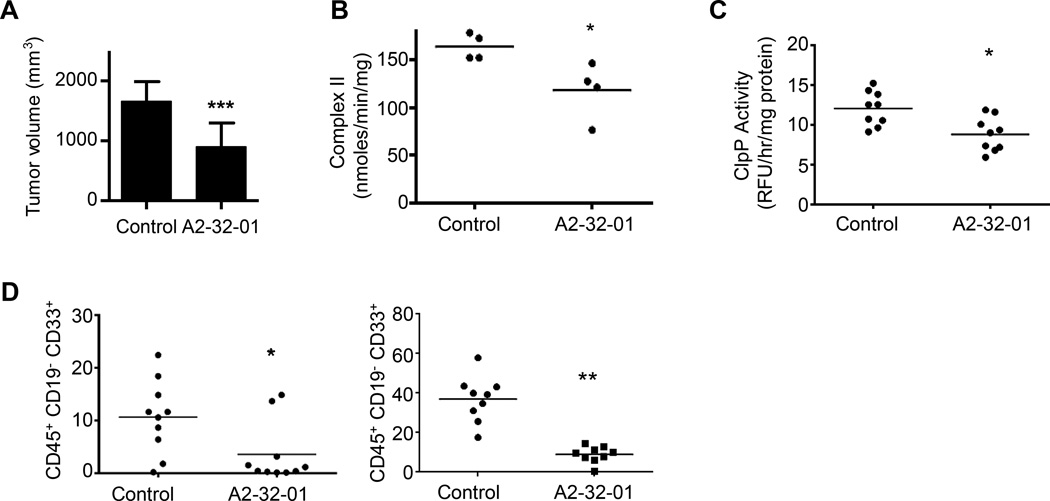
Finally, we evaluated the anti-leukemia efficacy and toxicity of A2-32-01 in mouse models of leukemia. Dissolving A2-32-01 in corn oil preserved its stability while in solution and the pharmacokinetics of A2-32-01 was determined after intraperitoneal injection of the compound into SCID mice (Figure S5A–S5B). Next, we evaluated the efficacy and toxicity of A2-32-01 in mice xenografted with OCI-AML2 cells. Daily treatment with A2-32-01 delayed the growth of OCI-AML2 cells in SCID mice without evidence of liver, muscle or renal toxicity (Figure 7A and Figure S5C–S5D). Moreover, tumors excised from mice treated for five days with A2-32-01 had reduced respiratory chain complex II activity compared to vehicle-treated controls (Figure 7B) and reduced enzymatic activity of ClpP (Figure 7C). Thus, A2-32-01 inhibits growth and mitochondrial function of this AML cell line in vivo.
Figure 7. A2-32-01 shows anti-AML activity in xenograft models of human leukemia.
(A) Effect of i.p. injection of 300 mg/kg of A2-32-01 or corn oil vehicle control twice daily for 5 of 7 days on tumor volume when OCI-AML2 cells were xenografted into SCID mice (n = 10 mice per group). Mean ± SD. *** p < 0.001 by t-test.
(B) Respiratory chain complex II activity in tumors from SCID mice xenografted with OCI-AML2 cells and treated with i.p. injection of 300 mg/kg A2-32-01 or corn oil vehicle control daily for 5 days (n = 4 mice per group). * p = 0.03, by t-test.
(C) ClpP activity measured by the cleavage of the fluorogenic substrate casein-FITC in mitochondrial lysates in tumors from SCID mice xenografted with OCI-AML2 cells and treated with i.p. injection of 300 mg/kg A2-32-01 or corn oil vehicle control daily for 4 days (n = 9 mice per group). * p = 0.005, by t-test.
(D) Human leukemia cell engraftment in sublethally irradiated female NOD/SCID mice and treated with A2-32-01 (300 mg/kg by i.p. injection daily) or vehicle control (n = 10 per group) 3 of 7 days for 4 weeks was measured by flow cytometric analysis. The panels show results from two independent experiments. * p = 0.02, ** p < 0.0001, as determined by t-test.
Horizontal bars indicate the mean in panels B–D.
See also Figure S5.
We also assessed the efficacy and toxicity of A2-32-01 in NOD-SCID mice engrafted intra-femorally with primary AML cells. A2-32-01 treated mice had significantly lower levels of leukemic engraftment compared to control-treated mice without evidence of toxicity (Figure 7D). Thus, inhibiting ClpP may be an effective strategy for a subset of AML patients.
Discussion
The therapy for AML has remained largely unchanged over the last 20 years and the outcome for most patients is poor. In search of more effective approaches, we recently found that blocking mitochondrial protein translation was selectively cytotoxic to AML cells (Skrtic et al., 2011). This followed the discovery that a subset of AML cells have unique mitochondrial characteristics with increased mitochondrial biogenesis and increased reliance on oxidative phosphorylation that could be inhibited by targeting mitochondrial protein translation (Lagadinou et al., 2013; Skrtic et al., 2011). Here, we identify ClpP as a potential component of the mitochondrial proteome whose inhibition might be cytotoxic to AML cells. We found that ClpP is over-expressed in the leukemic cells from a group of AML patients and inhibition of ClpP selectively induces death in these cells without apparent toxicity to normal hematopoiesis. The fact that increased expression of ClpP occurred across all currently defined subtypes of AML based on their morphologic, cytogenetic or mutational features, including those with a poor prognosis, lends further support to the possibility that ClpP may be an important therapeutic target for the treatment of AML. In addition, ClpP expression may be a useful biomarker to identify those patients most likely to respond to ClpP inhibitors.
Little is known about the endogenous proteins that interact with ClpP in the mitochondria of mammalian cells. Here, we identified most of the proteins interacted with ClpP were components of the respiratory chain and mitochondrial metabolic enzymes. It is noteworthy that a similar set of ClpP substrates, including the respiratory chain complex component SDHA, were identified by Haynes et al (Haynes et al., 2010), in their studies of ClpP function in C. elegans. Thus, ClpP appears to function to maintain the integrity of the respiratory chain and mitochondrial metabolism.
In C. elegans, peptides generated by ClpP-mediated protein degradation are exported from the mitochondria via the transporter haf-1 and signal the mtUPR response in the nucleus (Haynes et al., 2007; Haynes et al., 2010). Upon mitochondrial stress, levels of ClpP and mtUPR-associated genes increase. To date, the mechanisms by which ClpP regulate the mtUPR in mammalian cells remain unknown and the mammalian homologue of haf-1 has not yet been defined.
In our study, gene expression analysis demonstrated that higher levels of ClpP were associated with greater expression of genes associated with the mtUPR. Due to increased mitochondrial mass, high metabolic and biosynthetic requirements, and imbalances between levels of nuclear and mitochondrial encoded proteins, we hypothesize that the mitochondria in a subset of AML and other cancer cells are under increased basal stress and have upregulated ClpP in response to these stressors. In support of this hypothesis, induction of miotochondrial protein aggregates through the over-expression of mutant ornithine transcarbamylase in COS-7 cells induced mtUPR and increased ClpP expression (Zhao et al., 2002).
Mechanisms that regulate ClpP expression in AML cells are unknown and are an important direction of future study. In COS-7 cells, the expression of ClpP may be regulated at the level of its promoter by a combination of CHOP, MURE1, and MURE2 sequences (Aldridge et al., 2007). However, it is unknown whether these sequences also regulate ClpP expression in AML cells and the transcription factors that bind these sites are unknown. In addition, regulation of ClpP may also occur at the level of protein translation and degradation.
We used the bacterial ClpP inhibitor A2-32-01 to examine the impact of inhibiting mitochondrial ClpP in AML as the compound cross-reacted with the mitochondrial isoform. However, it is important to note that the selectivity of A2-32-01 for mitochondrial ClpP over other mammalian targets including other mitochondrial proteases remains to be determined. In addition, A2-32-301 has poor stability in aqueous media, so this molecule would not have therapeutic applications and its ability to reduce large bulk disease may be limited. Developing potent, stable, and specific ClpP inhibitors based on alternate scaffolds is a priority in order to fully assess the efficacy and toxicity of ClpP inhibition in vivo.
In AML cells, loss of viability after inhibition of ClpP was associated with an impairment of mitochondrial metabolism and oxidative phosphorylation. This finding is consistent with prior studies demonstrating that AML cells and stem cells have an increased reliance on oxidative phosphorylation (Skrtic et al., 2011) (Lagadinou et al., 2013). The fact that elevated ClpP expression was not associated with any known molecular or clinical feature of AML is consistent with this variable phenotype and suggests that increased ClpP expression may be a consequence of multiple dysregulated pathways that converge to impact mitochondrial function and metabolism.
In addition to ClpP, our shRNA screen of the mitochondrial proteome identified three other mitochondrial proteases whose knockdown reduced the growth and viability of K562 cells (NLN, PARL, PITRM1). Little is known about mitochondrial proteases as cancer targets and the impact of inhibiting select mitochondrial proteases in malignant cells. Potentially, these proteases could also be important therapeutic targets for AML given their role in regulating mitochondrial function. For example, PARL regulates PINK1 turnover, and knockdown of PARL promotes accumulation of PINK1 on the mitochondrial surface leading to increased mitophagy (Greene et al., 2012). However, further studies will be necessary to fully understand the efficacy and toxicity of inhibiting PARL and other mitochondrial proteases in vitro and in vivo.
Thus, in summary, while AML may have significant molecular and genetic heterogeneity, the dysregulation of some biological pathways such as mitochondrial function and metabolism remain common and may be amenable to therapeutic exploitation.
Experimental Procedures
See Supplemental Material for additional Experimental Procedures
shRNA screen to identify targets in the mitochondrial proteome
To identify shRNA that reduced the growth and viability of AML cells, we performed a pooled lentiviral shRNA screen in K562 cells similar to previously described (Ketela et al., 2011). K562 cells were selected due to their robust transduction efficiencies and their reliance on BCR-ABLl signaling would serve as a positive control for the screen. K562 leukemia cells were transduced with a pooled lentiviral library, consisting of 54,021 shRNA in bar-coded lentiviral vectors with puromycin selection markers targeting 11,255 nuclear-encoded genes. Of these 34,600 were associated with known genes and transcripts. These sequences were considered in the analysis. Cells were transduced at a multiplicity of infection of 0.3, ensuring that each cell receives at most 1 shRNA. The day after infection, cells were treated with puromycin to select infected cells. Resistant clones, containing a lentiviral vector and its associated shRNA, were then grown in puromycin-containing medium for 21 days. After 21 days, cells were harvested, genomic DNA isolated and the abundance of bar coded sequences were identified by microarray analysis. In conducting our lentiviral shRNA, we performed two independent experiments utilizing the same amount of virus for transduction (inter-experiment or biological replicates). Each trial was analyzed in duplicate on arrays (intra-experiment or technical replicates), leading to the generation of 4 replicates (2 biological, 2 technical) for the 0 and 21 day time points.
Log2 background-corrected fluorescence signal intensities were compared pairwise for all intra- and inter-experimental replicates at 0 and 21 days to ensure technical and experimental reproducibility. Correlation coefficients for all pairwise comparisons were r2 > 0.97. Thus, all four technical and experimental replicates for each time point were averaged to compare the 0 and 21 day time points. Fold changes were computed by calculating the difference in mean Log2 (background-corrected fluorescence signal intensities) for each individual shRNA, as follows:
Log2(fold-change) = Log2(mean intensityt = 21 days) - Log2(mean intensityt = 0 days)
Negative values indicated potential depletion of the individual shRNA. A percent depletion value was also calculated: % Depletion = (1–2Log2(fold-change)) × 100%.
shRNAs in the screen were ranked based on Log2(fold-change) and % depletion values. shRNAs were also grouped by target gene to assess whether multiple shRNAs targeting the same gene had high % depletion scores. Genes encoding the mitochondrial proteome were identified using MitoCarta (http://www.broadinstitute.org/pubs/MitoCarta/).
Reverse phase protein array
A reverse phase protein array (RPPA) was generated by using samples from patients with AML, MDS, and normal CD34+ cells as described previously (Kornblau and Coombes, 2011; Kornblau et al., 2011; Kornblau et al., 2010; Pierce et al., 2013; Tibes et al., 2006). The ClpP antibody was validated for use in the RPPA assay as previously described (Kornblau and Coombes, 2011; Kornblau et al., 2011; Kornblau et al., 2010; Pierce et al., 2013; Tibes et al., 2006). Briefly, the rabbit monoclonal ClpP antibody (clone, ab124822, Abcam, Toronto, ON) was validated to recognize only a single band corresponding to the expected molecular weight by immunoblotting. In addition, sensitivity of the antibody in the RPPA assay was validated using an array of cell line lysates seeded in serials dilutions. The RPPA signal was linear over the range of dilutions. In addition, the signal obtained with cell lysate dilutions by RPPA correlated with the signal intensity by observed by immunoblotting (R2 = 0.75). Once the RPPA was optimized for detection of ClpP expression, patient and normal samples were printed in five serial dilutions onto slides along with normalization and expression controls. Slides were probed with anti-ClpP primary antibody (Abcam, 124822) at 1:1000 and a secondary antibody followed by colorimetric assay. Stained slides were analyzed using Microvigene® software (Vigene Tech, Carlisle, MA).
Primary AML and normal hematopoietic cells
Primary mononuclear cells were isolated from peripheral blood samples from consenting patients with AML whose blast count was 80% or higher by low-density cells isolated by Ficoll density centrifugation. G-CSF mobilized peripheral blood stem cells were obtained from healthy consenting volunteers donating peripheral blood stem cells for allogeneic stem cell transplantation. All primary cells were cultured at 37 °C in IMDM, supplemented with 20% FBS, and penicillin-streptomycin. Informed consent was obtained from all subjects for the collection and use of samples. The collection and use of human tissue for this study were approved by the University Health Network institutional review board and Review Ethics Board of the University of British Columbia.
shRNA knockdown of ClpP
Construction of hairpin-pLKO.1 vectors (carrying a puromycin antibiotic resistance gene) containing shRNA sequences and production of short hairpin RNA viruses was performed as previously described (Simpson et al., 2012). The shRNAs targeting the ClpP (Accession No.NM_003321) coding sequence are as follows: ClpP shRNA1 5’-GCCCATCCACATGTACATCAA -3’, ClpP shRNA2 5’-CACGATGCAGTACATCCTCAA -3’, ClpP shRNA3 5’-GCTCAAGAAGCAGCTCTATAA -3’.
Lentiviral transduction was performed essentially as described (Xu et al., 2010). Briefly, cells (5 × 106) were centrifuged and re-suspended in 5 mL of medium containing protamine sulfate (5 µg/mL). 0.5 mL of virus (for adherent cells) and 2 mL of virus (for suspension cells) was added to the cells followed by overnight incubation (37 °C, 5% CO2). Cells were centrifuged, washed and fresh medium with puromycin (1 µg/mL) was added the following day. Three days later, cells were cultured to measure viability and growth.
Equal number of TEX cells transduced with shRNA targeting ClpP or control sequences (2×105) were injected into the right femur of sublethally irradiated NOD/SCID-GF (Nonobese diabetic/severe combined immunodeficiency-growth factor) mice with human transgenes producing interleukin-3 (IL-3), granulocyte/macrophage-stimulating factor (GM-CSF) and Steel factor (SF) (n = 10 per group) (Nicolini et al., 2004). 6 weeks after injection, mice were sacrificed and the percentage of human CD45+ cells in the non-injected left femur was determined by flow cytometry.
Evaluation of hematopoietic progenitors in WT and Clpp−/− mice
Heterozygous Clpp knockout mice (C57BL/6N background) were obtained from TIGM (Texas A&M Institute for Genomic Medicine) and breeding pairs were established in our facility. Studies with Clpp−/− mice were carried out according to the regulations of the Canadian Council on Animal Care and with the approval of the Ontario Cancer Institute and BC Cancer Agency animal ethics review boards.
Hematopoietic cells were flushed from the femurs and tibias of Clpp+/+ and Clpp−/− mice using DMEM medium and 2% FCS and passed through a 70 µM strainer. The population was enriched for lineage negative (Lin−) progenitor cells were selected using magnetic bead separation (Miltenyi Lineage Cell Depletion Kit, Miltenyi, Cambridge, MA). Cells (1–2 × 106) were preincubated with an Fc block for 10 min at 4 °C and immunostained with antibodies recognizing Sca1 (D7) and cKit (2B8) (both from eBioscience, San Diego, CA). Flow cytometry data were acquired using FACSCanto II (BD) flow cytometer, and analyzed with FlowJo analysis program.
W41/W41 CD45.1 sublethally irradiated (400 rad) mice were transplanted with 106 CD45.2 hematopoietic cells from the femurs of WT or Clpp−/− mice along with 106 competitor CD45.1 hematopoietic cells. Cells from two Clpp+/+ WT mice were pooled and injected into 5 recipient CD45.1 mice. Cells from each of three Clpp−/− mice were injected into 4–6 CD45.1 recipients. Peripheral blood samples were collected from the tail vein of recipient mice 5, 8, and 12 weeks after transplantation. Following lysis of the red blood cells with ammonium chloride (StemCell), the white blood cells (WBCs) were stained with antibodies for donor and recipient CD45 allotypes; anti-CD45.1-PECy7 (eBioscience), and anti-CD45.2-APCef780 (eBioscience) plus CD19-BV421 (BioLegend) for B cells, Ly6G-PE and Mac-1-PE (both BD Pharmingen) for myeloid cells, and CD3e-APC (BioLegend) for T cells. To calculate repopulation levels, the contributions of the donor CD45.2 positive cells to the populations of circulating GM, B, T and total WBCs were calculated.
To examine T function in the Clpp−/− and Clpp+/+ mice, 5×106 splenocytes per well were seeded in wells coated with 3 µg/ml Hamster anti-mouse CD3 (clone 2C11-145 from BD) and 1 µg/ml Hamster anti-mouse CD28 (clone 37.51 from eBioscience) for 24 hours. For B cell activation, splenocytes (5×106/per) were cultured for 24 hours with 10 µg/ml goat anti-mouse IgM (F(ab')2 fragment (Jackson Immunoresearch). To assess activation status cells were stained with fluorescence-conjugated antibodies against surface markers. Flow cytometry data were acquired using FACSCanto II (BD) flow cytometer, and analyzed with FlowJo analysis program.
BioID-MS to identify ClpP interacting proteins
Stable 293 T-REx cell pools expressing tetracycline-regulated, BirA*-tagged WT or catalytically-inactive mutant ClpP proteins were generated. Cell pools expressing the BirA* epitope tag alone, or BirA* fused to the unrelated mitochondrial enzyme ornithine transcarbamoylase (OTC) were also created, for use as negative controls. 293 T-REx cells (at ~60% confluence) were treated with 1 µg/ml tetracycline and 50 µM biotin for 24 hours to induce transgene expression and effect biotinylation of nearby polypeptides. Cells were scraped into PBS, pooled, washed twice in 25 ml PBS, and collected by centrifugation at 1000 × g for 5 min at 4°C. Cell pellets were lysed in 5 mL ice-cold modified RIPA buffer. 250 U benzonase (EMD) was added, and biotinylated proteins isolated. The resulting supernatant was incubated with 30 µl of (RIPA-equilibrated) streptavidin-sepharose beads (GE) with end-over-end rotation for 2 hours at 4 °C. Beads were washed 7 times with 1 mL of 50 mM ammonium bicarbonate (pH 8.0) prior to tryptic digest.
Mass spectrometry to identify ClpP interacting proteins
One microgram of MS-grade TPCK trypsin (Promega, Madison, WI) dissolved in 70 µl of 50 mM ammonium bicarbonate (pH 8.3) was added to the streptavidin-sepharose beads and incubated at 37 °C overnight. The eluate was collected and beads washed twice in 100 µl of 50 mM ammonium bicarbonate. The combined eluate was lyophilized, and then resuspended in 0.1% formic acid. LC analytical columns (75 µm inner diameter) and pre-columns (150 µm inner diameter) were made in-house from fused silica capillary tubing from InnovaQuartz (Phoenix, AZ) and packed with 100 Å C18-coated silica particles (Magic, Michrom Bioresources, Auburn, CA). Peptides were subjected to nano LC-ESI-MS/MS, using a 95 min reversed phase (10–30% acetonitrile, 0.1% formic acid) buffer gradient running at 250 nL/min on a Proxeon EASY-nLC pump in-line with a hybrid linear quadrupole ion trap (Velos LTQ) Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). A parent ion scan was performed in the Orbitrap, using a resolving power of 60,000. Simultaneously, the twenty most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation) using standard CID fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same m/z (within a 10ppm window, exclusion list size 500) detected two times within 15 sec were excluded from analysis for 30 sec.
For protein identification, Thermo.RAW files were converted to the. mzXML format using Proteowizard as part of the Trans-Proteomic Pipeline (Keller and Shteynberg, 2011). For each search, the iProphet probability at 1% error rate was used as a cutoff value to generate SAINT-compatible input files. SAINT parameters were as follows: 5000 iterations, low mode off, minFold 1 and normalization on (Choi et al., 2011).
Via the ProHits suite (Liu et al., 2012; Liu et al., 2010), MS data were analyzed using the X!Tandem database search algorithm, and the resulting peptide identifications subjected to iProphet analysis (Shteynberg et al., 2011) within the Trans-Proteomic Pipeline (Keller and Shteynberg, 2011). Using BirA* only cell data as a control for endogenous biotinylated proteins, and polypeptides that interact non-specifically with the solid phase support, the BirA*-ClpP data were subjected to Statistical Analysis of INTeractomes (SAINT) (Choi et al., 2011) to identify bona fide interacting partners. Polypeptides identified with an iProphet confidence value >0.8 (reflecting a ~1% error rate) and assigned a maximum SAINT confidence value >0.80 are reported in Supplemental Figure 3A.
The network of proteins that interacted with WT or catalytically inactive ClpP preferentially over ornithine carbamoyl transferase or empty vector were defined using GeneMANIA 3.3.1, Cytoscape 3.1.0 (Montojo et al., 2010). Preferential interactions compared to ornithine transcarbamoylase or empty vector were identified in 293 T-REx cells using the BirA* tagging method.
Bacterial and human ClpP activity
Bacterial ClpP was purified from E. coli as previously described (Kimber et al., 2010). To measure ClpP peptidase activity, 1 µM ClpP (monomer concentration) was dissolved in assay buffer (50 mM Tris-Cl pH 8.0, 200 mM KCl, 1 mM DTT) with 0.5 mM Suc-LY-AMC. This mixture was then plated at 50 µL/ well in a 96-well plate and read for fluorescence at 2 minute intervals for 1 hour at 360/460 nm. The rate of ClpP was determined by calculating the slope (in the linear range).
Human recombinant ClpP (1.2 µM) and ClpX (1.0 µM) were combined and dissolved in assay buffer consisting of: 25 mM HEPES pH 7.5, 5 mM MgCl2, 5 mM KCl, 0.03% Tween 20, 10% glycerol with an ATP regeneration system of 16 mM creatine phosphate and 13 U/ml creatine kinase. 3 mM ATP was added along with the ClpX substrate, casein-FITC (4.5 µM). All the components were mixed and covered from light at 37 °C for 5 minutes. The mixture was then plated at 200 µL per well in triplicates in a 96-well plate with or without ClpP inhibitors. Fluorescence at 485/535 nm was measured at 37 °C, every 10 minutes for 6 hours. The rate of ClpP activity was determined by calculating the slope (in the linear range).
Purification of human ClpP and ClpX
Plasmids encoding human ClpP and ClpX were transformed into B21 Gold DE3 pLys and B21 Gold DE3 bacteria, respectively. Transformed bacteria were plated on LB-Agar plates containing 100 µg/mL Ampicilin and incubated overnight at 37 °C. The following day individual colonies were grown overnight at 37 °C in LB medium containing 100 µg/mL Ampicilin (ClpP) or 85 µg/mL Chloramphenicol and 100 µg/mL Ampicilin (ClpX). ClpP and ClpX expression were induced with 1 mM IPTG in Terrific medium without antibiotics. Bacteria were collected after 4 hr and the pellet frozen at −70 °C. ClpP and ClpX proteins were purified on Ni-Agarose using lysis buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% Glycerol, 5 mM mercaptoethanol and 10 mM imidazole), wash buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% Glycerol, 5 mM mercaptoethanol and 50 mM imidazole), and elution buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% Glycerol, 5 mM mercaptoethanol and 250 mM imidazole). The proteins were dialyzed for 4 hours at 4 °C in the dialysis buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 25mM MgCl2, 10% g1ycerol, 1 mM DTT and 0.1 mM EDTA). The recombinant proteins were stored at −70 °C.
ClpP activity in isolated mitochondria
Intact mitochondria from cell lines were isolated as described above. The fluorogenic substrate Suc-LY-AMC (0.5 mM) was added to mitochondrial lysates in ClpP peptidase assay buffer (50 mM Tris-Cl pH 8.0, 200 mM KCl, 1 mM DTT, 0.1% DDM (n-dodecyl β-D-maltoside) and 2 mM ATP). This mixture was plated at 50 µL/well in a 96-well plate and fluorescence was measured at 2 minute intervals for 1 hour (360/460nm). The rate of ClpP activity was determined by calculating the slope (in the linear range).
Assessment of anti-leukemia activity of A2-32-01 in mouse models of human leukemia
OCI-AML2 human leukemia cells (5 × 105) were injected subcutaneously into the flanks of male SCID mice (Ontario Cancer Institute, Toronto, ON). When the tumors were palpable, mice were treated with A2-32-01, or vehicle control (corn oil) by i.p. injection (300 mg/kg twice daily for 5 of 7 days) for 10 days (n= 10 per group). Tumor volume was measured using caliper measurements at the end of the experiment. Mice were sacrificed, organs harvested, fixed with 10% buffered formalin, embedded, sectioned and stained with hematoxylin and eosin. The stained samples were scanned using Aperio Scanscope XT at 10× magnification, and analyzed using Aperio ImageScope.
To assess the effect of A2-32-01 on complex II activity in vivo, OCI-AML2 human leukemia cells (5 × 105) were injected subcutaneously into the flanks of male SCID mice (Ontario Cancer Institute, Toronto, ON). When the tumors were palpable, mice were treated with A2-32-01, or vehicle control (corn oil) by i.p. injection (300 mg/kg daily) for 5 days (n= 4 per group). After 5 days, tumors were excised and complex II activity was measured as above.
To assess A2-32-01 in mouse models of primary AML engraftment, a frozen aliquot of AML cells was thawed, counted and re-suspended in PBS and 2.5 × 106 viable trypan blue-negative cells were injected into the right femur of 10 week-old female NOD-SCID mice that had been irradiated 24 hours previously with 208 rad from a 137Cs source, and injected with 200 µg anti-mouse CD122. Two weeks after injection of AML cells, mice were treated with A2-32-01 (300 mg/kg by i.p. injection) or vehicle control (n=10 per group) 3 of 7 days for four weeks. Mice were then sacrificed, and the cells were flushed from the femurs. Engraftment of human AML cells into the marrow of the non-injected left femur was assessed by enumerating the percentage of human CD45+CD33+CD19− cells by flow cytometry using the BD FACS Calibur. Data were analyzed with FlowJo version 7.7.1 (TreeStar).
All animal studies were carried out according to the regulations of the Canadian Council on Animal Care and with the approval of the Ontario Cancer Institute animal ethics review board.
Supplementary Material
Significance.
Perturbations in the cytoplasmic and nuclear proteasome are known to contribute to the acquisition of malignant properties, but the importance of ClpP, a mitochondrial protease with structural homology to the proteasome, has not been established. We have now shown that loss of ClpP does not impact normal hematopoiesis, whereas ClpP inhibition kills human leukemic cells, including those with stem and progenitor activity, which may be attributable to their higher mitochondrial stress and a greater reliance on oxidative phosphorylation. These results suggest targeting mitochondrial ClpP may be an attractive therapeutic strategy for a subset of human acute leukemias.
Acknowledgements
We thank Urlich Baxa (Electron Microscopy Laboratory, NCI (National Institutes of Health)) for help with electron microscopy, Aisha Shamas-Din (Princess Margaret Cancer Center) for help with preparing the manuscript, and Jill Flewelling (Princess Margaret Cancer Center) for administrative assistance. This work was supported by the Canadian Stem Cell Network, the Leukemia and Lymphoma Society, the National Institutes of Health (NCI 1R01CA157456), Canadian Institutes of Health Research, a Terry Fox Research Foundation Program Project Group Grant to CJE, MaRS Innovation, the Ontario Institute for Cancer Research with funding provided by the Ontario Ministry of Research and Innovation, the Princess Margaret Cancer Centre Foundation, and the Ministry of Long Term Health and Planning in the Province of Ontario. SB was supported by a Vanier Scholarship award from CIHR. DVJ is a Fonds de recherche du Québec – Santé (FRQS) postdoctoral scholar. ADS holds the Barbara Baker Chair in Leukemia and Related Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
The microarray data from the screen is deposited with the following accession number: GenomeRNAi: GR00343-S.
References
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corydon TJ, Bross P, Holst HU, Neve S, Kristiansen K, Gregersen N, Bolund L. A human homologue of Escherichia coli ClpP caseinolytic protease: recombinant expression, intracellular processing and subcellular localization. Biochem J. 1998;331(Pt 1):309–316. doi: 10.1042/bj3310309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan S, Naito M, Tsuruo T. Selective induction of apoptosis in Philadelphia chromosome-positive chronic myelogenous leukemia cells by an inhibitor of BCR - ABL tyrosine kinase, CGP 57148. Cell Death Differ. 1998;5:710–715. doi: 10.1038/sj.cdd.4400400. [DOI] [PubMed] [Google Scholar]
- de Sagarra MR, Mayo I, Marco S, Rodriguez-Vilarino S, Oliva J, Carrascosa JL, Casta n JG. Mitochondrial localization and oligomeric structure of HClpP, the human homologue of E. coli ClpP. J Mol Biol. 1999;292:819–825. doi: 10.1006/jmbi.1999.3121. [DOI] [PubMed] [Google Scholar]
- Fisher RI, Bernstein SH, Kahl BS, Djulbegovic B, Robertson MJ, de Vos S, Epner E, Krishnan A, Leonard JP, Lonial S, et al. Multicenter phase II study of bortezomib in patients with relapsed or refractory mantle cell lymphoma. J Clin Oncol. 2006;24:4867–4874. doi: 10.1200/JCO.2006.07.9665. [DOI] [PubMed] [Google Scholar]
- Gispert S, Parganlija D, Klinkenberg M, Drose S, Wittig I, Mittelbronn M, Grzmil P, Koob S, Hamann A, Walter M, et al. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum Mol Genet. 2013;22:4871–4887. doi: 10.1093/hmg/ddt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goard CA, Schimmer AD. Mitochondrial matrix proteases as novel therapeutic targets in malignancy. Oncogene. 2013 doi: 10.1038/onc.2013.228. [DOI] [PubMed] [Google Scholar]
- Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson EM, Rehman AU, Walsh T, Clayton-Smith J, Lee K, Morell RJ, Drummond MC, Khan SN, Naeem MA, Rauf B, et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Dimitrova MN, Ortega J, Ginsburg A, Maurizi MR. Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J Biol Chem. 2005;280:35424–35432. doi: 10.1074/jbc.M507240200. [DOI] [PubMed] [Google Scholar]
- Keller A, Shteynberg D. Software pipeline and data analysis for MS/MS proteomics: the trans-proteomic pipeline. Methods Mol Biol. 2011;694:169–189. doi: 10.1007/978-1-60761-977-2_12. [DOI] [PubMed] [Google Scholar]
- Ketela T, Heisler LE, Brown KR, Ammar R, Kasimer D, Surendra A, Ericson E, Blakely K, Karamboulas D, Smith AM, et al. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics. 2011;12:213. doi: 10.1186/1471-2164-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimber MS, Yu AY, Borg M, Leung E, Chan HS, Houry WA. Structural and theoretical studies indicate that the cylindrical protease ClpP samples extended and compact conformations. Structure. 2010;18:798–808. doi: 10.1016/j.str.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Kornblau SM, Coombes KR. Use of reverse phase protein microarrays to study protein expression in leukemia: technical and methodological lessons learned. Methods Mol Biol. 2011;785:141–155. doi: 10.1007/978-1-61779-286-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, Qiu YH, Zhang N, Singh N, Faderl S, Ferrajoli A, York H, Qutub AA, Coombes KR, Watson DK. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–5612. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau SM, Singh N, Qiu Y, Chen W, Zhang N, Coombes KR. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin Cancer Res. 2010;16:1865–1874. doi: 10.1158/1078-0432.CCR-09-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O'Dwyer KM, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang J, Choi H, Lambert JP, Srikumar T, Larsen B, Nesvizhskii AI, Raught B, Tyers M, Gingras AC. Using ProHits to store, annotate, and analyze affinity purification-mass spectrometry (AP-MS) data. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al] 2012;8(8):16. doi: 10.1002/0471250953.bi0816s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang J, Larsen B, Stark C, Breitkreutz A, Lin ZY, Breitkreutz BJ, Ding Y, Colwill K, Pasculescu A, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol. 2010;28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolini FE, Cashman JD, Hogge DE, Humphries RK, Eaves CJ. NOD/SCID mice engineered to express human IL-3, GM-CSF and Steel factor constitutively mobilize engrafted human progenitors and compromise human stem cell regeneration. Leukemia. 2004;18:341–347. doi: 10.1038/sj.leu.2403222. [DOI] [PubMed] [Google Scholar]
- O'Connor OA, Stewart AK, Vallone M, Molineaux CJ, Kunkel LA, Gerecitano JF, Orlowski RZ. A phase 1 dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematologic malignancies. Clin Cancer Res. 2009;15:7085–7091. doi: 10.1158/1078-0432.CCR-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, Whetton AD, Meyer S, Ravandi-Kashani F, Borthakur G, Coombes KR, Zhang N, Kornblau S. Transglutaminase 2 expression in acute myeloid leukemia: association with adhesion molecule expression and leukemic blast motility. Proteomics. 2013;13:2216–2224. doi: 10.1002/pmic.201200471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352:2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. M111 007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CD, Hurren R, Kasimer D, MacLean N, Eberhard Y, Ketela T, Moffat J, Schimmer AD. A genome wide shRNA screen identifies alpha/beta hydrolase domain containing 4 (ABHD4) as a novel regulator of anoikis resistance. Apoptosis. 2012;17:666–678. doi: 10.1007/s10495-012-0723-4. [DOI] [PubMed] [Google Scholar]
- Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Vij R, Wang M, Kaufman JL, Lonial S, Jakubowiak AJ, Stewart AK, Kukreti V, Jagannath S, McDonagh KT, Alsina M, et al. An open-label, single-arm, phase 2 (PX-171-004) study of single-agent carfilzomib in bortezomib-naive patients with relapsed and/or refractory multiple myeloma. Blood. 2012;119:5661–5670. doi: 10.1182/blood-2012-03-414359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, Gronda M, Skrtic M, Li X, Hurren R, et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood. 2010;115:2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler E, Korotkov VS, Lorenz-Baath K, Bottcher T, Sieber SA. Development and characterization of improved beta-lactone-based anti-virulence drugs targeting ClpP. Bioorg Med Chem. 2012;20:583–591. doi: 10.1016/j.bmc.2011.07.047. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. Embo J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.