SUMMARY
Significance of ERG in human prostate cancer is unclear because mouse prostate is resistant to ERG-mediated transformation. We determined that ERG activates the transcriptional program regulated by YAP1 of the Hippo signaling pathway and found that prostate-specific activation of either ERG or YAP1 in mice induces similar transcriptional changes and results in age-related prostate tumors. ERG binds to chromatin regions occupied by TEAD/YAP1 and transactivates Hippo target genes. In addition, in human luminal type prostate cancer cells, ERG binds to the promoter of YAP1 and is necessary for YAP1 expression. These results provide direct genetic evidence of a causal role for ERG in prostate cancer and reveal a connection between ERG and the Hippo signaling pathway.
INTRODUCTION
The development of effective prevention and treatment strategies for prostate cancer requires understanding the critical molecular alterations that drive the initiation of neoplasia and subsequent development of malignant characteristics. The notable finding that recurrent chromosomal recombination events result in ERG oncogene overexpression in prostate cancers provides compelling evidence supporting the hypothesis that ERG, and potentially other ETS-family transcription factors, function as key drivers of prostate carcinogenesis (Tomlins et al., 2005). ETS-family gene rearrangements occur in 20-50% of all human prostate adenocarcinomas, depending on racial background, and are found in precursor lesions and across all histological grades and tumor stages (Sreenath et al., 2011; Tomlins et al., 2005).
Cause and effect studies of phenotypic changes resulting from high ERG activity have been conducted using a spectrum of cell lines, xenografts, and genetically engineered mouse (GEM) models. Knockdown of ERG in VCaP prostate adenocarcinoma cells, a line that harbors a functional TMPRSS2-ERG rearrangement (Tomlins et al., 2005) substantially reduces cell invasion and attenuates proliferation (Gupta et al., 2010; Tomlins et al., 2008; Wang et al., 2008). Consistent with loss-of-function studies in VCaP cells, overexpression of ERG or other ETS family genes in immortalized prostate epithelial cell lines results in substantial increase in cell invasion (Klezovitch et al., 2008; Tomlins et al., 2007). At the molecular level, ERG has been shown to influence androgen receptor signaling, induce a repressive epigenetic program via activation of EZH2, activate Wnt pathway signaling, and promote NFκB-mediated transcription (Chen et al., 2013; Gupta et al., 2010; Wang et al., 2011; Yu et al., 2010).
To confirm causal role for ERG in the genesis of prostate cancer, several GEM models have been constructed that express ERG specifically in prostate epithelial cells. These models are notable for a range of relatively subtle phenotypic changes that include the partially penetrant formation of focal precancerous lesions or focal hyperplasia (Baena et al., 2013; Chen et al., 2013; Klezovitch et al., 2008; Tomlins et al., 2008), or a complete absence of any discernable phenotype (Carver et al., 2009a; Carver et al., 2009b; King et al., 2009). In contrast to the minimal oncogenic effects observed in ERG transgenic mice, when combined with Tp53 loss or Pten inactivation, ERG promotes invasive and metastatic phenotypes. The differences in ERG-mediated effects between human and mouse cells, and between the different GEM models can be potentially explained by the level of transgene expression; however, the relative levels of ERG expression in many transgenic models have either not been reported (Carver et al., 2009a; Carver et al., 2009b; King et al., 2009), or found to be low in comparison to levels in human prostate cancer (Baena et al., 2013; Casey et al., 2012). To better understand the importance of ERG and determine the mechanism(s) by which it can promote neoplasia we analyzed transgenic mice expressing ERG in prostate epithelium at levels comparable to those found in ERG-rearranged primary human prostate cancers in vivo.
YAP1 is a component of the canonical Hippo signaling pathway that comprises a cascade of kinases that includes the Hippo/MST1-2 kinases, the adaptor Sav1, and the LATS1/2 kinases. Hippo signaling culminates in the phosphorylation and consequent inactivation of the transcriptional co-activators YAP1 and TAZ by LATS1/2, which suppresses the TEAD-dependent expression of a network of genes that promote cell proliferation and survival. Studies in mouse models have shown that LATS1/2 kinases exert tumor suppressive effects and YAP1 functions as an oncogene (Pan, 2010). Hippo pathway activity is strongly implicated in the pathogenesis of human medulloblastomas, oral squamous-cell carcinomas, and carcinomas of the lung, pancreas, esophagus, liver, and mammary gland (Pan, 2010). While previous studies have determined that the Hippo kinases MST1/2 and LATS1/2 are downregulated and YAP is upregulated in a subset of primary human prostate cancers (Cinar et al., 2007; Steinhardt et al., 2008; Zhao et al., 2012), the causes and consequences of YAP1 activation have not been defined, nor have causal roles for Hippo signaling in the genesis of prostate cancer been established.
RESULTS
Age-dependent prostate tumors develop in transgenic mice expressing high levels of ERG
To determine the consequences of ERG overexpression in prostate epithelium in vivo, we performed extensive longitudinal analyses of a GEM model, Tg(Pbsn-ERG)1Vv, designed to express high levels of ERG in prostate epithelium (Klezovitch et al., 2008). This mouse line integrated the transgene in an intergenic region on chromosome 1, expresses ERG at levels comparable to primary human prostate cancers with ERG rearrangements, and displays transcriptional changes that significantly correlate with those identified in ERG+ human prostate tumors (Figure 1A; Figure S1, Table S1A-C). We found that aged Tg(Pbsn-ERG)1Vv mice develop prostate tumors and have a shorter life-span than wild-type littermates (Figure 1B). Overall, approximately 50% of Tg(Pbsn-ERG)1Vv transgenic mice aged 2 years and older developed prostate tumors, while none were found in wild-type littermates (Table 1). This is consistent with a lack of spontaneous prostate cancer in aged wild-type mice (Sharma and Schreiber-Agus, 1999). The level of ERG transgene expression was important, as we found no prostate tumors in aged mice from a different line Tg(Pbsn-ERG)8Vv (n=9), which expressed substantially lower levels of ERG (Figure S1A).
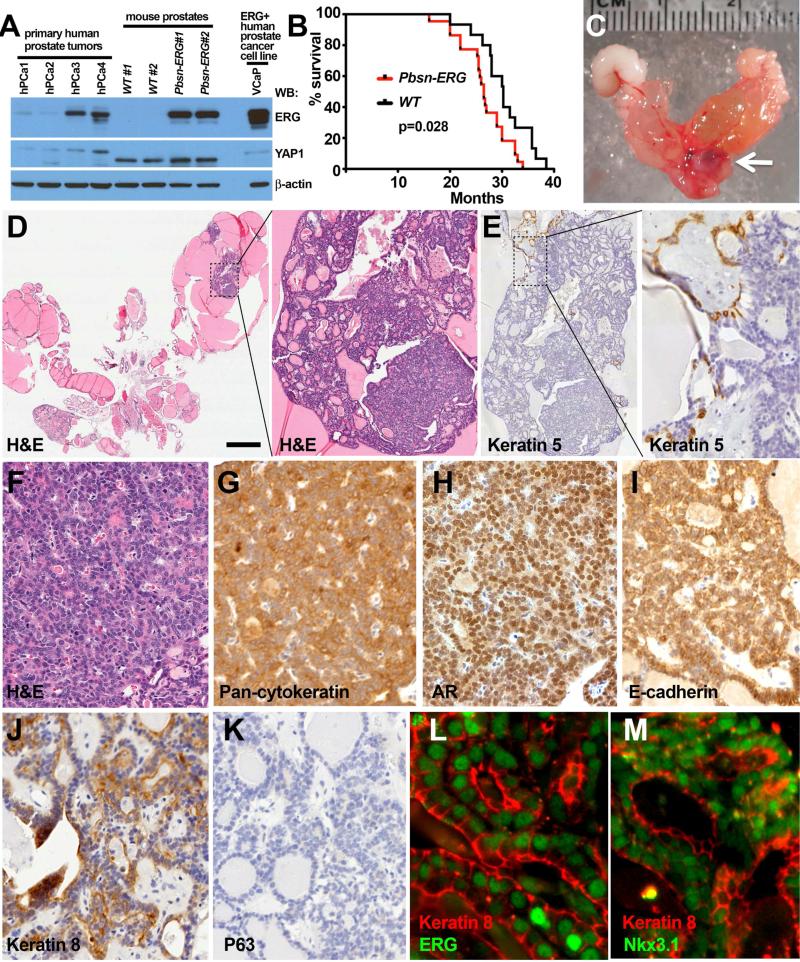
Figure 1. Prostate tumors in aged Tg(Pbsn-ERG)1Vv mice.
(A) Western blot (WB) analysis of ERG protein expression in primary human prostate tumors, ventral prostates from 8.5 month-old wild-type (WT) and Tg(Pbsn-ERG)1Vv (Pbsn-ERG) mice and the VCaP cell line.
(B) Kaplan-Meier survival curves of Tg(Pbsn-ERG)1Vv (Pbsn-ERG) and control wild-type littermate mice. n=15 for wild-type. n=24 for Pbsn-ERG. Logrank test.
(C) Gross appearance of urogenital tract organs in 20 month-old Pbsn-ERG male. Arrow points to the primary prostate tumor.
(D-M) Hematoxylin and Eosin (H&E, D, F), immunohistochemical (E, G-K) and immunofluorescence (L-M) staining of prostatic adenocarcinoma in 20 month-old Tg(Pbsn-ERG)1Vv (Pbsn-ERG) male. Areas outlined in D (left panel) and E (left panel) are shown at higher magnification in right panels in D and E. Staining was performed with indicated antibodies. Bar in D represents 2 mm in D left panel, 0.14 mm in D right panel, 0.2 mm in E left panel, 45 μm in E right panel, 36 μm in F-K, 12 μm in L-M. See also Figure S1, Table S1.
Table 1.
Prostate tumor incidence and tumor types in Tg(Pbsn-ERG)1Vv and control littermate mice.
| Genotype | Prostate gland tumor | Adenocarcinoma | Sarcomatoid carcinoma | Stromal tumor |
|---|---|---|---|---|
| Tg/+ Tg(Pbsn-ERG)1Vv | 16/25*** | 5/25 | 3/25 | 8/25 |
| +/+ Wild-type littermates | 0/15 | 0/15 | 0/15 | 0/15 |
Numbers denote mice with indicated tumors relative to the number of mice analyzed. The prostate glands were analyzed in mice euthanized at morbidity. The ages varied between 16 and 30 months in Tg(Pbsn-ERG)1Vv and between 16 and 38 months in wild-type littermate mice. Sigificance of association between prostate tumor development and genotype was tested using two-tailed Fisher's exact test.
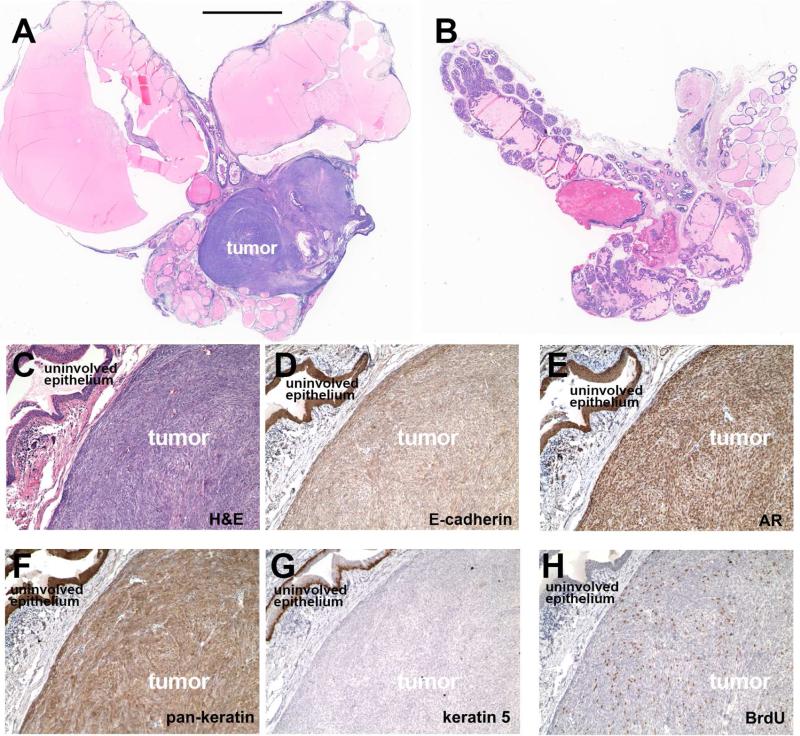
We observed several prostate tumor phenotypes in aged Tg(Pbsn-ERG)1Vv mice, including adenocarcinomas, sarcomatoid carcinomas (epithelial-mesenchymal transition (EMT)-like epithelial tumor, confirmed by showing pan-keratin expression), as well as stromal-type tumors (Table 1, Figures 1-2; Figure S1D-F). Similar to most human prostate cancers, adenocarcinomas in the Tg(Pbsn-ERG)1Vv mice are composed primarily of luminal (Keratin8+, Androgen Receptor high, Nkx3.1+) cells; however, small numbers of basal cells (Keratin5+) were present close to the margins of these tumors (Figure 1E-M; Figure S1G-M). The sarcomatoid carcinomas in the Tg(Pbsn-ERG)1Vv mice were usually quite large and contained many actively dividing (BrdU+) cells (Figure 2). We conclude that the expression of ERG in prostate epithelial cells in vivo, at levels that are clinically-relevant, is sufficient to cause the slow and partially penetrant emergence of age-related prostate tumors, a disease course that parallels prostate cancer development in men.
Figure 2. Large sarcomatoid carcinoma tumor in prostate gland of Tg(Pbsn-ERG)1Vv mice.
(A) Histology of prostate gland from 26-month-old Tg(Pbsn-ERG)1Vv mouse.
(B) Histology of prostate gland from 38-month-old wild-type mouse.
(C-H) Serial sections of the tumor and the adjacent, uninvolved epithelium from Tg(Pbsn-ERG)1Vv mouse stained with hematoxylin & eosin (C), anti-E-cadherin (epithelial cell marker, D), anti-Androgen Receptor (AR, luminal cell marker, E), anti-pan-cytokeratin (epithelial cell marker, F), anti-keratin 5 (basal epithelial cell marker, G) and anti-BrdU (proliferating cell marker, H). Bar in A represents 4 mm in A, B; 80 μm in C-H.
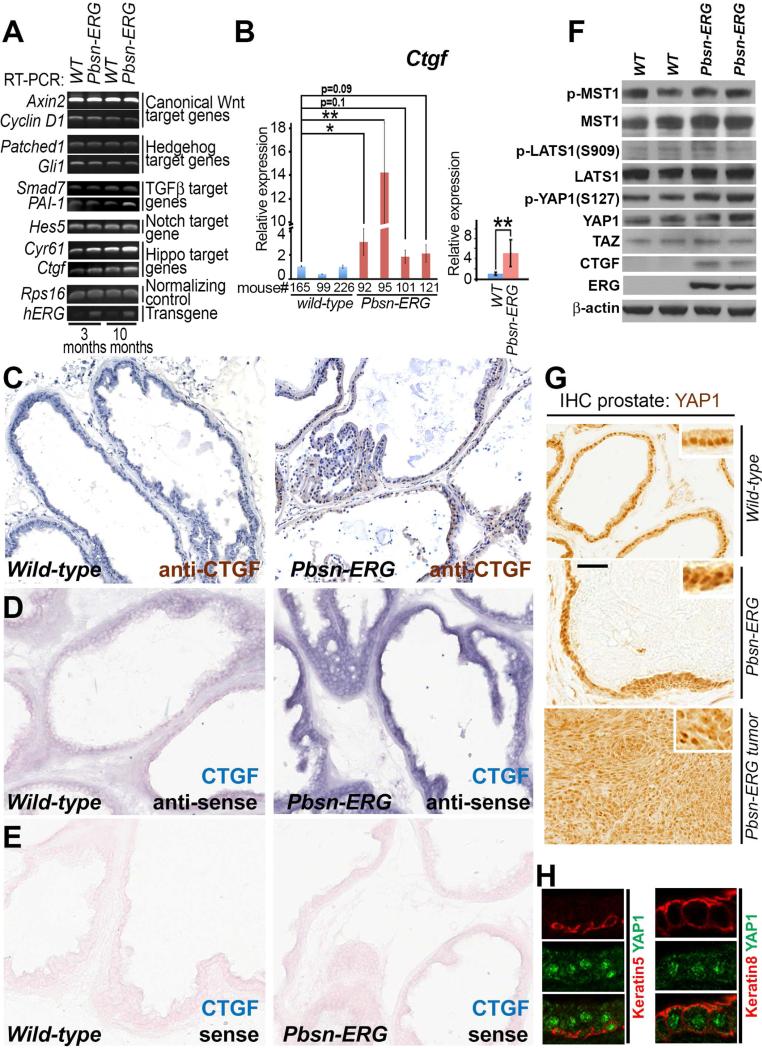
To determine the primary mechanisms responsible for the tumorigenic function of ERG, we analyzed gene expression changes in the prostates of Tg(Pbsn-ERG)1Vv mice prior to tumor development. Analyses of well-established gene targets of the canonical Wnt, Hedgehog, TGF[.beta], Notch, and Hippo signaling pathways revealed epithelium-specific activation of Ctgf, a gene directly regulated by the Hippo pathway transcriptional activator YAP1 (Figure 3A-E). In the canonical Hippo signaling pathway, YAP1 activity is negatively regulated by phosphorylation at S127 by the LATS1/2 kinases, which results in cytoplasmic retention and inactivation of YAP1 (Zhao et al., 2011). We hypothesized that as yet undefined targets of ERG reduced YAP1 S127 phosphorylation resulting in elevated YAP1 activity and consequent upregulation of Ctgf. However, we observed no reduction in YAP1 S127 phosphorylation or LATS1/2 inactivation in Tg(Pbsn-ERG)1Vv prostates compared to controls (Figure 3F). Moreover, we found that YAP1 was prominently expressed and localized to the nucleus in luminal epithelial cells in both wild-type and ERG transgenic mice (Figure 3G-H). These experiments indicate that the enhanced Ctgf expression in Tg(Pbsn-ERG)1Vv prostates is not due to decreased activity of LATS1/2 of the Hippo signaling pathway, though the findings did not preclude alternative mechanisms leading to the augmentation of YAP1 signaling and consequent Ctgf upregulation.
Figure 3. Upregulation of Hippo pathway target gene Ctgf in prostates of Tg(Pbsn-ERG)1Vv mice.
(A) RT-PCR analysis of major gene targets of the canonical Wnt, Hedgehog, TGF[.beta], Notch, and Hippo signaling pathways in prostate glands from 3- and 10-month-old Tg(Pbsn-ERG)1Vv (Pbsn-ERG) and wild-type littermate (WT) mice.
(B) qRT-PCR analysis of direct Hippo pathway gene target Ctgf in prostate glands from 10 month-old Pbsn-ERG and wild-type littermate mice. Individual mice (left) and wild-type and Pbsn-ERG group (right) comparisons. Data represent mean ± standard deviation. Student's t-test.
(C) Immunohistochemical analysis of CTGF expression in prostate glands from 5 month-old wild-type (left panel) and Pbsn-ERG (right panel) littermate mice.
(D-E) In situ hybridization analysis of Ctgf expression in prostate glands from 5 month-old wild-type (left panels) and Pbsn-ERG (right panels) littermate mice using anti-sense (D) and control sense (E) probes. For C-E, tissue sections from two WT and two Pbsn-ERG animals were placed on the same slide and all the incubation times and treatments were identical for both genotypes. (F) Western blot analysis of total protein extracts from the prostates of 11 month-old wild-type (WT) and Pbsn-ERG mice with anti-phospho(S127)-YAP1 (P-YAP1), anti-Yap1, anti-TAZ, anti-CTGF, anti-phospho(Thr183)-MST1 (P-MST1), anti-MST1, anti-phospho(S909)-LATS1 (PLATS1), anti-LATS1, anti-ERG, and anti-β-actin antibodies.
(G) Immunohistochemical staining of ventral prostates and a prostate tumor from 2.5 year-old wild-type and Pbsn-ERG mice with anti-YAP1 antibodies.
(H) Confocal sections of ventral prostate from 5 month-old wild-type mouse immunostained with rabbit anti-YAP1, guinea pig anti-keratin 5, and rat anti-keratin 8. Bar in G corresponds to 40 μm in C-E’, G, and 8.5 μm in H.
Overexpression of ERG in non-tumorigenic prostate epithelial cells activates YAP1 target genes and increases YAP1-mediated transcriptional output
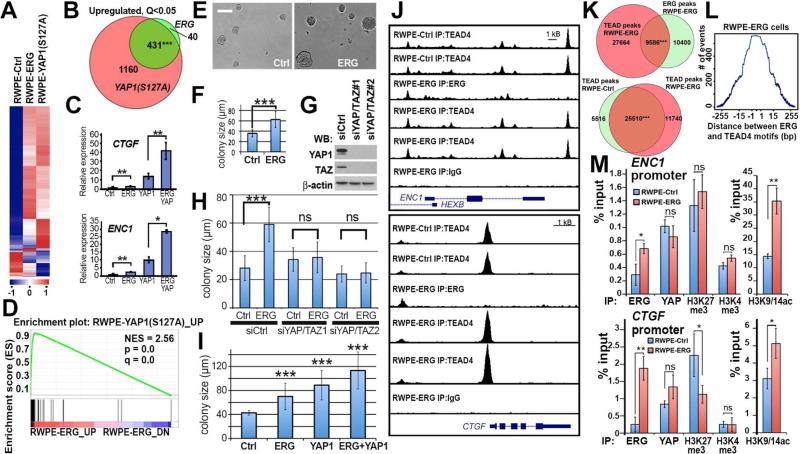
To investigate the role of ERG in regulation of Hippo signaling in prostate epithelial cells in culture using gain-of-function experiments, we engineered RWPE-1 cells to stably express ERG, constitutively active YAP1 (Yap1S127A, (Dong et al., 2007)), or both ERG and YapS127A (Figure 4A-D, Figure S2A). RWPE-1 cell line is a model of immortalized non-tumorigenic human basal epithelial cells, which do not express endogenous ERG (Bello et al., 1997; Litvinov et al., 2006). RNA-Seq analysis identified a significant overlap between genes upregulated through ERG expression and those regulated through Yap1S127A (p<0.001, Figure 4A-B, Tables S2-S3). Similarly, gene set enrichment analysis (GSEA) confirmed prominent similarities between ERG and Yap1S127A transcriptional programs (Figure 4D). qRT-PCR analysis not only confirmed ERG- mediated upregulation of the “Hippo pathway down” signature genes CTGF and ENC1 (Mohseni et al., 2014), but also demonstrated that ERG was able to increase their expression levels even in cells expressing constitutively-active Yap1S127A, which cannot be inactivated by phosphorylation at S127 by LATS1/2 (Figure 4C). Moreover, analyses of YAP1 S127 phosphorylation and the Hippo pathway kinases, MST1 and LATS1, demonstrated that ERG does not significantly impact their levels or activity (Figure S2A-B).
Figure 4. ERG binds to TEAD4-interacting regions of chromatin and transactivates Hippo pathway target genes in immortalized prostate epithelial cells.
(A-B) Significant overlap of gene expression changes induced by overexpression of ERG and constitutively active YAP1. Gene expression in RWPE-1 cells transduced with empty vectors (RWPE-Ctrl), ERG (RWPE-ERG) or YAP1S127A (RWPE-YAP1S127A) was analyzed by RNASeq. n=2 per each genotype. (A) Heat-map; (B) overlap between significantly (Q<0.05) upregulated genes. Two tailed Chi-square with Yates correction.
(C) qRT-PCR analyses of two Hippo pathway target genes CTGF and ENC1 in RWPE-Ctrl (Ctrl), RWPE-ERG (ERG), RWPE-YAP1S127A (YAP1) and RWPE-ERG and YAP127A (ERG YAP) expressing RWPE-1 cells. Data represent mean ± standard deviation (n = 3) from one of three independent experiments. Student's t test.
(D) GSEA analysis of similarities between ERG-mediated gene expression changes and YAP1S127A upregulated genes.
(E-F) Brightfield images (E) and size quantitation (F) of RWPE-Ctrl (Ctrl) and RWPE-ERG (ERG) cell colonies formed after 6 days in 3D organoid culture system. The graph shows mean ±SD. n≥40. Student's t-test. Bar correspond to 100 μm.
(G) Western blot (WB) analyses of RWPE-1 cells transfected with indicated siRNA oligos and analyzed with indicated antibodies.
(H) Colony size quantitation of RWPE-Ctrl (Ctrl) and RWPE-ERG (ERG) cells in 3D organoid culture system transfected with indicated siRNA oligos. The graph shows mean ±SD. n≥60. Student's t-test.
(I) Colony size quantitation of RWPE-Ctrl (Ctrl), RWPE-ERG (ERG), RWPE-YAP1S127A (YAP1) and RWPE-ERG + YAP127A (ERG + YAP1) cells in 3D organoid culture system. The graph shows mean ±SD. n≥44. Student's t-test.
(J) UCSC genome browser views of ChIP-Seq data from RWPE-Ctrl and RWPE-ERG cells using IgG, anti-ERG and anti-TEAD4 antibodies.
(K) Highly significant overlap between positions of TEAD4 and ERG peaks in RWPE-ERG cells (hypergeometric p-value=3.761525e-263 calculated by Bioconductor package “ChIPPeakAnno”). (L) Tendency toward close juxtaposition of ERG (A/C)GGAA(G/A) and TEAD4 GGAAT(G/T)(T/C) recognition motifs in ERG-TEAD4 overlapping peaks in RWPE-ERG cells.
(M) qPCR analysis of ChIP experiments from RWPE-Ctrl and RWPE-ERG cells using indicated antibodies. Data represent mean ± standard deviation (n = 4) from one of three independent experiments. Student's t test. See also Figure S2, Tables S2 and S3.
The actin cytoskeleton can in some cases regulate YAP1 activity independently from the Hippo/LATS phosphorylation cascade (Dupont et al., 2011). As expected, disruption of the actin cytoskeleton prominently downregulated expression of CTGF in RWPE-1 cells; however, this did not erase or decrease the differences in CTGF expression levels between the control and ERG expressing cells, indicating that ERG does not influence YAP1 activity via the actin cytoskeleton (Figure S2C-E).
YAP1/TAZ are necessary for ERG-mediated transformation and invasion of nontumorigenic prostate epithelial cells
We next analyzed the functional importance of YAP1/TAZ in mediating ERG-associated malignant phenotypes in RWPE-1 cells. Recently developed 3D organoid prostate culture system faithfully recapitulates the in vivo phenotypes and enables mechanistic studies of prostate development, homeostasis, and cancer (Karthaus et al., 2014). We found that overexpression of ERG significantly promotes growth of RWPE-1 cells (RWPE-ERG) in 3D organoid prostate culture system (Figure 4E-F). Importantly, YAP1/TAZ were necessary for this ERG-mediated transforming phenotype, because the knockdown of YAP1 and TAZ using two independent sets of siRNA oligos erased the growth differences between RWPE-Crtl and RWPE-ERG cells (Figure 4G-H). Similarly, the stable knockdown of YAP1 using two independent lentiviral shRNA constructs also erased the growth differences between RWPE-Crtl and RWPE-ERG cells in this culture system (Figure S2F-G). We also found that gain-of-function of YAP1 (YAP1S127A) was sufficient for stimulation of growth of RWPE-1 (RWPE-YAP1) cells in 3D organoid prostate culture system (Figure 4I). Moreover, ERG and YAP1 prominently cooperated with each other in stimulation of growth of RWPE-1 (RWPE-ERG+YAP1) cells in this culture system (Figure 4I). These data indicate that YAP1/TAZ were both necessary and sufficient for ERG-mediated transformation of RWPE-1 cells.
In addition to promoting the growth of RWPE-1 cells in 3D organoid prostate culture system, overexpression of ERG prominently stimulated formation of RWPE-1 cell colonies in soft agar assays in media containing 1% fetal bovine serum (Figure S2H). The stable knockdown of YAP1 using two independent shRNA constructs erased the differences between RWPE-Crtl and RWPE-ERG cells, confirming that YAP1 was necessary for ERG-mediated transformation of RWPE-1 cells (Figure S2H).
ERG has been shown to promote the invasive behavior of human prostate epithelial cells (Brenner et al., 2011; Klezovitch et al., 2008; Tomlins et al., 2008). Interestingly, constitutively-active YAP1 also promotes EMT and cell invasion across a spectrum of tumor types (Lamar et al., 2012; Overholtzer et al., 2006). Therefore, we asked whether ERG utilizes YAP1/TAZ in the development of this phenotype. As expected, overexpression of ERG promoted matrigel invasion of RWPE-1 (RWPE-ERG) cells (Figure S2I). Importantly, knockdown of YAP1 and TAZ erased the differences in invasion between RWPE-Ctrl and RWPE-ERG cells, indicating that YAP1/TAZ are necessary for ERG-mediated stimulation of cell invasion (Figure S2I).
ERG interacts with TEAD-occupied chromatin regions, increases histone acetylation, and promotes YAP1-dependent transcription
Since ERG and YAP1 both localize to the nucleus (Figures 1L, 3G-H), we hypothesized that ERG directly binds and activates YAP1 target genes. YAP1 associates with several distinct transcription factors, though members of the TEAD family are most directly responsible for the proto-oncogenic function of YAP1 (Liu-Chittenden et al., 2012; Zhao et al., 2008). ChIP-Seq experiments with RWPE-Ctrl and RWPE-ERG cells identified a highly significant overlap between TEAD4 and ERG peaks in RWPE-ERG cells with closely positioned ERG-TEAD4 recognition motifs (Figure 4J-L). Of overlapping ERG-TEAD4 peaks in RWPE-ERG cells, 75% were already occupied by TEAD4 in control RWPE-Ctrl cells, indicating that ERG does not function as a pioneering factor for TEAD4 chromatin binding. Analyses of potential changes in histone modifications in shared ERG-TEAD4 peaks at the promoters of Hippo gene targets, CTGF and ENC1, revealed an ERG-dependent increase in H3K9/14 acetylation, a chromatin mark highly localized to the 5′ regions of transcriptionally active human genes (Liang et al., 2004) (Figure 4M).
To determine whether ERG acts directly on the promoters of Hippo target genes, we performed luciferase assays with a CTGF promoter. ERG significantly increased YAP1/TEAD4-mediated CTGF transcription, and both ERG-binding motifs in the CTGF promoter and ERG's ability to bind to DNA were required for this transcriptional activation (Figure S2J-L). Overall, we conclude that in prostate epithelial cells ERG binds to many TEAD-interacting regions of chromatin, increases the histone H3K9/14 acetylation and promotes the transcriptional activity of YAP1 target genes.
ERG maintains YAP1 expression in neoplastic luminal prostate epithelial cells by controlling H3K9/14 acetylation of the YAP1 promoter
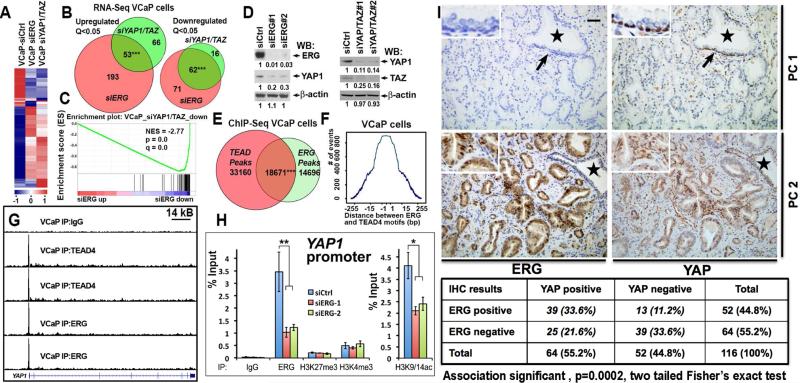
To complement the gain-of-function studies, we evaluated the effects of suppressing ERG and YAP1/TAZ activity in a human luminal epithelial line, VCaP, that endogenously expresses high ERG levels by virtue of a TMPRSS2-ERG rearrangement (Tomlins et al., 2005). RNA-Seq analysis of VCaP with siRNAs targeting ERG or YAP1 identified a highly significant overlap between genes altered by loss of ERG and those altered by loss of YAP1/TAZ activity (p<0.001, Figure 5A-D, Tables S4-S5). ChIP-Seq analysis of ERG and TEAD4 chromatin-binding sites confirmed a highly significant overlap between ERG and TEAD4 peaks in VCaP cells, with closely positioned ERG-TEAD4 recognition motifs (Figure 5E-F).
Figure 5. ERG maintains YAP1 expression in human prostate cancer cells.
(A-C) Significant concordance between ERG and YAP1/TAZ gene expression programs in VCaP cells. Gene expression determined by RNA-Seq of VCaP cells transiently transfected with non-targeting siRNA (siCtrl, n=2), siRNAs targeting ERG (siERG, n=2, using 2 independent siRNA oligos) and siRNAs targeting YAP1/TAZ (siYAP1/TAZ, n=2, using 2 independent siRNA oligo mixtures). (A) Heat-map; (B) overlap between significantly (Q<0.05) upregulated and downregulated genes; and (C) GSEA analysis of overlap between siERG-mediated gene expression changes and siYAP/TAZ downregulated genes. Two tailed Chi-square with Yates correction in B.
(D) Western blot (WB) analysis of VCaP cells transfected with indicated siRNA oligos. Numbers indicate relative expression values, with value in siCtrl adjusted to 1.
(E) Highly significant overlap between positions of TEAD4 and ERG peaks in VCaP cells (hypergeometric p-value=0 calculated by Bioconductor package “ChIPPeakAnno”).
(F) Tendency toward close juxtaposition of ERG (A/C)GGAA(G/A) and TEAD4 GGAAT(G/T)(T/C) recognition motifs in ERG-TEAD4 overlapping peaks in VCaP cells.
(G) UCSC genome browser views of ChIP-Seq data from VCaP cells using IgG, anti-ERG and anti-TEAD4 antibodies.
(H) qPCR analysis of ChIP experiments from siCtrl, siERG-1 or siERG-2 oligo transfected VCaP cells using indicated antibodies. Data represent mean±standard deviation (n = 4) from one of three independent experiments. Student's t test.
(I) Significant correlation between ERG expression and presence of nuclear YAP1 in primary human prostate tumors. Immunohistochemical (IHC) staining of prostate cancers (PC1 and PC2) with ERG and YAP1 antibodies. Note, YAP1 is strongly expressed in benign basal but not benign luminal epithelial cells (arrows), but is present in neoplastic luminal cells in a subset of human prostate carcinomas. Star indicates histologically benign prostatic gland. Bar corresponds to 50 μm. See also Figure S3, Tables S4 and S5.
Of interest, we found that suppressing endogenous ERG in VCaP cells by siRNAs significantly decreased the expression of endogenous YAP1, as determined by RNA-seq, qRT-PCR and Western Blot (Figure 5D, Figure S3A, Table S4). We found that both ERG and TEAD4 peaks were present at the transcriptional start site of YAP1, and ChIP-re-ChIP experiments revealed the co-occupancy of ERG and TEAD4 at the YAP1 promoter (Figure 5G, Figure S3B). Quantitative ChIP analyses demonstrated substantial downregulation of H3K9/14 acetylation at YAP1 and ENC1 promoters in siERG VCaP cells (Figure 5H, Figure S3C-D). These findings support a direct role for ERG in the maintenance of YAP1 expression in human luminal VCaP cells by controlling H3K9/14 acetylation of the YAP1 promoter.
ERG and YAP1 are co-expressed in a subset of primary human prostate cancers and YAP1 associates with adverse outcomes
To assess the relationships between ERG and YAP1 in human prostate cancer, we performed immunostaining of tissue microarrays (TMAs) with anti-ERG and anti-YAP1 antibodies. Surprisingly, in contrast to the mouse prostate, YAP1 was expressed in the basal, but not in luminal epithelial cells in normal human prostate glands (Figure 5I; Figure S3E-F). Significantly, in human luminal type prostate carcinomas, YAP1 protein was expressed in 55% (64 of 116) of the tumors evaluated (Figure 5I). In the analyzed cohort, 44% (52 of 116) of the cancers also exhibited high ERG expression and there was a highly significant overlap between those primary tumors expressing ERG and those expressing YAP1 (p=0.0002) (Figure 5I). Further, the expression of nuclear YAP1 in primary tumors was significantly associated with tumor recurrence after primary treatment (p=0.0002) (Figure S3F). We conclude that increased YAP1 expression in transformed luminal epithelium may be an important consequence of ERG activity in human prostate cancers.
YAP1 is necessary for ERG-mediated transformation and invasion of ERG-positive human prostate cancer cells
We next analyzed the functional importance of YAP1/TAZ in mediating ERG-associated malignant phenotypes in VCaP cells. Knockdown of ERG or YAP1/TAZ decreased the invasion of VCaP cells (Figure S4A). In addition, suppression of YAP1/TAZ in VCaP cells also decreased their anchorage-dependent and -independent growth (Figure S4B-D). Since ERG is necessary for the maintenance of YAP1 expression in VCaP cells, we analyzed whether restoration of YAP1 rescues phenotypes resulting from ERG suppression. Indeed, re-expression of YAP1 completely rescued the invasion and cell growth phenotypes in VCaP cells transduced with shRNAs targeting ERG (Figure S4E-G). shERG-mediated decrease in the soft-agar colony formation and orthotopic xenograft growth of VCaP cells were significantly, but partially rescued by re-expression of exogenous YAP1, suggesting that the direct control of YAP1 gene expression can account for many, but not all cancer relevant phenotypes of ERG in VCaP cells (Figure S4H-J).
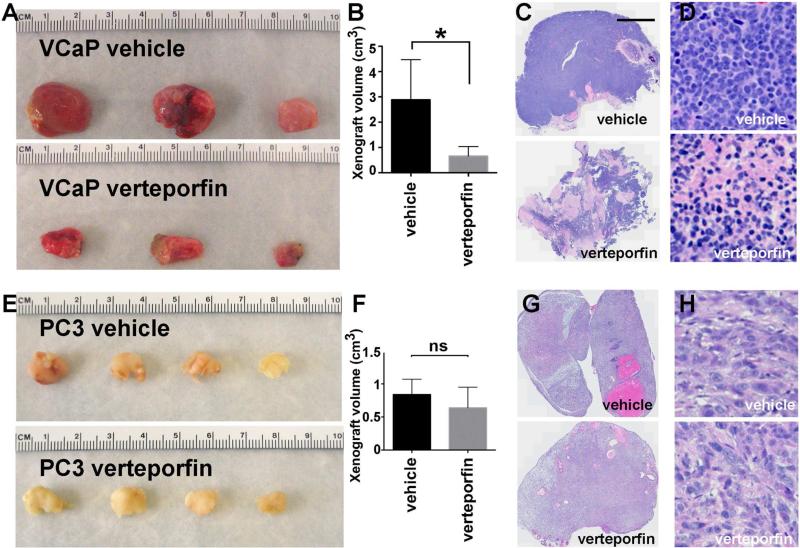
The in vitro studies indicated that YAP1 is responsible for a significant part of oncogenic programs activated by ERG in prostate cancer. To evaluate the in vivo effects of suppressing YAP1 in the context of ERG-driven cancer, we evaluated a small molecule FDA-approved drug Verteporfin, which was recently identified as a specific inhibitor of YAP1 (Liu-Chittenden et al., 2012). We found that the systemic administration of Verteporfin substantially inhibited the growth of pre-established orthotopic ERG-positive VCaP xenografts in vivo (Figure 6A-D). In contrast, Verteporfin treatment did not have statistically-significant impact on the growth of pre-established orthotopic ERG-negative PC3 xenografts (Figure 6E-H). As the direct activity of ERG and other transcription factors are challenging to inhibit in vivo, the suppression of YAP1 by Verteporfin may have clinical utility in treating prostate cancers with ERG rearrangements.
Figure 6. YAP1 inhibitor Verteporfin displays prominent negative impact on the growth of pre-established ERG-positive orthotopic human prostate tumors.
VCaP (ERG-positive) or PC3 (ERG-negative) human prostate cancer cells were injected into anterior lobe of prostate gland of NOD SCID mice and tumors were allowed to establish for 7 weeks without any treatment. Animals were separated into 2 random groups, which were treated with intraperitoneal injection of Verteporfin (or vehicle) every 2 days at 100mg/kg for 3 weeks. Resulting tumors were excised and analyzed.
(A) VCaP gross tumor appearance.
(B) Quantitation of VCaP tumor volume. Data represent mean±SD. Student's t-test.
(C-D) TissueFAX images of Hematoxylin & Eosin staining of VCaP tumor sections.
(E) PC3 gross tumor appearance.
(F) Quantitation of PC3 tumor volume. Data represent mean±SD. Student's t-test.
(G-H) TissueFAX images of Hematoxylin & Eosin staining of PC3 tumor sections. Bar in C corresponds to 2.5 mm in C and G, 50 μm in D and H. See also Figure S4.
Genetic activation of YAP1 in mouse prostate epithelium results in age-dependent development of prostate tumors
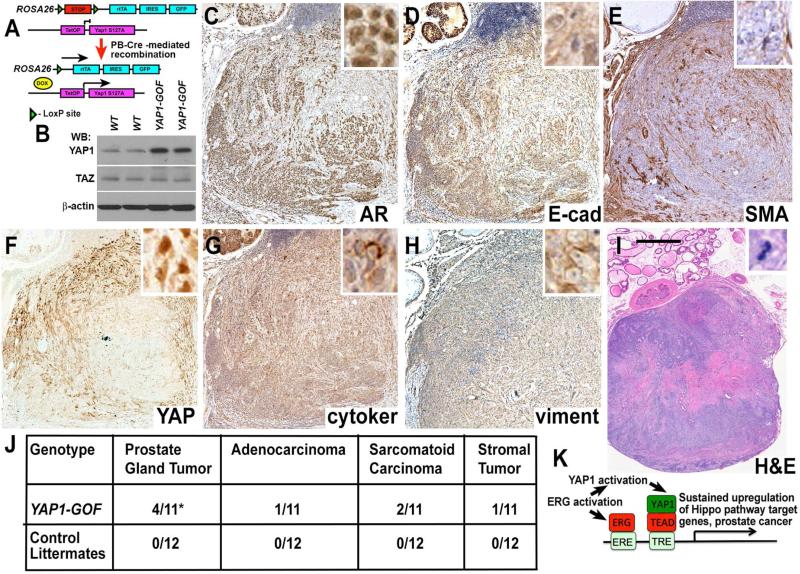
While ERG expression in prostate epithelial cells promotes a YAP1-mediated transcriptional program, the causal effects of activating this program in the context of prostate neoplasia in vivo are not known. To address this question, we utilized transgenic mice carrying a doxycycline (Dox)-inducible allele of constitutively active YAP1 (Yap1S127A) (Camargo et al., 2007). Prostate epithelium-specific Cre mice (Pbsn-Cre4 (Wu et al., 2001)) were utilized for Cre-mediated activation of Dox-dependent transactivator (rtTA) (Figure 7A-B). At three months of age, we exposed these Pbsn-Cre4/Rosa26-LSL-rtTA/TetOP-YAP1S127A (YAP1-GOF) mice to Dox-containing feed and examined their prostate glands at different time intervals thereafter. We found that similar to the prostate-specific ERG transgenic mice, young YAP1-GOF animals presented with minimal phenotypes, while aged YAP1-GOF animals developed prostate tumors with incidence rates and histological phenotypes similar to the prostate gland tumors in Tg(Pbsn- ERG)1Vv mice (Figure 7C-J). These data demonstrate that activation of a YAP1 transcriptional program in mouse prostate epithelium in vivo results in the development of partially penetrant age-related prostate tumors. Further, RNA-Seq analysis of prostate glands from YAP1-GOF mice revealed that transcriptional changes induced by YAP1 activation in prostate epithelium in vivo significantly overlapped with the transcriptional program activated by prostate epithelium-specific expression of ERG (Figure S5A-C, Tables S6-S8). Importantly, analyses of human prostate cancer datasets revealed a highly significant overlap between the murine prostate-gland specific YAP1-GOF gene signature and human ERG-positive prostate carcinomas (Figure S5D-F).
Figure 7. Activation of YAP1 in prostate epithelium in vivo is sufficient for the developmen of age-related prostate tumors.
(A) Model showing the development of prostate epithelium-specific YAP1-GOF mice.
(B) Western blot (WB) analysis of YAP1, TAZ and [.beta]-actin expression in total proteins extracted from ventral prostates of 20 month-old control (WT) and YAP1-GOF mice.
(C-I) Hematoxylin and Eosin (H&E, I) and immunohistochemical (C-H) staining of large prostatic sarcomatoid carcinoma in 14 month-old YAP1-GOF male. Staining with anti-androgen receptor (AR), anti-E-cadherin (E-cad), anti-smooth muscle actin (SMA), anti-YAP1 (YAP), anti-pancytokeratin (cytoker) and anti-Vimentin (viment) antibodies. Bar in I represents 1.4 mm in I, 0.4 mm in C-H, 40 μm in insets.
(J) Prostate tumor incidence and tumor types in YAP1-GOF and control littermate mice. YAP1-GOF-triple mutant Col1a1tm1(tetO-YAP1*)Fcam/Col1a1+, Gt(ROSA)26Sortm1(rtTA,EGFP)Nagy/Gt(ROSA)26Sor+, Tg(Pbsn-cre)4Prb/0. Control - double mutant Gt(ROSA)26Sortm1(rtTA,EGFP)Nagy/Gt(ROSA)26Sor+, Tg(Pbsn-cre)4Prb/0. Numbers denote mice with indicated tumors relative to the number of mice analyzed. The prostate glands were analyzed in mice euthanized at morbidity. The Dox exposure was started at 3 months of age and varied between 12 and 16 months. Two-tailed Fisher's exact test.
(K) Model showing the role of ERG and YAP1 in prostate cancer development. See also Figure S5, Tables S6-S8.
DISCUSSION
Chromosome rearrangements driving high levels of ERG expression occur frequently in prostate cancers, an observation that strongly implicates ETS-family oncogenes as major contributors to the genesis and potentially the progression of this disease. However, genetically engineered mouse strategies that have been quite useful in confirming cause and effect relationships for numerous other oncogenes and tumor suppressor pathways, have not identified clear mechanisms by which ERG contributes to prostate neoplasia. To date, the reported phenotypes resulting from engineered ERG expression in the mouse prostate have been quite modest (Baena et al., 2013; Carver et al., 2009a; Carver et al., 2009b; Chen et al., 2013; King et al., 2009; Klezovitch et al., 2008; Tomlins et al., 2008). One possibility for the lack of oncogenic activity hinges on the level of ERG expression. Dependence of a phenotype on the level of protooncogene expression is frequently seen in transgenic animals (Smith et al., 2006). The synthetic probasin promoter that confers prostate epithelial specificity provides relatively weak levels of prostate-specific ERG expression. For example, while we generated 14 Tg(Pbsn-ERG) founder lines, only 2 lines with highest levels of ERG expression display phenotypic changes and only the line with the highest level of ERG expression, which achieves levels seen in human prostate cancers with ERG rearrangements, develops overt tumors in the prostate gland.
A second consideration regarding prostate cancer phenotypes concerns the age-related dependency of prostate neoplasms. Human prostate cancer is highly associated with advanced age and is very infrequently diagnosed in men under age of 50. Mice do not develop spontaneous prostate cancer during their normal lifespan (Sharma and Schreiber-Agus, 1999). Thus, models that accurately reflect disease pathogenesis may exhibit long periods of latency. Consistent with these observations, the prostate tumors that developed in Tg(Pbsn-ERG)1Vv mice generally occurred between two and three years of age and were focal, involving subsets of epithelial cells with transgene expression. Therefore, even high levels of ERG in mouse luminal prostate epithelial cells are not sufficient to drive the rapid and consistent development of carcinomas, indicating that in addition to ERG expression other genetic or epigenetic changes must occur for frank tumor initiation.
A substantial proportion of the prostate tumors that developed in aged Tg(Pbsn-ERG)1Vv mice are either stromal tumors, which do not express epithelial cell markers, or sarcomatoid carcinomas expressing both epithelial and stromal cell markers. Since we did not detect ERG transgene expression in the prostate stromal cell compartment of Tg(Pbsn-ERG)1Vv mice (Klezovitch et al., 2008), the origin of the stromal tumors in these aged mice is not clear. Tumors expressing mesenchymal cell markers may originate either in stromal cells expressing very low levels of a transgene or in epithelial cells, which then undergo a prominent EMT and lose markers of epithelial differentiation. The presence of sarcomatoid carcinomas expressing both epithelial and stromal cell markers may represent an intermediate EMT phenotype. While sarcomatoid carcinoma is rarely seen in human prostate cancer, these tumors have epithelial origin (Rodrigues et al., 2014). Further, mouse prostate epithelial cells readily undergo EMT and present with sarcomatoid carcinomas, which is seen in mice with deletions of Tp53, or the activation of K-Ras or COUP-TFII in prostate epithelium (Martin et al., 2011; Mulholland et al., 2012; Qin et al., 2013; Vinall et al., 2012).
The generation of a GEM model that recapitulates the levels of ERG found in human prostate epithelium made it possible to establish a connection between ERG and the transcriptional output of the Hippo signaling pathway, which is controlled by YAP1/TAZ transcriptional co-activators. We identified two mechanisms by which ERG regulates Hippo pathway signaling (Figure 7K). First, we found that in prostate epithelial cells ERG binds to TEAD-occupied regions of chromatin, increases histone acetylation at these sites and amplifies the transcriptional output controlled by YAP1/TEAD. We also determined that ERG binds to the YAP1 promoter and is required for YAP1 expression in VCaP human luminal prostate cancer cells, identifying a second mechanism directly connecting ERG and YAP1-dependent regulation of transcription. This second mechanism is not apparent in mouse prostate epithelium and in basal type human prostate epithelial cells, which both already express ample levels of endogenous YAP1. However, it is readily detectable in human luminal type prostate epithelial cells, which do not express YAP1 under normal conditions and require high levels of ERG to drive YAP1 expression.
Mice with prostate epithelium-specific activation of YAP1 develop phenotypes that are remarkably similar to those observed in Tg(Pbsn-ERG)1Vv mice, suggesting that the activation of YAP1-mediated transcriptional output is an important part of ERG function in the prostate gland. While activation of YAP1 in variety of mouse organ systems causes dramatic stimulation of cell proliferation and growth, mouse prostate is resistant to YAP1-mediated transformation, with focal tumors appearing only in aged individuals. We found that, unlike in human prostate epithelium, endogenous nuclear YAP1 is abundantly present in mouse luminal prostate epithelial cells, thus potentially making the mouse prostate resistant to both ERG and YAP1-mediated transformations, since additional increase in YAP1-mediated transcriptional output is unlikely to have a profound physiological impact in this environment.
The finding that ERG and YAP1 are concordantly expressed in a subset of human prostate cancers provides support for the clinical relevance of the molecular interactions between these pathways. The tumors expressing YAP1 generally exhibit an unfavorable disease course and, consequently, YAP1 may serve as a marker of aggressive disease. It is not clear why certain tumors with high ERG levels did not express YAP1, though it is notable that none of such tumors analyzed in our study recurred after treatment. Though ERG is an attractive candidate for therapeutics due to the very high prevalence of ERG rearrangements in prostate cancers, in the past it has been challenging to develop agents capable of disrupting the activity of transcription factors. As YAP1 appears to regulate critical components of ERG-mediated neoplasia, inhibiting YAP1 is an attractive alternative. Our preclinical studies demonstrating substantial reductions in the growth of ERG-expressing prostate cancer by the FDA-approved YAP1 antagonist Verteporfin indicate that malignant phenotypes stemming from ERG activity can be suppressed by targeting YAP1, a finding that opens a more straightforward path for drug development and clinical applications.
Experimental Procedures
Mice
All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committees of Fred Hutchinson Cancer Research Center and followed NIH guidelines for animal welfare. Tg(Pbsn-ERG)1Vv mice (Klezovitch et al., 2008) on 129S1/SvlmJ genetic background were aged and analyzed at morbidity with wild-type littermates used as controls. For the generation of prostate-specific YAP1-GOF mice, double heterozygous Col1a1tm1(tetO-YAP1*)Fcam (Camargo et al., 2007) and Gt(ROSA)26Sortm1(rtTA,EGFP)Nagy (Belteki et al., 2005) mice were crossed with hemizygous mice expressing the prostate epithelium-specific Cre (Tg(Pbsn-cre)4Prb/0) (Wu et al., 2001). Double mutant Gt(ROSA)26Sortm1(rtTA,EGFP)Nagy /Gt(ROSA)26Sor, Tg(Pbsn-cre)4Prb/0 mice were used as controls. YAP1 GOF and control animals were switched to Doxycycline-containing food (www.bio-serv.com, #S3888) at 3 months of age and maintained on this diet until euthanasia. All animals were analyzed as heterozygous (knock-in) or hemizygous (transgene) mutants.
Mouse Prostate Gland Histo-pathology
The histological assessments of prostate tumors in Tg(Pbsn-ERG)1Vv mice was the result of online discussions with the Pathology Panel of NCI Mouse Models of Human Cancer Consortium (MMHCC) Prostate Cancer on 07/02/12, chaired by Prof. Robert D. Cardiff (UC Davis). Prostate gland pathology of YAP1-GOF mice was analyzed by Dr. Maria S. Tretiakova (a board certified pathologist with significant experience with the pathology of the prostate gland).
Gene Expression Analyses
Total RNA was extracted with TRIzol (Invitrogen) followed by RNAse-free DNAase treatment (Qiagen) and RNeasy RNA purification kit (Qiagen). Complementary DNA was prepared with SuperScript III First-Strand Synthesis kit (Invitrogen). qPCR was performed with Prism 7900HT (Applied Biosystems), platinum qPCR mix (Invitrogen), and Universal ProbeLibrary kit using the primers, probes, and PCR conditions recommended by the Universal ProbeLibrary assay center (http://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp). qPCR data were normalized to ribosomal protein Rps16. RNA-seq experiments on RNA from RWPE-1 cells were performed by Beijing Genomics Institute (BGI; Shenzhen, China). The total RNA samples were treated with DNase I, the mRNA was enriched by using the oligo(dT) magnetic beads, fragmented and used for cDNA synthesis. The library products were sequenced via Illumina HiSeq 2000. The primary reads were mapped using Tophat2 (Kim et al., 2013) and differential gene expression was analyzed using EdgeR Bioconductor software. RNA-Seq experiments on RNA from VCaP cells and mouse ventral prostate glands were performed using Encore Complete RNA-Seq Library Systems kit and the Illumina HiSeq 2500 system. The primary reads were mapped using Tophat2 (Kim et al., 2013) and differential gene expression was analyzed using CuffDiff (Trapnell et al., 2010) in Galaxy (Blankenberg et al., 2010). In situ hybridization was performed on frozen sections as described previously (Klezovitch et al., 2004)
Human Prostate Tumor Tissue Microarrays (TMAs) staining and analysis
Informed consent was obtained from all human subjects, and work was performed in accordance with institutional review board (IRB) approval (University of Washington IRB number 39053 and Fred Hutchinson IRB number 460). 127 specimens obtained from radical prostatectomies performed at the UW, including non-recurrent (n=64) and recurrent prostate cancer (n=63). TMAs were made with duplicate cores (1 mm) from paraffin embedded tissues. 4-micron TMA sections were deparaffinized, re-hydrated, rinsed and blocked for endogenous peroxidase on the automated immunostainer (Bond III, Leica Biosystems) using manufacturer recommended protocols and solutions. Then slides were incubated for 15 min with rabbit monoclonal ERG-antibody (Epitomics, clone EP111, 1:400) or rabbit polyclonal anti-YAP1 (Santa Cruz, #sc-15407, 1:100). This step was followed by 8 min with anti-rabbit Poly-HRP-IgG polymer detection reagent (Leica Biosystems), visualization with 3,3″-Diaminobenzidine chromogen, and counterstaining with hematoxylin. The TMA cores were scored by the pathologist (M.T.) blind to cancer outcomes. The tumors displaying positivity in >10% tumor nuclei were defined as either ERG-positive or YAP1-positive.
Statistical Analyses
Statistical significance was determined by Student’s, Fisher's exact, Mann-Whitney or Chi-square tests. p value is indicated by asterisks in the figures: *p < 0.05; **p < 0.01; ***p < 0.001. Differences at p = 0.05 and lower were considered statistically significant. In RNA-Seq and ChIP-Seq analyses, the differences with q<0.05 were considered statistically significant.
Supplementary Material
HIGHLIGHTS.
Upregulation of ERG is sufficient to cause age-dependent prostate tumors in mice
ERG transcriptional output is similar to the output of YAP1 of the Hippo pathway
ERG is necessary for YAP1 expression in luminal type human prostate cancer cells
Activation of YAP1 is sufficient to cause age-dependent prostate tumors in mice
SIGNIFICANCE.
Developing therapeutic approaches to inhibit ERG activity is challenged by a paucity of knowledge about the mechanisms responsible for ERG-mediated transformation. We found that ERG activates the transcriptional output regulated by YAP1 and determined that genetic activation of YAP1 in mouse prostate epithelium is sufficient to induce age-dependent prostate neoplasms that are similar to tumors caused by ERG upregulation. These results reveal an important connection between ERG and the Hippo pathway. Moreover, we demonstrate that pharmacological inhibition of YAP1 activity prominently inhibits the orthotopic growth of pre-established ERG-positive human prostate tumors, supporting the development of clinical strategies targeting YAP1 in tumors with ERG rearrangements.
Acknowledgements
We thank Dr. Robert Cardiff and all the MMHCC Pathology Panel members for help with histopathological analysis of prostate tumors in Tg(Pbsn-ERG)1Vv mice, Dr. Fernando Camargo for the gift of Col1a1tm1(tetO-YAP1*)Fcam/Gt(ROSA)26Sortm(rtTA,EGFP)Nagy mice, the Developmental Studies Hybridoma Bank for generous gift of antibodies, Drs. Robert K. Bradley and Jerry Davison for help with mapping the primary reads from the TCGA PCa dataset and in house NGS, Drs. Robert Vessella, Xiaolong Yang, Elaine Fuchs, Scott Lowe, Patrick Paddison, Julia Segre, Andrew Emili, Julian Kwan, Chuck Bieberich, Charles Sawyers, Celestia Higano, and Bruce Montgomery for gift of plasmids, antibodies and tissue samples. This work was supported in part by an award from the Prostate Cancer Foundation and by NCI grants R01CA176844, P01CA085859 and the Pacific Northwest Prostate Cancer SPORE P50CA097186.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
V.V., M.S.T., P.S.N., I. C., H. B., L.T.N., O.K., M.R.S., L.D.T. and J.L. designed the experiments and analyzed the data. L.T.N., M.S.T., M.R.S., V.V., O.K., J.L., C. M. and V.I.K. performed the experiments. V.V. and P.S.N. wrote the manuscript with all the authors contributing to writing and agreeing with the conclusions presented in the manuscript.
Accession Numbers
Data sets from RNA-seq and ChIP-Seq experiments were deposited at the Sequence Read
REFERENCES
- Baena E, Shao Z, Linn DE, Glass K, Hamblen MJ, Fujiwara Y, Kim J, Nguyen M, Zhang X, Godinho FJ, et al. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev. 2013;27:683–698. doi: 10.1101/gad.211011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Current protocols in molecular biology / edited by Frederick M Ausubel [et al] 2010;10:11–21. doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C, Gopalan A, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. ETS rearrangements and prostate cancer initiation. Nature. 2009a;457:E1. doi: 10.1038/nature07738. discussion E2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009b;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey OM, Fang L, Hynes PG, Abou-Kheir WG, Martin PL, Tillman HS, Petrovics G, Awwad HO, Ward Y, Lake R, et al. TMPRSS2-Driven ERG Expression In Vivo Increases Self-Renewal and Maintains Expression in a Castration Resistant Subpopulation. PLoS ONE. 2012;7:e41668. doi: 10.1371/journal.pone.0041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013 doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N, Rubin MA, Yelick PC, Freeman MR. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. The EMBO journal. 2007;26:4523–4534. doi: 10.1038/sj.emboj.7601872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer research. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nature genetics. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Chevillet J, Mirosevich J, Roberts RL, Matusik RJ, Vasioukhin V. Hepsin promotes prostate cancer progression and metastasis. Cancer Cell. 2004;6:185–195. doi: 10.1016/j.ccr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109:E2441–2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov IV, Vander Griend DJ, Xu Y, Antony L, Dalrymple SL, Isaacs JT. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res. 2006;66:8598–8607. doi: 10.1158/0008-5472.CAN-06-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes & development. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Liu YN, Pierce R, Abou-Kheir W, Casey O, Seng V, Camacho D, Simpson RM, Kelly K. Prostate epithelial Pten/TP53 loss leads to transformation of multipotential progenitors and epithelial to mesenchymal transition. The American journal of pathology. 2011;179:422–435. doi: 10.1016/j.ajpath.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohseni M, Sun J, Lau A, Curtis S, Goldsmith J, Fox VL, Wei C, Frazier M, Samson O, Wong KK, et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014;16:108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer research. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wu SP, Creighton CJ, Dai F, Xie X, Cheng CM, Frolov A, Ayala G, Lin X, Feng XH, et al. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues DN, Hazell S, Miranda S, Crespo M, Fisher C, de Bono JS, Attard G. Sarcomatoid carcinoma of the prostate: ERG fluorescence in situ hybridization confirms epithelial origin. Histopathology. 2014 doi: 10.1111/his.12493. [DOI] [PubMed] [Google Scholar]
- Sharma P, Schreiber-Agus N. Mouse models of prostate cancer. Oncogene. 1999;18:5349–5355. doi: 10.1038/sj.onc.1203037. [DOI] [PubMed] [Google Scholar]
- Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood. 2006;108:653–661. doi: 10.1182/blood-2006-01-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenath TL, Dobi A, Petrovics G, Srivastava S. Oncogenic activation of ERG: A predominant mechanism in prostate cancer. J Carcinog. 2011;10:37. doi: 10.4103/1477-3163.91122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Human pathology. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinall RL, Chen JQ, Hubbard NE, Sulaimon SS, Shen MM, Devere White RW, Borowsky AD. Initiation of prostate cancer in mice by Tp53R270H: evidence for an alternative molecular progression. Dis Model Mech. 2012 doi: 10.1242/dmm.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Shao LJ, Siddiqui J, Palanisamy N, Li R, Ren C, Ayala G, Ittmann M. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer research. 2011;71:1325–1333. doi: 10.1158/0008-5472.CAN-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mechanisms of development. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & development. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature cell biology. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.