Abstract
Exploitation of the zebrafish model in hematology research has surged in recent years, becoming one of the most useful and tractable systems for understanding regulation of hematopoietic development, homeostasis, and malignancy. Despite the evolutionary distance between zebrafish and humans, remarkable genetic and phenotypic conservation in the hematopoietic system has enabled significant advancements in our understanding of blood stem and progenitor cell (HSPC) biology. The strengths of zebrafish in hematology research lie in the ability to perform real-time in vivo observations of hematopoietic stem, progenitor and effector cell emergence, expansion and function, as well as the ease with which novel genetic and chemical modifiers of specific hematopoietic processes or cell-types can be identified and characterized. Further, a myriad of transgenic lines have been developed including fluorescent reporter systems to aid in the visualization and quantification of specified cell types of interest and cell-lineage relationships, as well as effector lines that can be used to implement a wide range of experimental manipulations. As our understanding of the complex nature of HSPC biology during development, in response to infection or injury, or in the setting of hematological malignancy, continues to deepen, zebrafish will remain essential for exploring the spatio-temporal organization and integration of these fundamental processes, as well as the identification of efficacious small molecule modifiers of hematopoietic activity. In this review, we discuss the biology of the zebrafish hematopoietic system, including similarities and differences from mammals, and highlight important tools currently utilized in zebrafish embryos and adults to enhance our understanding of vertebrate hematology, with emphasis on findings that have impacted our understanding of the onset or treatment of human hematologic disorders and disease.
Introduction
While the zebrafish (Danio rerio) was first suggested for use in hematology research in 19631, it is only in the last ~20 years that it has truly risen to prominence as one of the preeminent systems for hematopoietic stem cell (HSC) biology, particularly in the areas of development and regeneration. The zebrafish has emerged as a highly tractable model system for scientific research due in large part to the external fertilization of embryos and their optically clear development, allowing for real-time in vivo observation of developmental processes. Additionally, the ability of fecund females to lay hundreds of embryos per week enables rapid high-throughput experimentation and strong statistical analysis of phenotypes. Zebrafish are particularly useful for hematology research due to the high conservation of genetic factors regulating blood development as well as the structure and function of hematopoietic cell types, and the ability to visualize circulating erythrocytes with only a dissecting microscope.
Hematopoiesis is Highly Conserved in the Zebrafish Model
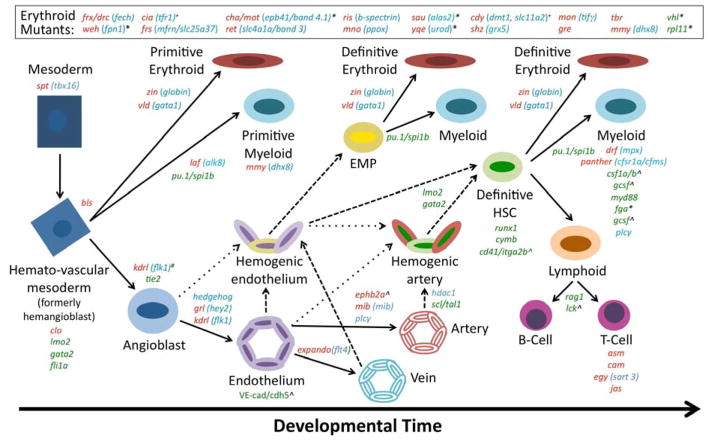
As in all other vertebrates analyzed to date, zebrafish hematopoiesis occurs in multiple phases (Figure 1). Primitive hematopoiesis, the first wave of blood development, occurs from ~12 to 24 hours post fertilization (hpf) in two anatomically distinct locations: a section of posterior lateral mesoderm called the inner cell mass gives rise primarily to cells of erythroid lineage2, while the rostral blood island in the anterior portion of the embryo gives rise to a primitive macrophage population3,4. More recent analysis have also suggested that neutrophils and thrombocytes are produced during the window of primitive hematopoiesis; however, their cellular origins and lineage relationships to the primitive erythrocyte and macrophage populations are currently unclear5,6. The process of erythropoiesis requires many of the same genes that are utilized during primitive hematopoiesis in other vertebrate species including scl7, gata18, and lmo29 while the generation of myeloid cells requires pu.1 and cebp110–12. A transient wave of definitive hematopoietic progenitors has also been recently identified, termed erythromyeloid precursors (EMPs), which are present in the embryo prior to the emergence of true multi-lineage HSCs13. These progenitors give rise to both definitive erythroid and myeloid (neutrophilic granulocytes, monocytes, and macrophage) colonies in culture. However, EMPs were never observed to populate the kidney marrow or thymus in vivo, indicating a lack of lymphoid potential and supporting their classification as hematopoietic progenitors rather than HSCs. Through a process which appears highly conserved among vertebrate species, definitive HSCs arise from hemogenic endothelium in the ventral wall of the dorsal aorta in the Aorta-Gonad-Mesonephros (AGM) beginning at ~30 hpf in zebrafish14,15, shortly after the onset of circulation16. Following their emergence in the AGM, HSCs migrate to the Caudal Hematopoietic Tissue (CHT), currently thought to be the maturational equivalent of the mammalian fetal liver, where phenotypically labeled cells expand in number. HSPCs subsequently seed the thymus and kidney marrow, the primary sites of adult hematopoiesis in the fish beginning at 3.5–4 dpf 17.
Figure 1. Models for investigating embryonic hematopoiesis in the Zebrafish.
Schematic representation of embryonic hematopoietic development showing lineage relationships between the hemato-vascular populations and available zebrafish mutant lines. Red type indicates mutants derived from forward genetic ENU-screening, with blue font illustrating the genetic lesion; green font represents targeted mutants. An * denotes mutants which recapitulate the disease phenotype associated with human gene mutation(s). A ^ indicates a targeted mutation in a conserved gene that has not yet been phenotypically validated. A # indicates a series of genetic alleles are available. A + notes mutants which exhibit an early embryonic phenotype but are viable as adults.
Many of the genetic factors that specify the emergence of definitive HSCs are well-elucidated, and are largely consistent with those required in mammalian systems, indicating a high degree of evolutionary conservation (Figure 1). A common cascade of transcription factors including sonic hedgehog, followed by Vascular Endothelial Growth Factor (VEGF)18,19, and Notch20,21 regulates the development of both arterial-specified endothelium as well as HSCs, reflecting the spatial emergence of HSCs from the ventral endothelial wall of the dorsal aorta in the developing embryo14,15. These factors lie upstream of both scl22 and the transcription factor runx1, also a key regulator of HSC development, in the fish23. In contrast, other signaling pathways appear to modulate scl/runx1+ HSC formation without impacting specification of the artery: both BMP-24 and Wnt-signaling25,26 appear to act in parallel to or intersect with the HH/VEGF/Notch cascade, indicating the process of HSC specification within a specific endothelial population is not a singular linear pathway, but the integrative activity of several regulatory cascades.
Not only are the genetic factors regulating HSC emergence maintained across species, the function of the different blood lineages appears to be highly conserved as well. Although zebrafish erythrocytes remain nucleated throughout their lifespan, they express the same globin genes that are found in mammals27, indicative of a similar function. Zebrafish also contain thrombocytes (platelets)28 that, as in mammals, play a role in blood clotting29. The cellular components of the innate immune system are also highly conserved; zebrafish contain granulocytes30 as well as macrophages4 and neutrophils31 in the myeloid lineage. Migration of myeloperoxidase+ granulocytes toward sites of injury and inflammation can be readily observed in vivo, confirming that these cells maintain their anti-inflammatory properties in the zebrafish30; similarly, macrophages possess the ability to phagocytose apoptotic cells and bacteria4. Conservation of myeloid function as well ex vivo development has also allowed exploitation of zebrafish as a model for infectious disease: in particular, the progression of tuberculosis, which is difficult to model in mice, has been demonstrated to be recapitulated in zebrafish embryos after infection with mycobacterium; embryos show development of macrophage aggregates and increased expression of Mycobacterium-associated genes32. Zebrafish also appear to possess a full component of cells of the adaptive immune system. Recombination Activating Gene (rag) expressing T-lymphocytes are found in the thymus as early as 3.5dpf by in situ hybridization33–35; B-cells have similarly been reported, although their site of maturation remains controversial36,37 and Natural Killer cells are also thought to exist38,39. Recent studies have indicated B cells possess similar function including proliferation in response to antigen exposure37. Zebrafish have recently been reported to possess dendritic cells that uptake antigens and stimulate T-cell proliferation, indicating conservation of function40. In sum, the high conservation of both genetics and function of the hematopoietic cells in zebrafish lends validity to their use in hematology research.
Genetic Manipulation in the Zebrafish Model
Beyond the strong conservation of factors regulating HSC development in the zebrafish, the system is also highly amenable to genetic manipulation, making it useful for identification of novel regulatory factors, as well as gain and loss-of-function studies. While homologous recombination akin to murine models remains difficult to achieve, knockdown of a given gene(s) in the zebrafish system is readily performed using morpholino technology. Morpholinos (MOs) are modified antisense oligonucleotides designed to block either splicing or translation of a specific mRNA, thereby resulting in a knockdown of function of the target gene due to a lack of translation. MOs are microinjected into zebrafish embryos at the 1-cell stage and they remain effective for the first ~5 days of development (depending on the dose)41, making the technology useful for its unique ability to rapidly assess the effects of loss of a given gene on vertebrate hematopoietic development. Other significant benefits of the MO system include the ability to titrate the dose of MO injected, making it possible to carefully control the degree of knockdown, often allowing one to bypass embryonic lethal mutations, as well the ease with which suppression of multiple genes can be examined at once via combinatorial injection. However, there are caveats to the use of MOs, including potential toxicity of the MO and/or injection procedure, off-target effects, and the inability to look at the consequences of long-term loss of function of a given gene(s). More recent technologies, including zinc finger nucleases, TALENS and the CRISPR/CAS system, have emerged as promising techniques to make specific, germline-transmissible mutations of genes of interest42–45. These techniques make it possible to generate full knockouts (null phenotypes) relatively quickly and will enable better assessment of genetic null phenotypes in the embryo as well as the adult, when MO technology is generally no longer useful. In addition to MO titration, zebrafish are also amenable to use of a variety of inducible promoter-driven transgenes including: heat shock inducible promoters46, the cre-lox system47,48, and the Gal4;UAS system49, that enable induction of gene expression at a specific time or location. More recently, tissue-specific ablation has also been achieved by driving expression of nitroreductase under a relevant promoter, followed by treatment with metronidazole during a time window(s) of interest50.
The previously discussed benefits of the zebrafish system including external transparent development and large clutch size have historically enabled the isolation of many hematopoietic mutant lines through large-scale mutagenesis screens. Significantly, zebrafish embryos are able to survive for up to 10 days in the absence of blood flow and heartbeat, enabling identification and analysis of mutations that would be impossible to study in mammalian systems due to early embryonic lethality51; additionally, as the liver is not a site of embryonic hematopoiesis17, mutations affecting the gastro-intestinal system will not necessarily impair hematopoietic development. One of the most common methods to introduce mutations into the germline of zebrafish involves the use of N-ethyl N-nitrourea (ENU)52–54, a mutagen which results in the induction of point mutations. The majority of the hematopoietic mutants isolated from the first ENU screens involved deficits in erythrocyte function (Figure 1). These mutants included well known master erythrocyte regulators, including gata1 in the vlad tepes line8 and globin in zinfandel zebrafish55, confirming the high conservation of hematopoietic regulation across species. More importantly, however, some of the mutations led to the identification of novel factors involved in erythrocyte maturation and function such as ferroportin56 and mitoferrin57; mutations in the ferroportin gene have subsequently been identified as one of the most common causes of iron overload in humans58, making it a prime example of the utility of zebrafish screening for the identification of causal genes relevant to human disease. The zebrafish is also an excellent model for several different forms of anemia, including Hereditary Elliptocytosis, Microcytic Anemia, and Congenital Dyserythropoietic Anemia, making them potentially useful for the identification of compounds or genes that alter the severity of anemic phenotypes59–61. Although the zebrafish genome was duplicated in recent evolutionary history62, resulting in partial redundancy, this can also be employed as a strength as the two paralogs of the duplicated gene often have similar but not identical expression and function.
Interestingly, zebrafish embryos can survive for several days as a haploid organism. As a result, UV inactivated sperm can be used to fertilize the eggs of females from the F1 generation of an ENU screen, thereby resulting in the formation of haploid embryos. Recessive phenotypes are visible in the F1 generation of haploid embryos, as they are not masked by the wild type copy of the mutated allele, thereby allowing assessment of phenotypes one generation prior to what would normally be possible in traditional zebrafish or mammalian screens63. This technique has revealed genes that are important in hematopoiesis including T-cell development64 and vasculogenesis, a process intimately tied to HSC specification and emergence65. Other methodologies, referred to as TILLING (Targeted Induced Local Lesions in Genomes) took advantage of the ease of ENU mutagenesis, combined with recent advances in sequencing and cloning of the zebrafish genome to identify specific mutations in hematopoietic regulatory factors of interest, such as rag166. Another common method of inducing mutations in the zebrafish genome involves retroviral insertional mutagenesis67,68, whereby retroviral DNA is randomly integrated into the genome, resulting in disruption of gene expression and/or function. Analysis of zebrafish mutants from one such insertional mutagenesis screened aided in the identification and sequential organization of a hierarchy of factors required for HSC emergence in the developing vertebrate embryo including VEGFAa, notch, hdac1, and runx169. Transposon-mediated gene mutation is also available in zebrafish70, although it is occasionally noted to exhibit integration biases71.
As discussed above, one of the most critical genes in the specification of HSCs during development is runx1. When runx1 was mutated in zebrafish, embryos failed to undergo AGM definitive hematopoiesis and showed an absence of definitive lineages at 5 days post-fertilization72. While most null animals died between days 11 and 20 post-fertilization, a small fraction survived until adulthood. The reasons for this are not well known, but could be due to the presence of normal erythromyeloid progenitors that fulfill some of the roles of HSCs enabling a portion of the null animals to survive. In addition to runx1, the gene cmyb is recognized as a conserved HSC marker73,74. cmybt25127 mutant zebrafish embryos show an absence of definitive hematopoiesis but again are able to survive into adulthood; it is believed this is likely to be due to diffusion of oxygen into the fish from the tankwater, making the requirement for oxygen-carrying erythrocyte capacity less critical75. However, an alternative cmyb mutant line is reported to be lethal by ~10dpf due to a failure in HSPC migration from the AGM76, suggesting that cmyb is indeed required for definitive hematopoietic development in the zebrafish. As the cmyb knockout mouse dies at E1573 prior to birth, direct comparisons of the zebrafish mutant lines may be particularly useful for analyzing the effects of varying degrees of loss of cmyb function on hematopoiesis during later developmental stages.
One of the most enigmatic mutants isolated in the large-scale ENU screens of the 1990s was cloche77. These embryos show broad hematovascular defects, with a strong reduction in the formation of both the blood and vasculature, as well as heart defects. cloche zebrafish are considered to be one of the strongest pieces of evidence for the “hemangioblast”, a theoretical bipotential cell population that supports hematopoietic and vascular development. While many attempts to clone the cloche gene have been made78,79, the identity of the exact genetic mutation underlying the cloche phenotype remains unknown, as loss of any single gene has been unable to replicate all the phenotypes observed in cloche mutant embryos. Beyond its location in a telomeric region, the difficulty of cloning cloche, which is recessive77, raises the intriguing possibility that the mutation may lie an intergenic or non-coding region, such as a miRNA or lncRNA that has the ability to impact the transcription or translation of multiple genes. While simply speculation, a mutation in one of these pleiotropic regulatory regions may explain why the cloche phenotype is so penetrant, leading to defects in multiple aspects of hemato-vascular development.
Screens to identify modifiers of hematopoietic phenotypes have also recently been performed using zebrafish; one of these screens utilized a MO-based “knockdown” approach to identify epigenetic factors with specific regulatory roles in hematopoiesis. This relatively high-throughput reverse-genetics screen resulted in the identification of several families of epigenetic regulators that impact either primitive or definitive hematopoiesis, or both, and provides insight into the role of epigenetic regulation in establishing hematopoietic commitment and expansion80. Zebrafish were also used in a genetic modifier screen to look for suppressors of the moonshine mutant, which has defects in erythropoiesis; this screen resulted in the identification of transcriptional elongation as a regulator of cell fate81. Together, these studies suggest that the zebrafish will continue to be useful for identifying novel regulatory factors that modify known hematopoietic phenotypes.
Real-time Imaging of Hematopoietic Stem Cell Emergence During Development
The strong genetic conservation in zebrafish has also enabled engineering of many transgenic reporter lines that express fluorescent proteins under the control of promoter regions of given hematopoietic genes of interest (Figure 2). These transgenic lines have been especially useful for quantifying alterations in hematopoietic cell number, particularly given the lack of cross-reactivity with many existing mammalian antibodies, and for imaging the emergence and migration of HSCs during development in vivo in real time, a process which is extremely difficult in mammalian systems 14,15,23. While many primitive hematopoietic and vascular reporter lines emerged in the late 1990s and early 2000s including gata182, lmo283, and fli1a84, the first transgenic HSC-reporter line to be developed was the Tg(-6.0itga2b (CD41):EGFP) line, which is commonly used to visualize HSCs after their emergence from the AGM28,85, particularly in the CHT and kidney marrow. Importantly, while also expressed at high levels on thrombocytes, the utility of CD41 as a blood stem cell marker appears to be conserved as it has also been reported to be present on HSCs during mammalian development86. Fluorescent reporters for runx187 and cmyb74 were subsequently created.
Figure 2. Optical clarity and transgenic tags allow in vivo visualization of hematopoiesis in zebrafish embryos.
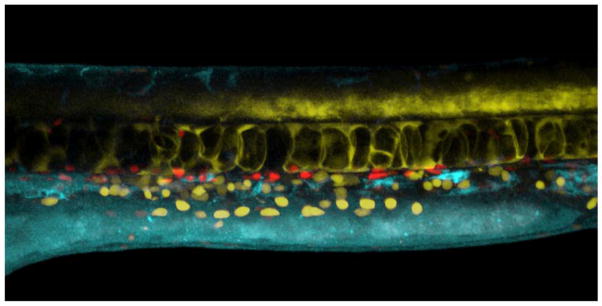
Trunk region of a live zebrafish embryo at 24hpf expressing cmyb:GFP (pseudocolored yellow), tp1:nuclear-mcherry (red), and flk1:ntr-cerulean (blue) (Unpublished image from A. Kim and D. Traver (UCSD); unpublished flk1:ntr-cerulean line from N. Chi (UCSD).
Over the past several years, elegant time-lapse imaging studies have enabled real-time, in vivo visualization of HSC emergence from hemogenic endothelium in the AGM. One recent study demonstrated the existence of hemogenic endothelium using an scl β-driven GFP construct and confirmed that scl is required for the birth of HSCs in the AGM88. Other studies visualized the birth of HSCs from hemogenic endothelium. Co-expression of cmyb and flk1 transgenes was shown by lineage tracing to mark budding HSCs in the vasculature14 and has been used to enable precise quantification and visualization of HSCs in the embryo. Simultaneously, time-lapse imaging of embryos expressing GFP under the control of the flk1 transgene was utilized to visualize the process of budding and egress of HSCs from the endothelium into the vasculature, now termed endothelial-to-hematopoietic transition (EHT)15. These studies both lent strong support to the theory that HSCs arise from a subset of endothelial cells, termed hemogenic endothelium, found in the vasculature, and the dorsal aorta in particular. This argument was further bolstered by visualization of runx1 transgenic reporter fish which indicated that runx1 was expressed in the AGM and in particular in the ventral wall of the dorsal aorta87, consistent with its expression in hemogenic endothelium in mammalian models89. The role of runx1 in EHT in zebrafish was functionally demonstrated in vivo via MO injection, as runx1 knockdown resulted in a lower rate of budding initiation, as well as cell disruption upon attempting to exit the aortic wall15, a finding consistent with observations from murine models 90. Interestingly, both zebrafish imaging studies indicated that HSCs, upon leaving the endothelium, enter into circulation via the vein rather than the artery, a process that appears to be unique in the fish14,15; the explanation for this difference could potentially result from the overlapping rather than continuous structure of the venous endothelium that enables easier intravasation of cells91 or may simply reflect the compact spatial alignment of the major trunk vessels in zebrafish compared with that of mammalian embryos, allowing local chemoattractants from the vein to reach newly produced HSCs immediately upon egress rather than in the circulation. In addition to in vivo documentation of the emergence of HSCs performed in the zebrafish system, concurrent studies using explant cultures have indicated a very similar process occurs in the mouse92, suggesting that this series of events is highly conserved. Transgenic HSC reporter lines can also be used to visualize activity in secondary sites of hematopoiesis including the CHT, thymus, and kidney, enabling their use for lineage tracing as well as analyzing migration, colonization and expansion of HSCs87,93. Photoconvertible lineage tracing can also be used to track progeny of a given cell of interest. For instance, use of a photoconvertible Notch Kaede reporter Tg(EPV.Tp1-Mmu.Hbb:Kaede) demonstrated that Notch signaling plays at least two temporally distinct roles in HSC development26. In the adult, use of GFP+ donor cells has enabled quantification of lineage contribution following kidney marrow transplantation, aiding monitoring and assessment of engraftment kinetics and efficiency94. In addition to genetic profiling of labeled embryonic blood populations79, our recent study has also indicated that transgenic lines (for example CD41) can be utilized to enrich for embryonic HSCs with adult kidney marrow repopulating activity by FACS sorting95. Together, these studies confirmed the in vivo functional potential of the labeled populations of interest and provide an additional resource for the prospective isolation and analysis of factors regulating HSCs during vertebrate development.
In Vivo High-Throughput Small Molecule Screening in the Zebrafish
Due the to the large numbers of embryos available from pair-wise mating(s) and the external development of the fish, it is relatively easy to do large-scale chemical screening for small molecule modifiers in zebrafish by simple addition of “test” compounds to the fish water. The first chemical screen was published in 2004 and was aimed at the identification of novel suppressors of the gridlock (grl) phenotype, caused by a mutation in hey2, that leads to aortic coarction and defects in arterial/venous development96. The compound discovered, GS4012, was a potent inducer of VEGF and was the first ”therapeutic” discovery in zebrafish, resulting in a reduction of the severity of a genetic disorder impacting the hematovascular system.
The first hematopoietic screen used the Tg(gata1:GFP) line to identify modifiers of primitive hematopoiesis97; subsequent screens have revealed other novel modifiers of both primitive and definitive hematopoiesis16,74,98. One screen hit that has shown direct therapeutic potential across species was Prostaglandin E2, which robustly increased the number of runx1+ HSCs in zebrafish and mice74, in part through modulation of the Wnt pathway25; following translational work using human cells99, PGE2 became the first compound derived from drug discovery in the zebrafish to reach FDA-approved clinical trials testing in human patients100. Other significant discoveries made via the chemical screening approach yielded compounds which could induce hematopoietic differentiation in the AML-ETO model of leukemia101,102. A subsequent screen also identified compounds that were specifically toxic to immature T-cells, such as Lenaldekar, a potential chemotherapeutic agent for patients with T-cell Acute Lymphoblastic Leukemia (T-ALL)103. While in vitro screening of cell populations of interest is certainly a valuable methodology for therapeutic compound identification, in vivo screening using the zebrafish model has the advantage of identifying drug toxicity “side effects” from the outset, as well as allow for production and evaluation of any compound metabolites that may be produced, boosting the translational potential of screening hits and speeding the time from discovery to application.
Adult Hematopoietic Assays Adapted for Use in the Zebrafish Model
While the bulk of the hematopoietic studies done in zebrafish have been performed using embryos, a growing number of techniques to study HSC biology in the adult animal are emerging. One of the primary challenges of working with zebrafish to study hematopoiesis in the adult is the lack of antibodies for cell surface markers to enable FACS analysis of HSCs and the different hematopoietic lineages. The first study to identify a simple way to isolate whole kidney marrow (WKM; the site of adult hematopoiesis) and analyze the cellular content of the marrow relied upon flow cytometry using cell size (forward scatter) and granularity (side scatter) to isolate the various lineage fractions104; while a similar technique has also been used in the mouse105, this was a huge advancement for studying hematopoietic cell production and homeostasis in zebrafish. Subsequent studies furthered the functional utility of the WKM dissection and analysis by introducing irradiation-mediated ablation of the hematopoietic system to both enable assessment of KM recovery after injury and to perform adult-to-adult HSC transplantation106, long considered the best technique to determine the presence of a true multipotent hematopoietic stem cell. Transplantation methodology was further enhanced by the generation of casper fish, which lack pigmentation, enabling real-time in vivo visualization of HSC homing and engraftment: for example, injection of beta-actin:GFP WKM donor cells in irradiated casper recipients enabled imaging of the donor cells from 2 hours until at least 5 weeks after transplantation107. Functional potential, including lineage tracing, can also be assessed by transplantation of donor stem cells that carry pan-hematopoietic (CD45) markers95. These methodologies have significant advantages over homing and expansion assays in mammalian systems where real-time visualization of is difficult due to the deep interior location of the bone marrow niche. Recent studies focused on the role of the Major Histocompatibility Complex (MHC) have further refined the transplantation protocol and indicated that matching the MHC type between donor and recipient can drastically improve rates of engraftment94. Regenerative assays using these techniques have identified several factors, including Notch signaling and PGE2/Wnt, which accelerate hematopoietic recovery after marrow injury, providing clinically relevant insight into the engraftment process following stem cell transplantation and strengthening the connection between regulators of embryonic development and tissue regeneration20,25. Importantly, while PGE2 was initially identified as a modifier of HSC regeneration in the zebrafish, it was simultaneously found to play a similar role in the mouse74, emphasizing the utility of the zebrafish to identify critical pathways in HSC regulation across vertebrates; the Notch pathway has likewise showed utility in transplantation therapies across species108. Although zebrafish cells in general have traditionally been difficult to maintain in vitro, recent studies indicate that long-term hematopoietic culture of isolated kidney marrow stem and progenitor cells is possible, as both erythroid and myeloid colonies were generated in a clonal methylcellulose assay109 following identification and isolation of zebrafish specific cytokines such as Erythropoietin (Epo) and Granulocyte Stimulating Colony Factor (GCSF). This advancement opens further avenues of functional analysis and manipulation of hematopoietic stem and progenitor cell biology in the zebrafish. Finally, while not yet examined in detail, the localization of the HSC niche within the kidney marrow rather than bone marrow niche, may itself prove advantageous to elucidation of cell autonomous and/or bone independent regulatory effects on HSCs and hematopoietic progenitors.
One of the most prominent areas of zebrafish hematology research is currently in cancer biology, particularly that of T-ALL, which was the first model of cancer in the fish110 and is the most frequent childhood hematological malignancy111. While the zebrafish has not been a long-standing cancer model, it is quickly gaining traction due to the ease of chemical and genetic screening for modifiers of cancer development or progression110,112. Recent studies have led to the identification of the COX/β-catenin102,113, PTEN114, S1P1 and ICAM1 pathways115 as playing key roles in cancer development and progression using zebrafish models of hematologic malignancy. In addition to the cyclooxygenase inhibitors mentioned above102 for the treatment of AML1-ETO associated AML, screening for chemical modifiers of T-ALL progression also led to the identification of perphenazine, an antipsychotic, as an inducer of apoptosis in malignant cells; importantly, this was true in zebrafish, mouse, and human T-ALL cells116, indicating the usefulness of the fish for identifying drugs that can impact the course of human cancer.
Concluding Remarks
Over the last 20 years, the use of zebrafish in the field of hematology has undergone a transformation from being an interesting alternative model to being one of the preeminent and tractable vertebrate systems for hematological studies. The amenity of the zebrafish to genetic manipulation together with the strong evolutionary conservation of the genetic regulation of hematopoietic development, homeostasis, and regeneration makes it a particularly useful system for blood research. However, some weaknesses with the zebrafish model system remain, including the lack of antibody cross reactivity with common cell surface markers for FACS, Western blots and immunohistochemistry104, differences in the anatomical location of some aspects of hematopoiesis and the evolutionary duplication of a portion of the zebrafish genome, resulting in genes with redundant functions. Nevertheless, great progress is being made in zebrafish biology to help alleviate, or even turn these concerns into strengths (for example, bone-independent HSC regulation), and to ensure the fish remains a useful system for future studies. One of the greatest advantages of the zebrafish system lies in its ability to act as an evolutionary intermediate between the more longstanding invertebrate animal models, such as Drosophila, and that of the mouse. The zebrafish maintains many of the advantages of lower organisms, including rapid external development and strong utility for forward genetic screening, while also being a vertebrate, with organ development and regeneration evolutionarily closer to humans at both the cellular and genetic level. Together, the zebrafish has many qualities that make it an extraordinarily useful model system to discover novel hematological findings that will continue to enhance our knowledge of basic HSC biology as well as provide key insights into human health and disease. The zebrafish is likely to be particularly adept in helping to delineate factors that regulate the developmental niche versus those that directly impact HSCs through the use of tissue and lineage specific promoters as well as real-time in vivo imaging, both of which are difficult to achieve in a mammalian system. One of the major goals of both the stem cell and regenerative biology fields has been the generation of a transplantable HSCs from either embryonic or iPSC sources for use in clinical applications. However, this goal has thus far proven elusive, suggesting that there is much more to be understood about the in vivo mechanisms that integrate to specific HSCs. Finally, the utility of the zebrafish for chemical screening in both a developmental and disease context is likely to continue to be one of the most useful aspect of the model system and will lead to the isolation of novel compounds that can bolster our understanding of embryonic hematopoiesis, as well as the onset and progression of hematological malignancies, and help identify potential therapeutics to aid in the treatment of human blood diseases.
Table 1.
Mutations in zebrafish hematopoietic and vascular genes shown in Figure 1 are detailed here. Reference listed is the first to clone the mutation. If the given mutation is unconfirmed, the reference is for the hematovascular expression of the conserved gene.
| Gene Name | Mutant (abbreviation) | Original Isolation Method | Cloning/Expression Reference |
|---|---|---|---|
| (unknown) | cloche (clo) | ENU screen | (Stainier et al., 1995) |
| (unknown) | bloodless (bls)/sort-of-bloodless (sob) | Insertional Mutagenesis | (Liao et al., 2002) |
| (unknown) | grenache (gre) | ENU screen | (Ransom et al., 1997) |
| (unknown) | thunderbird (tbr) | ENU screen | (Ransom et al., 1997) |
| (unknown) | assam (asm) | ENU-EP screen (targeted: T-cells) | (Trede et al., 2008) |
| (unknown) | camomile (cam) | ENU-EP screen (targeted: T-cells) | (Trede et al., 2008) |
| (unknown) | jasmine (jas) | ENU-EP screen (targeted: T-cells) | (Trede et al., 2008) |
| (unknown) | oolong (oln) | ENU-EP screen | (Trede et al., 2008) |
| activin A receptor, type 1 like (acvr1l; alk8) | lost-a-fin (laf) | ENU screen | (Bauer et al., 2001) |
| cadherin5/VE-Cadherin (cdh5) | (Helker et al., 2013) | ||
| colony stimulating factor 1a/b (csf1a/b)/macrophage colony stimulating factor (mcsf1/2) | Unconfirmed | csf1a (ENU screen); csf1b (Viral insertion) | (Wang et al., 2008) |
| colony stimulating factor 3 (csf3)/granulocyte colony stimulating factor (gcsf) | Unconfirmed | ENU screen | (Liongue et al., 2009) |
| csfr1a/cfms | panther | ENU screen | (Parichy et al., 2000) |
| dhx8 | mummy (mmy) | ENU screen | (English et al., 2012) |
| divalent metal transporter 1 (dmt1)/slc11a2 | chardonnay (cdy) | ENU screen | (Donovan et al., 2002) |
| epb41b/band4.1 | chablis (cha)/merlot (mot) | ENU screen | (Shafizadeh et al., 2002) |
| ephrin b2a | Unconfirmed | ENU screen | (Lawson et al., 2001) |
| ets variant gene 2 (etv2) | ENU screen | (Pham et al., 2007) | |
| ferrochetalase (fech) | frx/dracula (drc) | ENU screen | (Childs et al., 2000) |
| ferroportin1 (fpn1) | weissherbst (weh) | ENU screen | (Donovan et al., 2000) |
| fibrinogen, alpha chain (fga) | Unconfirmed | ENU screen | (Vo et al., 2013) |
| fli1a | (Liu and Patient, 2008) | ||
| flt4 | expando | ENU screen | (Hogan et al., 2009) |
| gata1a | vlad tepes (vld) | ENU screen TILLING |
(Lyons et al., 2002) |
| gata2a | Zinc-finger nuclease | (Zhu et al., 2011) | |
| globin (locus control region) | zinfandel (zin) | ENU screen | (Brownlie et al., 2003) |
| glutaredoxin 5 (grx5) | shiraz (shz) | ENU screen | (Wingert et al., 2005) |
| hairy/enhancer of split related with YRPW motif 2 (hey2) | gridlock (grl) | ENU Screen | (Zhong, 2000) |
| histone deacetylase 1 (hdac1) | ascending and descending (add)/colgate (col) | ENU Screen; retroviral insertion | (Nambiar et al., 2007; Yamaguchi et al., 2005) |
| IGM | Unconfirmed | Retroviral | (Danilova et al., 2005) |
| ikaros (ikzf1) | ENU screen | (Schorpp et al., 2006) | |
| integrin, alpha 2b (itga2b/CD41) | Unconfirmed | ENU Mutagenesis | (Ma et al., 2011) |
| kdrl/fetal liver kinase 1 (flk1)/VEGF receptor 2 (VEGFR2) | ENU screen | (Covassin et al., 2006) | |
| LIM domain only 2 (lmo2) | ENU screen (targeted) | (Weiss et al., 2012) | |
| lymphocyte specific protein tyrosine kinase (lck) | Unconfirmed | ENU Mutagenesis | (Murayama et al., 2006) |
| mindbomb | mindbomb (mib); white tail (wit) | ENU screen | (Itoh et al., 2003) |
| mitoferrrin/slc25a37 | frascati (frs) | ENU screen | (Shaw et al., 2006) |
| myeloid differentiation primary response gene 88 (myd88) | TILLING | (van der Vaart et al., 2013) | |
| myeloperoxidase (mpx/mpo) | durif (drf) | ENU screen | (Pase et al., 2012) |
| phospholipase C, gamma 1 (plcγ1) | deadbeat (ded) | 2004: retroviral insertion; 2005 and 2009: ENU screen | (Amsterdam et al., 2004; Covassin et al., 2009; Rottbauer et al., 2005) |
| protoporphyrinogen oxidase (ppox) | montalcino (mno) | ENU screen | (Dooley et al., 2008) |
| pu.1/spi1b | TILLING | (Jin et al., 2012) | |
| recombination activating gene 1 (rag1) | TILLING | (Wienholds et al., 2002) | |
| ribosomal protein L11 (rpl11) | Retroviral Insertion | (Danilova et al., 2011) | |
| runt-related transcription factor 1 (runx1) | TILLING | (Sood et al., 2010) | |
| slc4a1a/band3 | retsina (ret) | ENU screen | (Paw et al., 2003) |
| sonic hedgehog (shha) | sonic you (syu) | ENU Screen | (Schauerte et al., 1998) |
| squamous cell carcinoma antigen recognized by T-cells 3 (sart3) | earl grey (egy) | ENU-EP screen (targeted: T-cells); | (Trede et al., 2007) |
| T-box gene 16 (tbx16) | spadetail (spt) | ENU screen | (Griffin et al., 1998) |
| T-Cell acute lymphocytic leukemia 1 (tal1)/stem cell leukemia (scl) | ENU screen (targeted: vessels) | (Bussmann et al., 2007) | |
| TEK tyrosine kinase, endothelial (tie2/tek) | TILLING | (Gjini et al., 2011) | |
| tif1γ | moonshine (mon) | ENU screen | (Ransom et al., 2004) |
| transcription factor cmyb (cmyb) | 2010: ENU screen (targeted: T-cells); 2011: ENU screen | (Soza-Ried et al., 2010; Zhang et al., 2011) | |
| transducin (beta)-like 3 (tbl3) | ceylon (cey) | ENU-EP screen (targeted: T-cells) | (Hutchinson et al., 2012; Trede et al., 2008) |
| transferrin1 (tfr1) | chianti (cia) | ENU screen | (Wingert et al., 2004) |
| uroporphyrinogen decarboxylase (urod) | yquem (yqe) | ENU screen | (Wang et al., 1998) |
| v-ets erthroblastosis virus E26 oncogene homolog 2 (ets2) | Unconfirmed | Retroviral Insertion | (Liu and Patient, 2008) |
| Vascular Endothelial Growth Factor (VEGF) | (Lawson et al., 2002) | ||
| Von Hippel-Lindau (vhl) | TILLING | (van Rooijen et al., 2009) | |
| β-spectrin | riesling (ris) | ENU screen | (Liao et al., 2000) |
| δ-aminolevulinate synthase 2 (alas2) | sauternes (sau) | ENU screen | (Brownlie et al., 1998) |
References:
Amsterdam, A., Nissen, R.M., Sun, Z., Swindell, E.C., Farrington, S., and Hopkins, N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. U.S.a. 101, 12792–12797.
Bauer, H., Lele, Z., Rauch, G.J., Geisler, R., and Hammerschmidt, M. (2001). The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development 128, 849–858.
Brownlie, A., Donovan, A., Pratt, S.J., Paw, B.H., Oates, A.C., Brugnara, C., Witkowska, H.E., Sassa, S., and Zon, L.I. (1998). Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat. Genet. 20, 244–250.
Brownlie, A., Hersey, C., Oates, A.C., Paw, B.H., Falick, A.M., Witkowska, H.E., Flint, J., Higgs, D., Jessen, J., Bahary, N., et al. (2003). Characterization of embryonic globin genes of the zebrafish. Dev. Biol. 255, 48–61.
Bussmann, J., Bakkers, J., and Schulte-Merker, S. (2007). Early endocardial morphogenesis requires Scl/Tal1. PLoS Genetics 3, e140.
Childs, S., Weinstein, B.M., Mohideen, M.A., Donohue, S., Bonkovsky, H., and Fishman, M.C. (2000). Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Curr. Biol. 10, 1001–1004.
Covassin, L.D., Siekmann, A.F., Kacergis, M.C., Laver, E., Moore, J.C., Villefranc, J.A., Weinstein, B.M., and Lawson, N.D. (2009). A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev. Biol. 329, 212–226.
Covassin, L.D., Villefranc, J.A., Kacergis, M.C., Weinstein, B.M., and Lawson, N.D. (2006). Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc. Natl. Acad. Sci. U.S.a. 103, 6554–6559.
Danilova, N., Bussmann, J., Jekosch, K., and Steiner, L.A. (2005). The immunoglobulin heavychain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol 6, 295–302.
Danilova, N., Sakamoto, K.M., and Lin, S. (2011). Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. British Journal of Haematology 152, 217–228.
Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S.J., Moynihan, J., Paw, B.H., Drejer, A., Barut, B., Zapata, A., et al. (2000). Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403, 776–781.
Donovan, A., Brownlie, A., Dorschner, M.O., Zhou, Y., Pratt, S.J., Paw, B.H., Phillips, R.B., Thisse, C., Thisse, B., and Zon, L.I. (2002). The zebrafish mutant gene chardonnay (cdy) encodes divalent metal transporter 1 (DMT1). Blood 100, 4655–4659.
Dooley, K.A., Fraenkel, P.G., Langer, N.B., Schmid, B., Davidson, A.J., Weber, G., Chiang, K., Foott, H., Dwyer, C., Wingert, R.A., et al. (2008). montalcino, A zebrafish model for variegate porphyria. Experimental Hematology 36, 1132–1142.
English, M.A., Lei, L., Blake, T., Wincovitch, S.M., Sood, R., Azuma, M., Hickstein, D., and Liu, P.P. (2012). Incomplete splicing, cell division defects, and hematopoietic blockage in dhx8 mutant zebrafish. Dev. Dyn. 241, 879–889.
Gjini, E., Hekking, L.H., Küchler, A., Saharinen, P., Wienholds, E., Post, J.-A., Alitalo, K., and Schulte-Merker, S. (2011). Zebrafish Tie-2 shares a redundant role with Tie-1 in heart development and regulates vessel integrity. Dis Model Mech 4, 57–66.
Griffin, K.J., Amacher, S.L., Kimmel, C.B., and Kimelman, D. (1998). Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 125, 3379–3388.
Helker, C.S.M., Schuermann, A., Karpanen, T., Zeuschner, D., Belting, H.-G., Affolter, M., Schulte-Merker, S., and Herzog, W. (2013). The zebrafish common cardinal veins develop by a novel mechanism: lumen ensheathment. Development 140, 2776–2786.
Hogan, B.M., Herpers, R., Witte, M., Helotera, H., Alitalo, K., Duckers, H.J., and Schulte-Merker, S. (2009). Vegfc/Flt4 signalling is suppressed by Dll4 in developing zebrafish intersegmental arteries. Development 136, 4001–4009.
Hutchinson, S.A., Tooke-Locke, E., Wang, J., Tsai, S., Katz, T., and Trede, N.S. (2012). Tbl3 regulates cell cycle length during zebrafish development. Dev. Biol. 368, 261–272.
Itoh, M., Kim, C.-H., Palardy, G., Oda, T., Jiang, Y.-J., Maust, D., Yeo, S.-Y., Lorick, K., Wright, G.J., Ariza-McNaughton, L., et al. (2003). Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Developmental Cell 4, 67–82.
Jin, H., Li, L., Xu, J., Zhen, F., Zhu, L., Liu, P.P., Zhang, M., Zhang, W., and Wen, Z. (2012). Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 119, 5239–5249.
Lawson, N.D., Scheer, N., Pham, V.N., Kim, C.H., Chitnis, A.B., Campos-Ortega, J.A., and Weinstein, B.M. (2001). Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development 128, 3675–3683.
Lawson, N.D., Vogel, A.M., and Weinstein, B.M. (2002). sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Developmental Cell 3, 127–136.
Liao, E.C., Paw, B.H., Peters, L.L., Zapata, A., Pratt, S.J., Do, C.P., Lieschke, G., and Zon, L.I. (2000). Hereditary spherocytosis in zebrafish riesling illustrates evolution of erythroid beta-spectrin structure, and function in red cell morphogenesis and membrane stability. Development 127, 5123–5132.
Liao, E.C., Trede, N.S., Ransom, D., Zapata, A., Kieran, M., and Zon, L.I. (2002). Non-cell autonomous requirement for the bloodless gene in primitive hematopoiesis of zebrafish. Development 129, 649–659.
Liongue, C., Hall, C.J., O’Connell, B.A., Crosier, P., and Ward, A.C. (2009). Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood 113, 2535–2546.
Liu, F., and Patient, R. (2008). Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circulation Research 103, 1147–1154.
Lyons, S.E., Lawson, N.D., Lei, L., Bennett, P.E., Weinstein, B.M., and Liu, P.P. (2002). A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc. Natl. Acad. Sci. U.S.a. 99, 5454–5459.
Ma, D., Zhang, J., Lin, H.-F., Italiano, J., and Handin, R.I. (2011). The identification and characterization of zebrafish hematopoietic stem cells. Blood 118, 289–297.
Murayama, E., Kissa, K., Zapata, A., Mordelet, E., Briolat, V., Lin, H.-F., Handin, R.I., and Herbomel, P. (2006). Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25, 963–975.
Nambiar, R.M., Ignatius, M.S., and Henion, P.D. (2007). Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mechanisms of Development 124, 682–698.
Parichy, D.M., Ransom, D.G., Paw, B., Zon, L.I., and Johnson, S.L. (2000). An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 127, 3031–3044.
Pase, L., Layton, J.E., Wittmann, C., Ellett, F., Nowell, C.J., Reyes-Aldasoro, C.C., Varma, S., Rogers, K.L., Hall, C.J., Keightley, M.C., et al. (2012). Neutrophil-delivered myeloperoxidase dampens the hydrogen peroxide burst after tissue wounding in zebrafish. Curr. Biol. 22, 1818–1824.
Paw, B.H., Davidson, A.J., Zhou, Y., Li, R., Pratt, S.J., Lee, C., Trede, N.S., Brownlie, A., Donovan, A., Liao, E.C., et al. (2003). Cell-specific mitotic defect and dyserythropoiesis associated with erythroid band 3 deficiency. Nat. Genet. 34, 59–64.
Pham, V.N., Lawson, N.D., Mugford, J.W., Dye, L., Castranova, D., Lo, B., and Weinstein, B.M. (2007). Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 303, 772–783.
Ransom, D.G., Haffter, P., Odenthal, J., Brownlie, A., Vogelsang, E., Kelsh, R.N., Brand, M., van Eeden, F.J., Furutani-Seiki, M., Granato, M., et al. (1997). Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development 123, 311–319.
Ransom, D.G., Bahary, N., Niss, K., Traver, D., Burns, C., Trede, N.S., Paffett-Lugassy, N., Saganic, W.J., Lim, C.A., Hersey, C., et al. (2004). The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2, E237.
Rottbauer, W., Just, S., Wessels, G., Trano, N., Most, P., Katus, H.A., and Fishman, M.C. (2005). VEGF-PLCgamma1 pathway controls cardiac contractility in the embryonic heart. Genes & Development 19, 1624–1634.
Schauerte, H.E., van Eeden, F.J., Fricke, C., Odenthal, J., Strähle, U., and Haffter, P. (1998). Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development 125, 2983–2993.
Schorpp, M., Bialecki, M., Diekhoff, D., Walderich, B., Odenthal, J., Maischein, H.-M., Zapata, A.G., and Boehm, T. (2006). Conserved functions of Ikaros in vertebrate lymphocyte development: genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J. Immunol. 177, 2463–2476.
Shafizadeh, E., Paw, B.H., Foott, H., Liao, E.C., Barut, B.A., Cope, J.J., Zon, L.I., and Lin, S. (2002). Characterization of zebrafish merlot/chablis as non-mammalian vertebrate models for severe congenital anemia due to protein 4.1 deficiency. Development 129, 4359–4370.
Shaw, G.C., Cope, J.J., Li, L., Corson, K., Hersey, C., Ackermann, G.E., Gwynn, B., Lambert, A.J., Wingert, R.A., Traver, D., et al. (2006). Mitoferrin is essential for erythroid iron assimilation. Nature 440, 96–100.
Sood, R., English, M.A., Belele, C.L., Jin, H., Bishop, K., Haskins, R., McKinney, M.C., Chahal, J., Weinstein, B.M., Wen, Z., et al. (2010). Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood 115, 2806–2809.
Soza-Ried, C., Hess, I., Netuschil, N., Schorpp, M., and Boehm, T. (2010). Essential role of cmyb in definitive hematopoiesis is evolutionarily conserved. Proceedings of the National Academy of Sciences 107, 17304–17308.
Stainier, D.Y., Weinstein, B.M., Detrich, H.W., Zon, L.I., and Fishman, M.C. (1995). Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121, 3141–3150.
Trede, N.S., Medenbach, J., Damianov, A., Hung, L.-H., Weber, G.J., Paw, B.H., Zhou, Y., Hersey, C., Zapata, A., Keefe, M., et al. (2007). Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc. Natl. Acad. Sci. U.S.a. 104, 6608–6613.
Trede, N.S., Ota, T., Kawasaki, H., Paw, B.H., Katz, T., Demarest, B., Hutchinson, S., Zhou, Y., Hersey, C., Zapata, A., et al. (2008). Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev. Dyn. 237, 2575–2584.
van der Vaart, M., van Soest, J.J., Spaink, H.P., and Meijer, A.H. (2013). Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Model Mech 6, 841–854.
van Rooijen, E., Voest, E.E., Logister, I., Korving, J., Schwerte, T., Schulte-Merker, S., Giles, R.H., and van Eeden, F.J. (2009). Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood 113, 6449–6460.
Vo, A.H., Swaroop, A., Liu, Y., Norris, Z.G., and Shavit, J.A. (2013). Loss of fibrinogen in zebrafish results in symptoms consistent with human hypofibrinogenemia. PLoS ONE 8, e74682.
Wang, H., Long, Q., Marty, S.D., Sassa, S., and Lin, S. (1998). A zebrafish model for hepatoerythropoietic porphyria. Nat. Genet. 20, 239–243.
Wang, T., Hanington, P.C., Belosevic, M., and Secombes, C.J. (2008). Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J. Immunol. 181, 3310–3322.
Weiss, O., Kaufman, R., Michaeli, N., and Inbal, A. (2012). Abnormal vasculature interferes with optic fissure closure in lmo2 mutant zebrafish embryos. Dev. Biol. 369, 191–198.
Wienholds, E., Schulte-Merker, S., Walderich, B., and Plasterk, R.H.A. (2002). Target-selected inactivation of the zebrafish rag1 gene. Science 297, 99–102.
Wingert, R.A., Brownlie, A., Galloway, J.L., Dooley, K., Fraenkel, P., Axe, J.L., Davidson, A.J., Barut, B., Noriega, L., Sheng, X., et al. (2004). The chianti zebrafish mutant provides a model for erythroid-specific disruption of transferrin receptor 1. Development 131, 6225–6235.
Wingert, R.A., Galloway, J.L., Barut, B., Foott, H., Fraenkel, P., Axe, J.L., Weber, G.J., Dooley, K., Davidson, A.J., Schmid, B., et al. (2005). Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436, 1035–1039.
Yamaguchi, M., Tonou-Fujimori, N., Komori, A., Maeda, R., Nojima, Y., Li, H., Okamoto, H., and Masai, I. (2005). Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development 132, 3027–3043.
Zhang, Y., Jin, H., Li, L., Qin, F.X.-F., and Wen, Z. (2011). cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood 118, 4093–4101.
Zhong, T.P. (2000). gridlock, an HLH Gene Required for Assembly of the Aorta in Zebrafish. Science 287, 1820–1824.
Zhu, C., Smith, T., McNulty, J., Rayla, A.L., Lakshmanan, A., Siekmann, A.F., Buffardi, M., Meng, X., Shin, J., Padmanabhan, A., et al. (2011). Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development 138, 4555–4564.
Acknowledgments
We thank D. Traver, L.I. Zon and W. Goessling for helpful discussions and assistance with this manuscript. We thank A. Kim, D. Traver, H. Yang, and N. Chi for the unpublished image shown in Figure 2. KJC is supported by a National Science Foundation Graduate Research Fellowship, and a Harvard University Vranos Family Graduate Research Fellowship in Developmental Biology. TEN is supported by grants from the NIH NIDDK (1R01DK098241, 5K01DK080226, 5R03DK096156) and NIEHS (5R21OD012227) and the American Society of Hematology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.COLLE-VANDEVELDE A. Blood anlage in teleostei. Nature. 1963;198:1223. doi: 10.1038/1981223a0. [DOI] [PubMed] [Google Scholar]
- 2.Detrich HW, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci USA. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieschke GJ, et al. Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274–295. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- 4.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 5.Le Guyader D, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 6.Warga RM, Kane DA, Ho RK. Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Developmental Cell. 2009;16:744–755. doi: 10.1016/j.devcel.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dooley KA, Davidson AJ, Zon LI. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev Biol. 2005;277:522–536. doi: 10.1016/j.ydbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Lyons SE, et al. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc Natl Acad Sci USA. 2002;99:5454–5459. doi: 10.1073/pnas.082695299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson LJ, et al. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 2007;109:2389–2398. doi: 10.1182/blood-2006-02-003087. [DOI] [PubMed] [Google Scholar]
- 10.Lyons SE, et al. Molecular cloning, genetic mapping, and expression analysis of four zebrafish c/ebp genes. Gene. 2001;281:43–51. doi: 10.1016/s0378-1119(01)00774-0. [DOI] [PubMed] [Google Scholar]
- 11.Su F, et al. Differential regulation of primitive myelopoiesis in the zebrafish by Spi-1/Pu.1 and C/ebp1. Zebrafish. 2007;4:187–199. doi: 10.1089/zeb.2007.0505. [DOI] [PubMed] [Google Scholar]
- 12.Hsu K, et al. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104:1291–1297. doi: 10.1182/blood-2003-09-3105. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand JY, et al. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertrand JY, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 16.North TE, et al. Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murayama E, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Gering M, Patient R. Hedgehog Signaling Is Required for Adult Blood Stem Cell Formation in Zebrafish Embryos. Developmental Cell. 2005;8:389–400. doi: 10.1016/j.devcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Developmental Cell. 2002;3:127–136. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 20.Burns CE. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes & Development. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson ND, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 22.Kim PG, et al. Signaling axis involving Hedgehog, Notch, and Scl promotes the embryonic endothelial-to-hematopoietic transition. Proceedings of the National Academy of Sciences. 2013;110:E141–50. doi: 10.1073/pnas.1214361110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalev-Zylinska ML, et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson RN, et al. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Developmental Cell. 2009;16:909–916. doi: 10.1016/j.devcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goessling W, et al. Genetic Interaction of PGE2 and Wnt Signaling Regulates Developmental Specification of Stem Cells and Regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clements WK, et al. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474:220–224. doi: 10.1038/nature10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganis JJ, et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366:185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin HF. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jagadeeswaran P, Sheehan JP, Craig FE, Troyer D. Identification and characterization of zebrafish thrombocytes. British Journal of Haematology. 1999;107:731–738. doi: 10.1046/j.1365-2141.1999.01763.x. [DOI] [PubMed] [Google Scholar]
- 30.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98:3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- 31.Bennett CM, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 32.Davis JM, et al. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 33.Willett CE, Cortes A, Zuasti A, Zapata AG. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn. 1999;214:323–336. doi: 10.1002/(SICI)1097-0177(199904)214:4<323::AID-AJA5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 34.Willett CE, Zapata AG, Hopkins N, Steiner LA. Expression of zebrafish rag genes during early development identifies the thymus. Dev Biol. 1997;182:331–341. doi: 10.1006/dbio.1996.8446. [DOI] [PubMed] [Google Scholar]
- 35.Keightley MC, Layton JE, Hayman JW, Heath JK, Lieschke GJ. Mediator subunit 12 is required for neutrophil development in zebrafish. PLoS ONE. 2011;6:e23845. doi: 10.1371/journal.pone.0023845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danilova N, Steiner LA. B cells develop in the zebrafish pancreas. Proc Natl Acad Sci USA. 2002;99:13711–13716. doi: 10.1073/pnas.212515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page DM, et al. An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood. 2013;122:e1–11. doi: 10.1182/blood-2012-12-471029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoder JA, et al. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc Natl Acad Sci USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoder JA, et al. Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci USA. 2004;101:15706–15711. doi: 10.1073/pnas.0405242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lugo-Villarino G, et al. Identification of dendritic antigen-presenting cells in the zebrafish. Proceedings of the National Academy of Sciences. 2010;107:15850–15855. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 42.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cade L, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halloran MC, et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 47.Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS ONE. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thummel R, et al. Cre-mediated site-specific recombination in zebrafish embryos. Dev Dyn. 2005;233:1366–1377. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- 49.Scheer NN, Campos-Ortega JAJ. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mechanisms of Development. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- 50.Curado S, Stainier DYR, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stainier DY, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 52.Driever W, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 53.Weinstein BM, et al. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–309. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- 54.Ransom DG, et al. Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development. 1997;123:311–319. doi: 10.1242/dev.123.1.311. [DOI] [PubMed] [Google Scholar]
- 55.Brownlie A, et al. Characterization of embryonic globin genes of the zebrafish. Dev Biol. 2003;255:48–61. doi: 10.1016/s0012-1606(02)00041-6. [DOI] [PubMed] [Google Scholar]
- 56.Donovan A, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 57.Shaw GC, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 58.Pietrangelo A. The ferroportin disease. Blood Cells, Molecules, and Diseases. 2004;32:131–138. doi: 10.1016/j.bcmd.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Shafizadeh E, et al. Characterization of zebrafish merlot/chablis as non-mammalian vertebrate models for severe congenital anemia due to protein 4.1 deficiency. Development. 2002;129:4359–4370. doi: 10.1242/dev.129.18.4359. [DOI] [PubMed] [Google Scholar]
- 60.Paw BH, et al. Cell-specific mitotic defect and dyserythropoiesis associated with erythroid band 3 deficiency. Nat Genet. 2003;34:59–64. doi: 10.1038/ng1137. [DOI] [PubMed] [Google Scholar]
- 61.Donovan A, et al. The zebrafish mutant gene chardonnay (cdy) encodes divalent metal transporter 1 (DMT1) Blood. 2002;100:4655–4659. doi: 10.1182/blood-2002-04-1169. [DOI] [PubMed] [Google Scholar]
- 62.Postlethwait JH, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 63.Walker C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- 64.Trede NS, et al. Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev Dyn. 2008;237:2575–2584. doi: 10.1002/dvdy.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Covassin LD, et al. A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev Biol. 2009;329:212–226. doi: 10.1016/j.ydbio.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wienholds E, Schulte-Merker S, Walderich B, Plasterk RHA. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- 67.Amsterdam A, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes & Development. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaiano N, et al. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 69.Burns CE, et al. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 2009;113:5776–5782. doi: 10.1182/blood-2008-12-193607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genomewide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sood R, et al. Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood. 2010;115:2806–2809. doi: 10.1182/blood-2009-08-236729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mucenski ML, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 74.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proceedings of the National Academy of Sciences. 2010;107:17304–17308. doi: 10.1073/pnas.1004640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Jin H, Li L, Qin FXF, Wen Z. cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood. 2011;118:4093–4101. doi: 10.1182/blood-2011-03-342501. [DOI] [PubMed] [Google Scholar]
- 77.Stainier DY, Weinstein BM, Detrich HW, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 78.Xiong JW, Yu Q, Zhang J, Mably JD. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circulation Research. 2008;102:1057–1064. doi: 10.1161/CIRCRESAHA.107.163907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weber GJ, et al. Mutant-specific gene programs in the zebrafish. Blood. 2005;106:521–530. doi: 10.1182/blood-2004-11-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang HT, et al. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nature Publishing Group. 2013;15:1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bai X, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Long Q, et al. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 83.Zhu H, et al. Regulation of the lmo2 promoter during hematopoietic and vascular development in zebrafish. Dev Biol. 2005;281:256–269. doi: 10.1016/j.ydbio.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 84.Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 85.Ma D, Zhang J, Lin HF, Italiano J, Handin RI. The identification and characterization of zebrafish hematopoietic stem cells. Blood. 2011;118:289–297. doi: 10.1182/blood-2010-12-327403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mikkola HKA, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 87.Lam EYN, Hall CJ, Crosier PS, Crosier KE, Flores MV. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood. 2010;116:909–914. doi: 10.1182/blood-2010-01-264382. [DOI] [PubMed] [Google Scholar]
- 88.Zhen F, Lan Y, Yan B, Zhang W, Wen Z. Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development. 2013;140:3977–3985. doi: 10.1242/dev.097071. [DOI] [PubMed] [Google Scholar]
- 89.North T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 90.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kissa K, et al. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- 92.Boisset JC, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 93.Yi Ni Lam E, et al. Zebrafish runx1 promoter-EGFP transgenics mark discrete sites of definitive blood progenitors. Blood. 2009;113:1241–1249. doi: 10.1182/blood-2008-04-149898. [DOI] [PubMed] [Google Scholar]
- 94.de Jong JLO, et al. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood. 2011;117:4234–4242. doi: 10.1182/blood-2010-09-307488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harris JM, et al. Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood. 2013;121:2483–2493. doi: 10.1182/blood-2012-12-471201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peterson RT, et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 97.Shafizadeh E, Peterson RT, Lin S. Induction of reversible hemolytic anemia in living zebrafish using a novel small molecule. Comparative Biochemistry and Physiology, Part C. 2004;138:245–249. doi: 10.1016/j.cca.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 98.Paik EJ, de Jong JLO, Pugach E, Opara P, Zon LI. A chemical genetic screen in zebrafish for pathways interacting with cdx4 in primitive hematopoiesis. Zebrafish. 2010;7:61–68. doi: 10.1089/zeb.2009.0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goessling W, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8:445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cutler C, et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood. 2013;122:3074–3081. doi: 10.1182/blood-2013-05-503177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yeh JRJ, et al. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2007;135:401–410. doi: 10.1242/dev.008904. [DOI] [PubMed] [Google Scholar]
- 102.Yeh JRJ, et al. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5:236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ridges S, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119:5621–5631. doi: 10.1182/blood-2011-12-398818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Traver D, et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 105.Dzierzak E, de Bruijn M. Isolation and analysis of hematopoietic stem cells from mouse embryos. Methods Mol Med. 2002;63:1–14. doi: 10.1385/1-59259-140-X:001. [DOI] [PubMed] [Google Scholar]
- 106.Traver D, et al. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood. 2004;104:1298–1305. doi: 10.1182/blood-2004-01-0100. [DOI] [PubMed] [Google Scholar]
- 107.White RM, et al. Transparent Adult Zebrafish as a Tool for In Vivo Transplantation Analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Delaney C, et al. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16:232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stachura DL, et al. Clonal analysis of hematopoietic progenitor cells in the zebrafish. Blood. 2011;118:1274–1282. doi: 10.1182/blood-2011-01-331199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langenau DM, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 111.Kaatsch P. Epidemiology of childhood cancer. Cancer Treat Rev. 2010;36:277–285. doi: 10.1016/j.ctrv.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 112.Langenau DM, et al. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, et al. AML1-ETO mediates hematopoietic self-renewal and leukemogenesis through a COX/β-catenin signaling pathway. Blood. 2013;121:4906–4916. doi: 10.1182/blood-2012-08-447763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gutierrez A, et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med. 2011;208:1595–1603. doi: 10.1084/jem.20101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feng H, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18:353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gutierrez A, et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J Clin Invest. 2014;124:644–655. doi: 10.1172/JCI65093. [DOI] [PMC free article] [PubMed] [Google Scholar]