Abstract
Respiratory syncytial virus (RSV) is the leading cause of hospitalization for children under five years of age. We sought to engineer a viral antigen that provides greater protection than currently available vaccines and focused on antigenic site Ø, a metastable site specific to the prefusion state of the RSV fusion (F) glycoprotein, as this site is targeted by extremely potent RSV-neutralizing antibodies. Structure-based design yielded stabilized versions of RSV F that maintained antigenic site Ø when exposed to extremes of pH, osmolality, and temperature. Six RSV F-crystal structures provided atomic-level data on how introduced cysteine residues and filled hydrophobic cavities improved stability. Immunization with site Ø-stabilized variants of RSV F in mice and macaques elicited levels of RSV-specific neutralizing activity many times the protective threshold.
Respiratory syncytial virus (RSV) is one of the last remaining highly prevalent childhood diseases without an approved vaccine. It is estimated to be responsible for 6.7% of deaths in children 1 month to 1 year of age and causes excess mortality in the elderly at levels comparable to influenza virus (1). Although RSV infection does not induce fully protective immunity, antibodies against the RSV fusion (F) glycoprotein can prevent severe disease in humans as demonstrated by passive prophylaxis with the F-directed antibody, palivizumab (Synagis®) (2).
The proven success of palivizumab (3) has spurred vaccine efforts aimed at eliciting protective RSV F-directed antibodies. These efforts have been complicated by the conformational diversity of RSV F (4-8), a type I fusion protein that merges virus and host-cell membranes by using the difference in folding energy between two substantially different states: a metastable state adopted prior to virus-cell interaction (prefusion) and a stable state that occurs after merging of virus and cell membranes (postfusion). Both states exhibit epitopes targeted by neutralizing antibodies, and postfusion RSV F is being developed as a vaccine candidate (6, 9). Recently, however, the major target of RSV-neutralizing antibodies elicited by natural infection was found to reside primarily on the prefusion conformation of RSV F (10). Antibodies such as 5C4 (7), AM22, and D25 (11, 12) are substantially more potent than palivizumab and target antigenic site Ø, a metastable site located at the membrane-distal apex of the prefusion RSV F trimer (7).
To enhance elicitation of similarly potent antibodies, we engineered soluble variants of RSV F with stably exposed antigenic site Ø. These variants were characterized antigenically and crystallographically and tested for immunogenicity in mice and non-human primates (rhesus macaques).
Structure-based Vaccine Strategy
We and others have engineered antigenicity (13-17) through structure-based design of the epitopes recognized by template neutralizing antibodies. For example, the crystal structure of motavizumab (a variant of palivizumab) bound to its F glycoprotein epitope (18) allowed us to create epitope scaffolds, which stably presented the motavizumab epitope on heterologous proteins (19). Although motavizumab-epitope scaffolds could elicit immune responses that recognized F, substantial neutralizing activity was not induced (19). We hypothesized that instead of a single epitope recognized by a single template antibody, it would be advantageous to present a “supersite” (20), comprising a collection of overlapping epitopes recognized by multiple antibodies. Even more preferable would be for such a site to be ultra-sensitive to neutralization. These considerations led to a “neutralization-sensitive site” strategy: (i) to identify a viral site targeted by multiple antibodies with extremely potent neutralizing activity, (ii) to determine the structure of the site in complex with a representative antibody, (iii) to engineer the stable presentation of the site in the absence of recognizing antibody, and (iv) to elicit high titer protective responses through immunization with engineered antigens that stably present the neutralization-sensitive site (fig. S1).
Engineering of RSV F Antigens
Antigenic site Ø was chosen as the target site because of its recognition by RSV-neutralizing antibodies that are 10-100-fold more potent than palivizumab (7, 11, 12). We previously determined the structure of antigenic site Ø in complex with the D25 antibody (7). Structure determination involved appending the T4-phage fibritin trimerization domain (“foldon”) (21, 22) to the C terminus of the RSV F ectodomain (5) and binding of the prefusion-specific D25 antibody. Although these approaches stabilized antigenic site Ø, D25 binding sterically occluded the target site. To stably present antigenic site Ø in the absence of D25, we retained the C terminal trimerization domain and combined it with other means of stabilization, including the introduction of cysteine pairs or cavity-filling hydrophobic substitutions.
The β-carbons of serine residues 155 and 290 are 4.4 Å apart in the D25-bound RSV F structure (7) and 124.2 Å apart in the postfusion structure (5) (Fig. 1 and fig. S2). A S155C-S290C double mutant (DS) formed stable RSV F trimers, expressed at 1.4 mg/L, retained antigenic site Ø, and was homogeneous as judged by negative stain-electron microscopy (Table 1, fig. S3) (23, 24). Other intra-chain cysteine modifications, such as those between regions of RSV F that do not rearrange between pre- and postfusion states (e.g. S403C and T420C), did not stabilize antigenic site Ø (Table 1). We also tested potential inter-chain double cysteine modifications, but none expressed at levels sufficient for ELISA detection (table S1) (25).
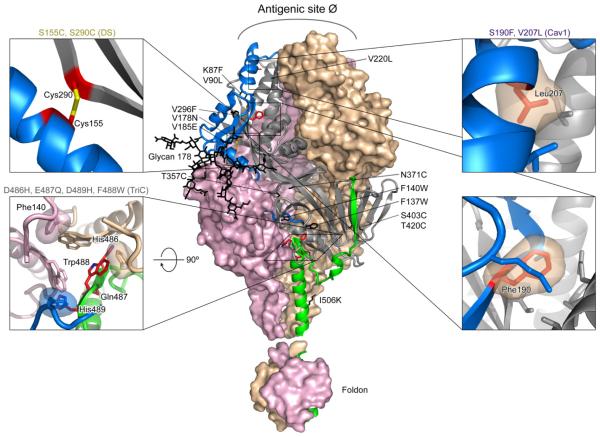
Fig. 1. Design of soluble site Ø-stabilized RSV F trimers.
Over 100 variants of RSV F containing the T4 fibritin-trimerization domain (foldon) were designed to provide greater stability to antigenic site Ø (table S1). Shown here is the structure of the RSV F trimer in its D25-bound conformation with modeled C-terminal appended foldon. The trimer is displayed with two of the three F1F2protomers in molecular surface representation (colored tan and pink), and the third F1F2 protomer in ribbon representation. The ribbon is colored gray in regions where it is relatively fixed between pre- and postfusion, while the N- and C-terminal residues that move more than 5 Å between pre- and postfusion conformations are colored blue and green, respectively. Mutations compatible with RSV F expression and initial D25 recognition (table S1), but insufficiently stable to allow purification of RSV F as a homogenous trimer (Table 1), are labeled and shown in stick representation (colored black). Insets show enlargements of stabilizing mutations in stick representation (colored red) for DS, Cav1 and TriC variants, all of which sufficiently stabilize antigenic site Ø to allow purification as a homogeneous trimer (Table 1).
Table 1.
Antigenic and physical characteristics of engineered RSV F glycoprotein variants.
|
Mechanism of
stabilization |
RSV F variant |
Oligomeric
state# |
Yield
(mg/L)* |
Antibody KD value (nM)@ |
Physical characterization
(fractional D25 reactivity) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||||
| Site Ø | Site I | Site II$ | Site IV | Temp (°) | pH |
Osmolality
(mM) |
Freeze-
thaw |
||||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|||||||
| D25 | AM22 | 5C4 | 131-2a | Paliv | Mota | 101F | 50 | 70 | 3.5 | 10.0 | 10 | 3000 | 10& | ||||
| Cavity filling | K87F-V90L | Aggregate | 0.3 | >1000 | >1000 | >1000 | 7.6 | 1.7 | 0.17 | 1.6 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| F137W-F140W | N.D. | <0.1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| F137W-F140W-F488W | N.D. | <0.1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| S190F-V207L (Cav1) | Trimer | 2.2 | 0.23 | <0.10 | 9.3 | >1000 | 43 | <0.01 | 2.9 | 0.8 | 0.1 | 0.7 | 0.8 | 1.0 | 0.7 | 0.6 | |
| S190F-V296F | Aggregate | 0.4 | >1000 | >1000 | >1000 | 4.2 | 1.7 | <0.01 | 1.6 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| V207L-V220L | Aggregate | 0.4 | >1000 | >1000 | >1000 | 2.8 | 0.99 | 0.014 | 0.69 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
|
D486H-E487Q-F488W-
D489H (TriC) |
Trimer | 0.8 | 0.012 | 1.0 | 34 | >1000 | 31 | 0.48 | 4.0 | 0.8 | 0.1 | 0.1 | 0.8 | 1.3 | 0.6 | 0.1 | |
| F488W | Trimer | 1.7 | 0.087 | 0.25 | 27 | >1000 | 32 | 0.1 | 4.7 | 0.9 | 0.1 | 0.1 | 0.7 | 1.1 | 0.5 | 0 | |
|
|
|||||||||||||||||
| Disulfide formation | S155C-S290C (DS) | Trimer | 1.4 | 0.29 | <0.01 | 35 | 3.4 | 2.8 | 0.043 | 2.2 | 0.3 | 0 | 0.1 | 0.8 | 1.3 | 0.8 | 0.3 |
| T357C-N371C | N.D. | <0.1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| S403C-T420C | Aggregate | 0.3 | >1000 | >1000 | >1000 | 3.05 | 3.3 | 0.05 | 1.85 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
|
|
|||||||||||||||||
| N-Glycan addition | V178N | Aggregate | <0.1 | >1000 | >1000 | >1000 | 7.2 | 3.1 | 0.11 | 1.64 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
|
|
|||||||||||||||||
| Postfusion destabilization |
V185E | Aggregate | <0.1 | >1000 | >1000 | >1000 | 3.5 | 1.7 | 0.11 | 1.64 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| I506K | Aggregate | <0.1 | >1000 | >1000 | >1000 | 4.6 | 1.7 | 0.054 | 1.39 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
|
|
|||||||||||||||||
| Acid patch neutralization |
D486H-E487Q-D489H | Aggregate | 0.1 | >1000 | >1000 | >1000 | >1000 | 9.5 | 0.57 | 12.7 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
|
|
|||||||||||||||||
| Double combinations | DS-Cav1 | Trimer | 1.9 | 0.15 | <0.01 | 13 | >1000 | 23 | 0.041 | 3.2 | 0.9 | 0 | 0.8 | 0.9 | 1.0 | 0.8 | 0.7 |
| DS-TriC | Trimer | 0.6 | 0.17 | <0.01 | 33 | 2.0 | 4.8 | 0.055 | 3.1 | 0.9 | 0 | 0.3 | 0.9 | 0.5 | 0.9 | 0.5 | |
| Cav1-TriC | Trimer | 0.2 | 0.99 | 0.086 | 5.1 | >1000 | 33 | 0.17 | 3.1 | 0.9 | 0.1 | 0.3 | 0.8 | 0.6 | 0.5 | 0 | |
|
|
|||||||||||||||||
| Triple combinations | DS-Cav1-TriC | Trimer | 1.3 | 0.17 | 0.02 | 18 | >1000 | 20 | 0.10 | 3.2 | 0.9 | 0.1 | 0.6 | 0.9 | 0.6 | 0.6 | 0 |
Variants were often observed to exist in a mixture of oligomeric states on size chromatography. If a measureable trimeric fraction was observed, then this is indicated by the label “Trimer”, and the various properties of the trimer fraction are listed. If no trimeric fraction was observed, then the oligomeric state of the dominant oligomeric species is provided. If the total yield prior to size chromatography was <0.1 mg/l, then oligomeric state is listed as not determined (N.D.).
Yield shown is for the specified oligomeric state.
When no binding was observed for 1 μM Fab, the KD is shown as >1000 nM; when total yield was <0.1 mg/l, then the value for KD is not applicable (N/A).
Palivizumab (Paliv); motavizumab (Mota).
Ten cycles of freeze-thaw.
We analyzed the D25-bound RSV F structure for hydrophobic cavities unique to the D25-bound conformation of RSV F that abutted regions that differed in the prefusion and postfusion states (26). Several such cavities were identified in the membrane-distal “head” of the prefusion structure, close to the binding site of D25, and we engineered hydrophobic substitutions to fill these cavities. S190F and V207L substitutions were predicted to adopt prevalent side chain conformations with minimal clashes, while K87F, V90L, V220L and V296F showed less steric compatibility. We assessed the impact of filling these cavities with pairs of changes. A S190F-V207L pair (Cav1) (Fig. 1), formed stable RSV F trimers, expressed at 2.2 mg/l, and retained antigenic site Ø (Table 1). Meanwhile, K87F-V90L, S190F-V296F and V207L-V220L variants showed enhanced retention of D25 recognition, but expressed at less than 0.1 mg/l (Table 1).
Other cavities we identified towards the center of prefusion RSV F were close to the fusion peptide, the trimer axis, and an acidic patch comprising residues Asp486, Glu487, and Asp489. We modeled several cavity-filling substitutions including F137W, F140W, and F488W, and analyzed these substitutions in combination with D486H, E487Q, and D489H (Table 1). Of the six combinations tested, only two (F488W and D486H-E487Q-F488W-D489H) expressed levels of purified RSV F trimer at greater than 0.1 mg/l and retained D25 recognition. The D486H-E487Q-F488W-D489H variant (TriC) formed stable RSV F trimers, expressed at 0.8 mg/l, and retained antigenic site Ø (Fig. 1 and Table 1).
We also tested the impact of destabilizing the postfusion conformation on the preservation of antigenic site Ø. The substitution V178N, which is predicted to introduce an N-linked glycan compatible with the prefusion but not the postfusion conformation of F, did not appear to stabilize antigenic site Ø, nor did V185E or I506K, which were predicted to place a glutamic acid or a lysine into the interior of the postfusion six-helix bundle (Table 1). In all, over 100 RSV F variants were constructed (table S1), expressed in a 96-well transfection format (27) (fig. S4), and tested by ELISA of the culture supernatants for binding to D25 and motavizumab (figs. S5 and S6). Fifteen constructs were compatible with D25 binding, six of which retained D25 recognition for at least 7 days at 4°C, and three of these could be purified as homogeneous trimers that retained antigenic site Ø (Table 1 and fig. S7). Overall, we observed strong correlation between retention of D25 binding for at least 7 days at 4°C in 96-well supernatants and yield of purified trimers after large scale expression and purification (Fig. S5).
Combinatorial Optimization of Site Ø Stability
DS, Cav1, and TriC variants displayed a variety of physical and antigenic properties. The DS variant was the least stable to pH and temperature variation, but was more permanently stabilized in the trimeric state, while a low level of continual conversion from trimer to aggregate was observed for Cav1 and TriC on size-exclusion chromatography. Combinations of the three variants improved retention of D25 reactivity to physical extremes, a characteristic helpful to manufacturing. Overall, the DS-Cav1 combination appeared optimal in terms of trimer yield and physical stability to extremes of temperature, pH, osmolality, and freeze-thaw (Table 1), and was homogeneous as judged by negative stain electron microscopy (fig. S2).
Crystallographic Analysis
To provide atomic-level information, we determined crystal structures of site Ø-stabilized variants of RSV F (Fig. 2). The DS, Cav1, DS-Cav1 and DS-Cav1-TriC variants all crystallized in similar 1.5 M tartrate pH 9.5 conditions, and these cubic crystals diffracted X-rays to resolutions of 3.2 Å, 3.1 Å, 3.1 Å and 2.8 Å, respectively (table S2). Molecular replacement solutions were obtained by using the D25-bound RSV F structure as a search model, and these revealed a single RSV F protomer in the asymmetric unit, with the trimeric F axis aligned along the crystallographic threefold. Tetragonal crystals of Cav1 and cubic crystals of DS-Cav1 were also obtained from 1.7 M ammonium sulfate pH 5.5 conditions, and these diffracted to resolutions of 2.4 Å and 3.0 Å, respectively (table S2). Molecular replacement revealed the tetragonal lattice to have a full RSV F trimer in the asymmetric unit, and to be highly related to the tartrate cubic lattices. Overall these structures revealed the engineered RSV F variants to be substantially in the D25-bound conformation (28).
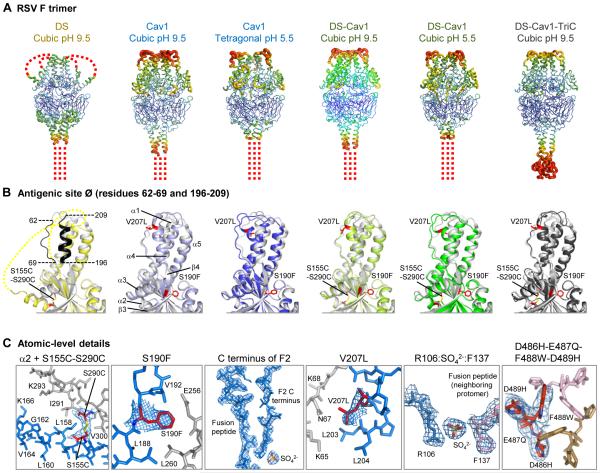
Fig. 2. Crystal structures of RSV F trimers, engineered to preserve antigenic site Ø.
(A-C) Six structures for RSV F variants are shown, labeled by stabilizing mutation (DS, Cav1, DS-Cav1, and DS-Cav1-TriC) and the crystal lattice (cubic or tetragonal). (A) RSV F trimers are displayed in Ca-worm representation, colored according to atomic mobility factor, with regions of higher flexibility in warmer colors (red) and regions of lower flexibility in cooler colors (blue). Missing regions are shown as dotted lines. These occur at the C-terminal membrane-proximal region, where the foldon motif is not seen, except in the DS-Cav1-TriC structure (far right). In the DS structure, two loops in the head region are also disordered. (B) Antigenic site Ø of a RSV F protomer is displayed in ribbon diagram, with the structure of D25-bound RSV F in gray and different variants colored yellow (DS), light and dark blue (Cav1), light and dark green (DS-Cav1), and black (DS-Cav1-TriC). Residues corresponding to antigenic site Ø are highlighted in black on the image at far left, and secondary structural elements are shown on the second image from left. Stabilizing mutations are labeled and shown in stick representation (colored red). Perpendicular view presented in figure S8. (C) Atomic-level details are shown in stick representation, colored the same as in Fig. 2, with regions of RSV F that change conformation between prefusion and postfusion conformation in red and blue, and those that remain constant in gray. Stabilizing carbon atoms for stabilizing mutations are highlighted in red. In Cav1 (pH5.5) and in DS-Cav1 (pH5.5) novel features were observed involving the interaction of the C-terminus of the F2 peptide with a sulfate ion and the fusion peptide. In the DS-Cav1-TriC structure, the D486H-E487Q-F488W-D489H mutations interact with the two neighboring protomers (colored tan and pink) around the trimer axis.
Although the structure of the DS variant (Fig. 2, left column) revealed the cysteine-substituted residues at 155 and 290 formed a disulfide bond that prevented triggering to the postfusion state, much of the membrane-distal portion of the RSV F trimer, including antigenic site Ø, was either disordered (residues 63-72 and 169-216) or in a different conformation. Thus, for example, residues 160-168 in the DS structure extended the α2-helix instead of forming a turn and initiating the α3-helix, as in the D25-bound F structure (Fig. 2C, left panel). One possible explanation for the differences between the DS structure and the D25-bound RSV F structure is that antigenic site Ø in DS is flexible, and crystallized in a conformation that does not bind D25 (29). Overall, the DS variant retained many of the features of the prefusion state of RSV F, including the fusion peptide in the interior of the trimeric cavity; however, it failed to fix antigenic site Ø in its D25-bound conformation (fig. S8).
In comparison to DS, the Cav1 structure (Fig. 2, second and third columns) was more ordered at its membrane-distal apex, with α3-helix, β3/β4 hairpin, and α4-helix clearly defined. Residues 137-202, which contain the S190F substitution, had a Cα-root mean square deviation of 0.6 Å when compared to the D25-bound F structure. The higher degree of structural order was likely due to the S190F mutation that filled a cavity observed in the D25-bound F structure, and increased van der Waals contacts with residues Ile57, Lys176, Val192, Glu256, Ser259 and Leu260. The other cavity-filling mutation in Cav1, V207L, was shifted by 5.5 Å compared to the D25-bound F structure, with the C-terminal portion of the α4-helix kinking near Pro205 and adopting distinct conformations in the two crystallization conditions (Fig. 2, second and third panels from left), suggesting that the V207L mutation is unable to stabilize the α4-helix in the D25-bound conformation.
A striking feature of the Cav1 structure in the tetragonal crystal lattice is the C-terminus of F2, which is disordered in the D25-bound F structure, but in Cav1, tunnels into the trimeric cavity alongside the fusion peptide. Interestingly, the C terminus ends with Ala107, and not Arg109, as expected after cleavage of the furin site (Arg106-Ala107-Arg108-Arg109) (30). In the Cav1 structure, the positive charge of Arg106 is offset by an ordered sulfate ion (Fig. 2C). Biologically, the interior position of the F2 C-terminus may play a role in triggering the prefusion F conformation.
Comparison of the DS-Cav1 structures from the two cubic crystal forms (Fig. 2, second and third columns from right) to those of Cav1 revealed only minor differences (31). The largest differences occurred at the RSV F apex, including antigenic site Ø and specifically at residues 64-73 and 203-216. Notably, the atomic mobility (B-factor) was highest in this apex region for all of the site Ø-stabilized variants, perhaps indicative of intrinsic site Ø flexibility. Interestingly, however, site Ø has low atomic mobility when bound by D25, revealing the ability of D25 to stabilize both overall and local RSV F conformations.
The structure of the DS-Cav1-TriC triple combination (Fig. 2, far right column) was also highly similar to other Cav1-containing RSV F variant structures. One difference in the electron density corresponded to an expanse of weak density at the membrane-proximal region, which approximated the dimensions of the T4-fibritin trimerization domain (32), but is not visible in other crystallized RSV F structures containing this domain, including the D25-bound structure (7). Small structural differences in packing likely allowed for the partial ordering of this domain (and may also have accounted for the increased diffraction limit of the DS-Cav1-TriC crystals relative to the other cubic variants), rather than differences in the interaction between the DS-Cav1-TriC stabilized RSV F and the trimerization domain.
The critical F488W substitution in the TriC alteration of residues 486-489 packed directly against the F488W substitutions of neighboring protomers of the RSV F trimer. The indole side chain of Trp488 pointed towards the trimer apex and also formed ring-stacking interactions with the side chain of Phe140 of the fusion peptide (Fig. 2C, far right panel). This fusion peptide interaction, which is not observed in any of the Phe488-containing structures, likely inhibits the extraction of the fusion peptide from the prefusion trimer cavity, providing a structural rationale for the ability of the F488W substitution to stabilize the prefusion state of RSV F (Table 1).
Immunogenicity of Antigenic Site Ø-Stabilized RSV F
To assess the effect of site Ø-stabilization on the elicitation of RSV-protective humoral responses, we immunized CB6F1/J mice with various forms of RSV F, injecting each mouse with 10 μg RSV F combined with 50 μg poly I:C adjuvant at weeks 0 and 3, and measured the ability of week 5 sera to prevent RSV infection of HEp-2 cells. DS, Cav1, and TriC each elicited high titers of neutralizing activity (geometric mean 50% effective concentrations (EC50s) of 1826-2422). This level was ~4-fold higher than that elicited by postfusion F (504 EC50), and ~20-fold higher than the protective threshold (33). By comparison, DS-Cav1 elicited neutralizing activity of 3937 EC50, roughly 8-fold higher than postfusion F and 40-fold higher than the protective threshold (33) (Fig. 3A).
Fig. 3. Immunogenicity of engineered RSV F trimers.
RSV F proteins engineered to stably display antigenic site Ø elicit neutralizing titers significantly higher than those elicited by postfusion F. (A) Neutralization titers of sera from mice immunized with 10 μg of RSV F (left). Postfusion F, as well as RSV F bound by antibodies AM22 or D25, were administered at 20 μg of the RSV F-antibody complex per mouse (right). Titers from each mouse are shown as individual black dots, and geometric means are indicated by red horizontal lines. (B) Neutralization titers of sera from rhesus macaques immunized with 50 μg of RSV F protein variants. Titers from each macaque are shown as individual black dots, and geometric means are indicated by red horizontal lines. Protective threshold (33) is indicated by a dotted line, and p-values are provided for postfusion versus DS-Cav1 as assessed by two-tailed T-test.
To quantify antibody responses to different sites on prefusion RSV F, we used antigenic site Ø-occluded forms of RSV F. CB6F1/J mice immunized with 20 μg RSV F bound by antigenic site Ø-directed antibodies (comprising ~10 μg of RSV F and ~10 μg of the antigen-binding fragment of antibody) developed week 5 geometric mean neutralizing titers of 911 and 1274 EC50 for AM22 and D25 complexes, respectively, roughly double that of postfusion at 10 μg/ml and comparable to those elicited by postfusion at 20 μg/ml (Fig. 3A). These findings suggested that the very high titers elicited by immunization with RSV F variants stabilized in the prefusion state, especially DS-Cav1, were a result of antibodies targeting antigenic site Ø.
To examine the generality of site Ø elicitation, we immunized rhesus macaques with DS-Cav1, DS and postfusion forms of RSV F, injecting each macaque with 50 μg RSV F mixed with 500 μg poly I:C adjuvant at weeks 0 and 4 and measured the ability of week 2, 4, 6, 8, 10, and 14 sera to inhibit RSV infection. Formulated proteins retained their expected antigenic profiles as measured by D25 binding to material from the injectate (fig. S9). At week 6, DS and DS-Cav1 elicited geometric mean titers of 1222 and 2578 EC50, respectively, roughly 5- and 10-fold higher than postfusion F (287 EC50) (Fig. 4B). By week 8, geometric mean titers of DS-Cav1 were roughly 60-fold higher than those elicited by postfusion F against the homologous RSV subtype A (fig. S10) and more than 80-fold higher against RSV subtype B (table S4). Overall, these results demonstrate a conservation of the relative immunogenicity for the different forms of RSV F immunogen between mice and primates, with DS-Cav1 eliciting RSV-protective titers many times the protective threshold in a primate immune system against both major RSV subtypes.
Fig. 4. Informatics of site Ø-stabilized RSV F immunogens.
(A) Physical stability of site Ø versus immunogenicity. Physical stability (horizontal axis), as determined by the average of seven measurements of D25-retained activity in Table 1, is compared to elicited RSV-protective titers from Fig. 3 (vertical axis). (B) Structural mimicry of site Ø versus immunogenicity. Structural mimicry (horizontal axis) is the root-mean-square deviation of atom distances between different unbound RSV F structures (Fig. 2) and the D25-bound RSV F structure for all atoms within 10 Å of D25. This is compared to elicited RSV-protective titers from Fig. 3 (vertical axis). (C) Antigenic analysis of sera from immunized macaques. Binding of sera to sensor-tip immobilized DS-Cav1 (left), DS (middle), or postfusion F (right) was measured directly (black bars) or after incubation with excess postfusion F (light grey bars), DS (grey bars), or DS-Cav1 (dark grey bars). The mean responses of four macaque sera are shown, with standard deviations as error bars. Additional analysis of immunized sera is shown in fig. S12. (D) Elicited binding responses relative to surface areas and shared or unique portions of immunogens. Surface areas were calculated (fig. S11 and table S3) and compared to binding responses for the 36 response measurements in (C). (E) Correlation of immunogenicity and antigenic specificity of immunized macaque sera. The mean EC50 neutralization titers (week 6) of the four macaque sera in each group are plotted against the ratio of prefusion-specific/postfusion F-specific binding responses in (C) (37).
Optimization of RSV F protective responses
The interplay between design, physical and antigenic properties, atomic-level structure, and immunogenicity provides a basis for further optimization of RSV vaccine candidates (34). For example, to obtain insight into the relationship between various antigenic and physical properties of engineered RSV Fs and the elicitation of RSV-protective responses, one can correlate antigenic and physical properties (Table 1) with immunogenicity (Fig. 3). Such correlations indicate that increasing site Ø stability to physical extremes should increase protective titers elicited upon immunization (Fig. 4A) (35). Similarly, correlations between various conformational states or regions of RSV F (Fig. 2) and immunogenicity (Fig. 3) provide design insight into the conformation and regions of RSV F that provide the most protective responses. Specifically, enhancing structural mimicry of antigenic site Ø in its D25-bound conformation appears to be a promising strategy for improving protective antibody responses (Fig. 4B).
We should also be able to estimate the degree to which improvement can be obtained relative to a particular parameter. For example, once a correlation has been established between physical stability or structural mimicry and protective responses, physical stability can be maximized (e.g., to 100% retention of D25 binding) or structural mimicry idealized (e.g., to exact mimicry of the D25-bound conformation) to gain an estimate of the maximal improvement of the elicited protective response relative to that particular parameter. The results (Fig. 4A,B) suggest that increased structural mimicry would likely not have much effect on immunogenicity, but that additional physical stabilization of antigenic site Ø might improve the protective titers by up to 4-fold. Independent parameters such as adjuvant, multimerization, delivery vehicle, or immunization regimen offer further avenues for improvement of the elicited response (36).
To quantify elicited binding responses, we coupled different variants of RSV F to an Octet biosensor tip and measured their reactivity with elicited sera (Fig. 4C). “Self-reactive” binding responses (with DS-Cav1, DS or postfusion F on the biosensor tip and sera from animals immunized with DS-Cav1, DS, or postfusion F, respectively) were comparable for DS-Cav1, DS and postfusion F. To provide insight into the binding responses relative to surfaces that were either shared or unique to DS-Cav1, DS, and postfusion F, we measured “cross-reactive” binding responses, for example, with DS-Cav1 on the biosensor tip and sera from animals immunized with DS or postfusion F, as well as binding responses with “preabsorbed” sera, to which different forms of RSV Fs had been added (Fig. 4C). We calculated the shared and unique surface areas, computed from crystal structures of DS-Cav1, DS, and postfusion F (fig. S11 and table S3), and analyzed these relative to the 36 measured binding responses in Fig. 4C. Strong correlation was observed between binding responses and calculated surface areas (Fig. 4D). Several outliers involved surfaces unique to DS-Cav1, surfaces that likely comprise antigenic site Ø. Together the results indicate that the quantity of elicited binding antibody was, to a first approximation, proportional to the surface area of the immunogen, independent of whether that immunogen was DS-Cav1, DS or postfusion F; within this overall similarity, antigenic site Ø elicited ~2-fold higher titer of binding antibody relative to its surface area. Overall, elicited EC50 titers did not correlate with antigenic responses measured against either prefusion or postfusion forms of RSV F, but did correlate with the level of prefusion-specific responses, either measured as a difference or as a ratio (p=0.005) between prefusion-specific and postfusion-specific RSV F-directed responses (Fig. 4E) (37). Notably, the steep slope of this correlation likely reflects the substantially higher neutralization potency observed for prefusion-specific antibodies that target antigenic site Ø (7, 38).
Structural Vaccinology
Structural biology can be used in vaccine development to identify key sites by which to disable a pathogen, to pinpoint residue substitutions that inactivate toxoids, to stabilize select conformations of a subunit antigen, and to determine antibody-antigen complexes, which serve as the basis for epitope-specific strategies of elicitation. Importantly, atomic-level control of antigenicity increasingly appears to be within the reach of structure-based design (16, 39, 40). With complex pathogens like malaria or meningoccocus, antibody-mediated neutralizing activity can be directed against many potential antigens, and vaccine discovery has focused on identifying the best target antigen by methods including genomic analysis or “reverse vaccinology” (41-44). However, for pathogens such as HIV-1, influenza, or RSV, where neutralizing antibodies act almost exclusively through one or two envelope glycoproteins, a site- or supersite-specific strategy might have advantages. Specifically, a viral site of vulnerability to neutralizing antibody may be more amenable to structure-based design than a viral subunit, while retaining a higher level of immunogenicity than an epitope-specific scaffold (45).
With site-specific vaccine strategies, selection of an appropriate target site is of crucial importance. Elicitation frequency, breadth, and potency are key considerations. Antigenic sites that comprise multiple overlapping epitopes of frequently elicited antibodies may be good targets, because of their expected capacity to elicit high levels of reactive antibodies in the general population. In terms of breadth, site Ø is among the most divergent of the antigenic sites on RSV; however, site Ø-directed antibodies such as D25 are capable of neutralizing most human RSVs, likely because of the relatively low genetic diversity of F among circulating human RSV strains, and both subtype A and B neutralizing activity was potently induced by the stabilized RSV Fs reported here (table S4). Potency played a key role in our selection of site Ø as a vaccine target, and antigenic analysis of the elicited response (Fig. 4E) indicated potency of elicited antibodies to be crucial to the success of our site Ø-focused approach (46). Whether the “neutralization-sensitive site” strategy employed here will have success in other contexts thus depends on virus-specific factors such as circulating viral diversity and required level of neutralizing activity to achieve protection. Nonetheless, many of the lessons learned from our efforts with RSV, such as the importance of examining natural human immune responses and of selecting the appropriate target site, are generally applicable.
Supplementary Material
Acknowledgments
We thank J. Stuckey for assistance with figures, I.S. Georgiev and M. Pancera for assistance with immunogen design, G.J. Nabel, R.A. Seder, L. Shapiro, and members of the Structural Biology Section, Structural Bioinformatics Core Section, and Viral Pathogenesis Laboratory at the Vaccine Research Center for helpful comments, and J. Chrzas, J. Gonczy, U. Chinte, and staff at SER-CAT (Southeast Regional Collaborative Access Team) for help with X-ray diffraction data collection. The data presented in this manuscript are tabulated in the main paper and supplementary materials. Atomic coordinates and structure factors of the reported crystal structures have been deposited in the Protein Data Bank under accession codes 4MMQ-4MMV. Support for this work was provided by the Intramural Research Program (National Institute of Allergy and Infectious Diseases) and the National Natural Science Foundation of China (81161120419). Use of insertion device 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38. H.S and T.B. are inventors on an international patent application describing the technology used to isolate D25 and AM22 (Means and methods for influencing the stability of antibody producing cells, WO2007067046A1). A.Q.B and T.B. are inventors on international patent applications for antibody D25 (RSV-specific binding molecules and means for producing them, WO2008147196A2) and antibody AM22 (RSV-specific binding molecule, WO2011043643A1). J.S.M., M.G.J, B.Z., B.S.G., and P.D.K are inventors on U.S. patent applications describing the use of prefusion-stabilized RSV F glycoproteins as vaccine antigens (Prefusion RSV F proteins and their use, 61/798,389 and 61/863,909). J.S.M, M.C., Z.Z., N.X., and B.S.G. are inventors on a Chinese patent application for antibody 5C4 and its epitope (Anti-RSV high neutralization antibody and the peptide can be used for prevention of RSV infection and related disease, 201310082338.1). Reagents from the NIH are subject to non-restrictive materials transfer agreements.
References and Notes
- 1.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 3.The IMpact-RSV Study Group Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 4.Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson LJ, et al. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine. 2013;31(Suppl 2):B209–215. doi: 10.1016/j.vaccine.2012.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magro M, et al. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spits H, Beaumont T. RSV-specific binding molecules and means for producing them. 2010 Patent Application 12/600,950. [Google Scholar]
- 12.Beaumont T, Bakker AQ, Yasuda E. RSV specific binding molecule. 2012 Patent Application 12/898,325. [Google Scholar]
- 13.Burton DR. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 14.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jardine J, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuire AT, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLellan JS, et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat. Struct. Mol. Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLellan JS, et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J. Mol. Biol. 2011;409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong L, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efimov VP, et al. Fibritin encoded by bacteriophage T4 gene wac has a parallel triple-stranded alpha-helical coiled-coil structure. J. Mol. Biol. 1994;242:470–486. doi: 10.1006/jmbi.1994.1595. [DOI] [PubMed] [Google Scholar]
- 22.Miroshnikov KA, et al. Engineering trimeric fibrous proteins based on bacteriophage T4 adhesins. Protein engineering. 1998;11:329–332. doi: 10.1093/protein/11.4.329. [DOI] [PubMed] [Google Scholar]
- 23. Information on materials and methods is available on Science Online.
- 24. RSV F variants were assessed by transient expression in 96-well format (fig. S4). If supernatants showed retained reactivity with antibodies motavizumab and D25 after 1 week at 4° C, they were expressed by transient transfection of 1 l of Expi293F cells, purified by use of appended His-tag and StreptagII, and analyzed by size-exclusion chromatography (fig. S7)
- 25. The inability to express potential inter-chain double cysteine substitutions, despite reasonable modeling in the mature prefusion F1F2 structure, may indicate that RSV F0 protomers, prior to cleavage and removal of peptide 27, adopt substantially different inter-protomer conformations.
- 26. Cavities in the D25-bound RSV F structure were visualized with PyMol using the “Cavities & Pockets Only” option for Surface settings. Amino acid substitutions designed to fill the resulting cavities were identified using the Mutagenesis wizard.
- 27.Pancera M, et al. N332-Directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS One. 2013;8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. The engineered RSV F variants had Cα-root mean square deviations from the D25-bound conformation of 0.7-1.5 Å and from the postfusion conformation of approximately 30 Å.
- 29. Crystallized DS retained ~20% of its D25 recognition relative to crystallized Cav1 (normalized to motavizumab recognition) after 4 months at 20°C, thereby indicating that the crystallized DS was both capable of recognizing D25 as well as converting to a conformation incompatible with D25 binding. By contrast, soluble DS lost all recognition of D25 after 2 months of incubation at 4°C in PBS. We were unable to crystallize DS, which had been heat triggered at 50°C, despite the heat-triggered DS retaining a trimeric state on size exclusion chromatography.
- 30. Observing Ala107 rather than Arg109 at the C terminus of F2 is not totally unexpected: Garten and Klenk (47) found that an arginine at the cleavage site of influenza hemagglutinin was trimmed by a cellular carboxypeptidase activity.
- 31. Cα-root mean square deviations of 0.86 Å for 447 residues between Cav1 and DS-Cav1 grown in ammonium sulfate condition; Cα-root mean square deviations of 0.47 Å for 447 residues of DS-Cav1 in cubic lattices.
- 32.Stetefeld J, et al. Collagen stabilization at atomic level: crystal structure of designed (GlyProPro)10foldon. Structure. 2003;11:339–346. doi: 10.1016/s0969-2126(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 33. When palivizumab (Synagis®) is dosed at a concentration of 15 mg/kg, serum levels at trough are ~40 μg/ml, which provides protection in infants from severe disease and protection in cotton rats from RSV infection. In our neutralization assay, ~40 ug/ml of palivizumab yields an EC50 of 100.
- 34.Nabel GJ. Philosophy of science. The coordinates of truth. Science. 2009;326:53–54. doi: 10.1126/science.1177637. [DOI] [PubMed] [Google Scholar]
- 35. By contrast, affinity of RSV F variants for D25 antibody did not show correlation with elicitation of protective titers. Yield of RSV F trimers also did not correlate with protective titers.
- 36. Flexibility of an antigenic site may increase its immunogenicity by allowing the site to conform to a wider diversity of antibodies. We note in this context that the atomic-mobility factors of antigenic site Ø were among the highest in the RSV F ectodomain.
- 37. For the “prefusion” form of RSV F, we used the DS-Cav1 stabilized variant of RSV F.
- 38. It should be possible to deconvolute the elicited response, by using structurally defined probes, as shown with D25 and motavizumab-bound RSV F in fig. S12.
- 39.Dormitzer PR, Grandi G, Rappuoli R. Structural vaccinology starts to deliver. Nat. Rev. Microbiol. 2012;10:807–813. doi: 10.1038/nrmicro2893. [DOI] [PubMed] [Google Scholar]
- 40.Azoitei ML, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 41.Delany I, Rappuoli R, Seib KL. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb. Perspect. Med. 2013;3:a012476. doi: 10.1101/cshperspect.a012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moriel DG, et al. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9072–9077. doi: 10.1073/pnas.0915077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mora M, Donati C, Medini D, Covacci A, Rappuoli R. Microbial genomes and vaccine design: refinements to the classical reverse vaccinology approach. Curr. Opin. Microbiol. 2006;9:532–536. doi: 10.1016/j.mib.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 45. While an epitope-scaffold strategy may not elicit titers as high as a neutralization-sensitive site strategy, for specific antibodies, such as antibody MPE8 (48), which is capable of neutralizing not only RSV, but other paramyxoviruses including human metapneumovirus, an epitope-specific strategy may suffice.
- 46. The magnitude of antibody activity is a crucial determinant of how well an individual will be protected following vaccination and how long protective responses will be maintained in infants who have passively acquired antibody from their mothers.
- 47.Garten W, Klenk HD. Characterization of the carboxypeptidase involved in the proteolytic cleavage of the influenza haemagglutinin. J. Gen. Virol. 1983;64:2127–2137. doi: 10.1099/0022-1317-64-10-2127. Pt 10. [DOI] [PubMed] [Google Scholar]
- 48.Corti D, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013 doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.