Abstract
Background
Early in the cell cycle a pre-replicative complex (pre-RC) is assembled at each replication origin. This process involves the sequential assembly of the Origin Recognition Complex (ORC), Cdc6, Cdt1 and the MiniChromosome Maintenance (Mcm2-7) proteins onto chromatin to license the origin for use in the subsequent S phase. Licensed origins must then be activated by S phase-inducing cyclin-dependent kinases (S-CDKs) and the Dbf4/Cdc7 kinase.
Results
We have cloned a Xenopus homologue of Dbf4 (XDbf4), the sequence of which confirms the results of Furukhori et al. We have analysed the role of XDbf4 in DNA replication using cell-free extracts of Xenopus eggs. Our results indicate that XDbf4 is the regulatory subunit of XCdc7 required for DNA replication. We show that XDbf4 binds to chromatin during interphase, but unlike XCdc7, its chromatin association is independent of pre-RC formation, occurring in the absence of licensing, XCdc6 and XORC. Moreover, we show that the binding of XCdc7 to chromatin is dependent on the presence of XDbf4, whilst under certain circumstances XDbf4 can bind to chromatin in the absence of XCdc7. We provide evidence that the chromatin binding of XDbf4 that occurs in the absence of licensing depends on checkpoint activation.
Conclusions
We have identified XDbf4 as a functional activator of XCdc7, and show that it is required to recruit XCdc7 to chromatin. Our results also suggest that XCdc7 and XDbf4 are differentially regulated, potentially responding to different cell cycle signals.
Background
The eukaryote genome is organised into multiple chromosomes, whose replication must be strictly controlled in order to ensure that the DNA is replicated once and only once in each cell cycle. DNA replication initiates from multiple origins, whose activation can be divided into two stages. In the first stage, pre-RCs are assembled at replication origins by the sequential binding of the origin recognition complex (ORC), Cdc6, Cdt1 and Mcm2-7 (the MCM/P1 proteins) [1,2]. This assembly takes places during late mitosis and G1, and results in the origin becoming "licensed" for DNA replication. The second stage occurs during S phase and involves the activation of licensed origins by the action of two S phase-promoting kinases: S-phase promoting CDKs and Cdc7/Dbf4, leading to the loading of Cdc45 and the initiation of a pair of replication forks.
Cdc7 is a serine/threonine kinase, conserved from yeast to humans that is required for the initiation of DNA replication (reviewed in [3-6]). Although Cdc7 protein levels are approximately constant throughout the cell cycle, Cdc7 kinase activity peaks at the G1/S transition [7,8]. This cyclic control of activity reflects changes in the abundance of its positive regulatory subunit Dbf4 [9-11], which is degraded in late mitosis and early G1 by the anaphase promoting complex [12-14]. The regulatory subunit Dbf4 has been identified in different organisms, and shows three stretches of conserved amino acids, called motifs-N, M and C [15,16]. The M and C-domains are both necessary for Cdc7 activation [17,18].
Cdc7 is required early in S phase for the initiation of early-origins as well as late in S phase for the initiation of late-firing origins [19,20]. This suggests, that instead of acting as a global activator of S phase, Cdc7 is required to promote initiation at individual origins. Consistent, with this proposal, Cdc7 and Dbf4 have been shown to associate with chromatin [13,21,22], and Dbf4 has been shown to localize to a replication origin using a one-hybrid assay [23]. In Saccharomyces cerevisiae, Dbf4 binds to chromatin at the G1/S transition and remains attached during S phase, whilst Cdc7 appears to remain chromatin-associated throughout the cell cycle [13]. The association of S. cerevisiae Dbf4 with chromatin depends on the presence of ORC, but does not require Cdc6 or Mcm2-7 [21]. In the Xenopus cell-free system, Cdc7 also binds to chromatin during G1 and S phase, but its association with chromatin appears different from yeast. The association of Xenopus Cdc7 with chromatin depends on the presence of Mcm2-7 (i.e. licensed origins), but does not require the continued presence of XORC or XCdc6 once they have fulfilled their essential role in licensing [22]. Recently, it has been described that human Cdc7 and ASK (a human Dbf4 homologue) show a different pattern of nuclear localization and chromatin binding [18]. Human Cdc7 binds to chromatin before ASK is loaded onto chromatin after nuclear re-location. The difference in chromatin binding profile of the catalytic and the regulatory subunits of the Cdc7/Dbf4 kinase suggest that its recruitment is under intricate control.
In order to understand the regulation of the XCdc7/XDbf4 kinase we have cloned a Xenopus homologue of Dbf4 (XDbf4). Whilst this work was in progress, a virtually identical Xenopus sequence was published [24]. A related gene, Drf1, which also binds and activates Cdc7 [25] has also been recently cloned from Xenopus [26]. We have used cell-free extracts of Xenopus eggs to study the requirement of XDbf4 for DNA replication and characterise the chromatin binding profile of XCdc7/XDbf4. Our results suggest that XDbf4 is essential for DNA replication. We show that XDbf4 binds to chromatin independently of pre-RC formation, and does not require the presence of XCdc7. In contrast, the chromatin binding of XCdc7 depends on XDbf4.
Results
Cloning of a Xenopus homolog of Dbf4
The cloning of homologs of Dbf4 from different species has revealed three conserved amino acid domains, named the N-, M- and C-domains [4,16]. We used degenerate PCR to clone a Xenopus Dbf4 homolog. Two degenerate primers were designed to match partially the human Dbf4 N- and C-domains, and we used a Xenopus cDNA library as template (Fig 1). Our approach using a PCR-touchdown protocol in the presence of both degenerate primers, allowed us to amplify a specific fragment with a molecular weight similar to that expected for mammalian Dbf4, which was not amplified when one of the primers were excluded from the PCR mix. This band was sequenced and showed homology to human Dbf4. Afterwards, two specific primers for the PCR product described above were designed and combined with primers specific for the vector library. The PCR reactions generated two partially overlapping PCR products corresponding to the 5' and 3' ends of the gene. These products were sequenced and new specific primers were designed to amplify a fragment containing a single reading frame that codes for a 661 amino acid protein.
Figure 1.
Amino acid sequence of XDbf4. Alignment of the full-length XDbf4 (GenBank accession number AY460183) and human Dbf4. The underlined, the double-underlined and the dash-underlined regions indicate the three conserved domains (N-, M- and C-domains). Arrows indicate the position of the two degenerate primers used for PCR amplification. The overall and the homology between domains are indicated.
The overall identity of human Dbf4 and the Xenopus homologue was low, however the similarity was higher in the previously described N-, M- and C-domains (Fig 1). Another Dbf4-related human protein, called Drf1, has been identified which binds and activates Cdc7 [25]. The protein described by us shows higher similarity to human Dbf4 than to Drf1. During the preparation of this manuscript, two Xenopus Cdc7 regulatory proteins were identified [24,26]. One of them was described by the authors as Xenopus homolog of Drf1 and the second one as a Xenopus homolog of Dbf4. The polypeptide identified in our study seems to correspond to the Dbf4 homologue described by Furukohri et al [24]. Therefore, based on the experimental proof presented below and the evidence described by Furukohri et al. [24], we conclude that we have cloned a Xenopus homolog of Dbf4 (XDbf4).
XDbf4 can support DNA replication
In order to characterise the role of XDbf4 in DNA replication, rabbit polyclonal antibodies were raised against recombinant XDbf4. These antibodies were used for Western blotting and for immunodepletion of XDbf4 from cell interphase extracts. The anti-XDbf4 polyclonal antibody recognised two bands in whole extract of ~95–105 kDa (Fig 2A). However, only the upper band associated with interphase chromatin, which also contains bound Cdc7 [22], and so most likely represents XDbf4. Consistent with previous reports [24], treatment of chromatin with phosphatase reduced the molecular weight of this protein to the size of the His-tagged recombinant protein (data not shown). To confirm this identification, we employed an immunodepletion strategy, using either anti-Cdc7, anti-XDbf4 or non-immune (NI) antibodies (Fig 2B). Immunodepletion of whole extract with anti-Cdc7 antibodies co-depleted the upper XDbf4-reactive band, consistent with this band representing XDbf4. Conversely, depletion of extracts with anti-XDbf4 antibodies depleted the upper XDbf4-reactive band whilst only partially depleting the lower band. Further, XCdc7 was partially co-depleted with the XDbf4 antibodies, consistent with the idea that the Dbf4 regulatory subunit is less abundant than the Cdc7 kinase which it regulates. This is also consistent with the quantification of XCdc7 and XDbf4 levels in whole extract described recently [24]. The excess of XCdc7 could potentially form complexes with the recently identified Drf1 or other unknown proteins.
Figure 2.
Reconstitution of XCdc7/XDbf4. (A) Whole Xenopus egg extract and chromatin isolated from interphase extract were immunoblotted with XDbf4, XCdc7 and XMcm7 antibodies. (B) Interphase Xenopus egg extracts were immunodepleted with antibodies against XDbf4 (XDbf4-) or XCdc7 (XCdc7-) antibodies or with an equal quantity of non-immune antibodies (NI-). Depleted extracts were immunoblotted with XDbf4, XCdc7 or XMcm7 antibodies. (C) Interphase extracts depleted with either XCdc7 antibodies or non-immune antibodies were supplemented with [α-32P]dATP and incubated for 2 hr with Xenopus sperm nuclei or licensed interphase chromatin, in the presence or absence of recombinant XDbf4 and/or XCdc7. After incubation, DNA synthesis was measured, and expressed as a percentage of that occurring in control extract.
Next, we examined the requirement for XDbf4 in DNA replication. Unfortunately, depletion with the XDbf4 antibody generated extracts with defects in nuclear envelope formation, an essential step required for DNA replication [27]. Even the addition of interphase chromatin could not rescue DNA replication in XDbf4-depleted extract (data not shown). Therefore, in order to study the role of XDbf4 in DNA replication we adopted another approach by taking advantage of the observation that XCdc7-depleted extracts contains neither XCdc7 nor XDbf4 (Fig 2B); they are also likely to be depleted of the XDrf1 homologue of XDbf4 [26]. XCdc7-depleted extracts are unable to support sperm DNA replication, but replication can be restored by addition of a Cdc7 kinase activity partially-purified from Xenopus egg extract [22]. We therefore examined DNA synthesis in XCdc7-depleted extract supplemented with recombinant XCdc7 and/or XDbf4 (Fig 2C). The addition of recombinant XCdc7 or XDbf4 alone was not able to rescue DNA replication when sperm DNA was incubated in XCdc7-depleted extract. However, when the extracts were supplemented with both XCdc7 and XDbf4 proteins we observed sperm DNA replication to levels similar to the ones achieved using interphase chromatin (which already contains active Cdc7 kinase [22]) (Fig 2C). This suggests that XDbf4 is required for DNA replication in Xenopus egg extracts.
XDbf4 chromatin binding differs from XCdc7
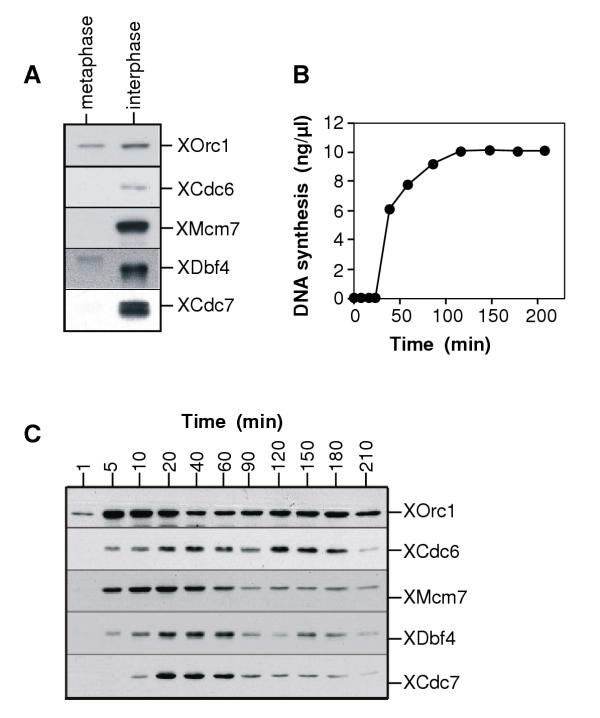
One of the crucial regulatory steps of DNA replication is the orderly recruitment of the different proteins involved in formation of the pre-RC and origin activation [27-29]. We and others have previously described that the association of XCdc7 with chromatin is dependent on the DNA being licensed (ie containing bound Mcm2-7) [22,30]. This differs from yeast, where it appears that the kinase complex is targeted to origins by the binding of Dbf4 to ORC [31]. Moreover, in yeast and humans it has been described that Cdc7 and Dbf4 show different patterns of chromatin binding at different stages of the cell cycle [13,18]. We therefore examined the pattern of XDbf4 chromatin association during the course of the Xenopus cell cycle. First, we checked whether XDbf4 binds to chromatin in metaphase arrested extracts. Fig 3A shows that in metaphase, XDbf4 associates only weakly, if at all, with chromatin. On exit into interphase, strong binding of XDbf4 and XCdc7 then occurs.
Figure 3.
The association of XDbf4 with chromatin differs from that seen with XCdc7. (A) Xenopus sperm nuclei were incubated in metaphase or interphase extract for 30 min. Chromatin was isolated and immunoblotted with antibodies specific for XOrc1, XDbf4, XCdc7, XMcm7 and XCdc6. (B, C) Sperm nuclei were incubated at 10 ng/μl in interphase Xenopus egg extract. At the indicated times, (B) samples were assayed for DNA synthesis by [α-32P]dATP incorporation, or (C) chromatin was isolated and immunoblotted with antibodies specific for XOrc1, XCdc6, XMcm7, XDbf4 and XCdc7.
We then investigated the chromatin binding during interphase. During the first 30–40 minutes the extracts assembled sperm chromatin into interphase nuclei, and during the next hour the DNA was replicated (Fig 3B). The extracts then arrested in a G2-like state because protein synthesis had been blocked by addition of cycloheximide. At the times indicated in Fig 3C, chromatin was isolated from the extracts and immunoblotted for XOrc1, XCdc6, XMcm7, XCdc7 and XDbf4. As has been described previously, the assembly of licensed chromatin by the binding of XORC, XCdc6 and XMcm2-7 was followed by the loading of XCdc7 onto chromatin. On progression through S phase, XMcm2-7 and XCdc7 were displaced from chromatin [22,32]. The binding of XDbf4 to chromatin in general resembled that of XCdc7, with levels peaking at the start of S phase and declining as the DNA was replicated. This is somewhat different from results previously reported by Furukohri et al [24], who saw no significant decrease in the levels of Dbf4 remaining on chromatin during progression through S phase. This may reflect a lowered efficiency of replication termination. However, consistent with the observations of Furukohri et al, early in the timecourse (5–10 min) we observed XDbf4 binding to chromatin in the apparent absence of XCdc7. This suggests that the two proteins could be differentially recruited to chromatin.
The loading of XDbf4 onto chromatin, unlike XCdc7, does not require pre-RC proteins
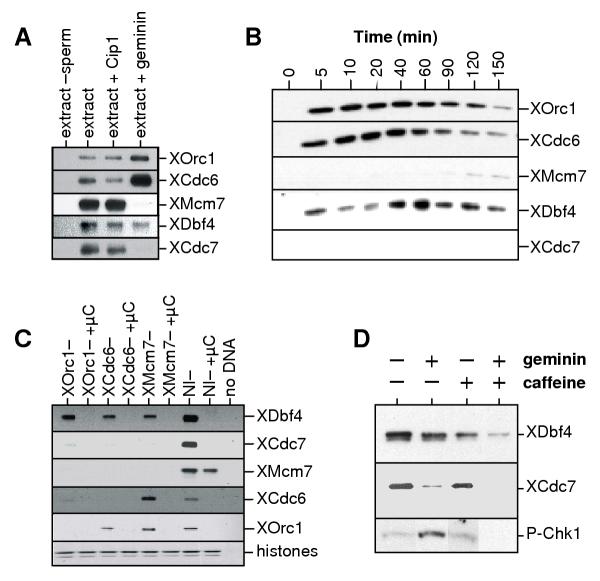
We and others have previously shown that XCdc7 chromatin binding is dependent on licensing [22,30]. This result apparently differs from results obtained in yeast which show that Dbf4 can be recruited to chromatin by ORC [21]. Since we had data suggesting that XDbf4 and XCdc7 could be differentially loaded onto chromatin, we examined the chromatin association of XDbf4 under a variety of different conditions. First we showed that the binding of XDbf4 to chromatin was not dependent on-cyclin-dependent kinase (CDK) activity, since it was not affected by the CDK inhibitor p21Cip1 (Fig 4A). This is consistent with the very early binding of XDbf4 to chromatin seen in Fig 3C (which occurs well before CDK activation), and is similar to the behaviour of Cdc7 [22,30]. In order to investigate whether XDbf4 chromatin association was dependent on licensing, sperm nuclei were incubated in extract treated with geminin, which blocks licensing and the assembly of Mcm2–7 onto chromatin [33-35]. In geminin-treated extracts, XMcm7 and XCdc7 did not associate to chromatin, but XDbf4 was still loaded to levels ~50% of that seen in untreated extracts (Fig 4A). This shows clearly that XDbf4 and XCdc7 can bind to chromatin in a different manner. During the course of the cell cycle in the presence of geminin we observed that XDbf4 binds at the same times as is observed in the absence of geminin, but the removal from chromatin is not so evident (Fig 4B).
Figure 4.
XDbf4 chromatin association is independent of a pre-replication complex. (A) Sperm nuclei were incubated for 30 min in interphase extracts minus or plus added Cip1 (extract+Cip1) or geminin (extract+geminin). Chromatin was isolated, subjected to SDS-PAGE and immunoblotted for XOrc1, XCdc6, XMcm7, XDbf4 and XCdc7. A control lacking added sperm is also shown. (B) Sperm nuclei were incubated at 10 ng/μl in interphase Xenopus egg extract in the presence of geminin. At the indicated times, chromatin was isolated and immunoblotted with antibodies specific for XOrc1, XCdc6, XMcm7, XDbf4 and XCdc7. Equivalent exposures to the time course of Fig 3 are shown. (C) Sperm nuclei were incubated for 40 min in interphase extracts previously immunodepleted with antibodies against XOrc1 (XOrc1-), XCdc6 (XCdc6-), and XMcm7 (XMcm7-), or with non-immune antibodies (NI-). The incubations also were performed in the presence of the phosphatase inhibitor microcystin (μC) or with a control lacking added sperm. Chromatin was then isolated and immunoblotted for XOrc1, XCdc6, XMcm7 and XCdc7. A Coomassie-stained image of the gel containing the histones is shown as loading control. (D) Sperm nuclei were incubated for 1 hr in extract supplemented with geminin and/or 5 mM caffeine. Chromatin was then isolated and immunoblotted with antibodies against XDbf4 and XCdc7. In parallel, nuclei were isolated and immunoblotted for phospho-Ser344-Chk1.
Next, we investigated whether XDbf4 loading was dependent on other pre-RC proteins (Fig 4C). Sperm nuclei were incubated in extracts that had been previously immunodepleted with antibodies specific for XOrc1 (which remove the entire ORC complex [36]), XCdc6, or XMcm7 (which remove the entire Mcm2-7 complex [37-39]). Chromatin was incubated in these extracts, then isolated and immunoblotted for bound proteins. As negative controls, extract was incubated in the absence of sperm, and all incubations were also performed in the presence of microcystin-LR ('μC' in Fig 4B), a phosphatase inhibitor which we had previously seen to disturb the association of many chromatin binding proteins (data not shown). Although microcystin reduced the binding of Orc1 and Cdc6 to chromatin, it did not prevent Mcm7 loading; this is consistent with previous reports showing that the high-affinity chromatin binding of ORC and Cdc6 is not required for functional licensing and stable Mcm2-7 chromatin loading [34,40]. Consistent with the results obtained in geminin-treated extracts, chromatin incubated in XMcm7-depleted extract showed binding of XOrc1, XCdc6 and XDbf4 but not XCdc7. In XCdc6-depleted extract, XOrc1 and XDbf4 associated with chromatin, but XMcm7 and XCdc7 did not. Even in XOrc1-depleted extract, where neither XCdc6, XMcm7, nor XCdc7 associated with chromatin, XDbf4 still showed strong chromatin association. However, XDbf4 binding to chromatin was not completely unspecific, because it could be abolished by treating the extracts with microcystin.
Since all the XDbf4 appears to be complexed with XCdc7 (Fig 2B), XDbf4 must be separated from XCdc7 when it loads onto chromatin when licensing is inhibited with geminin. Since previous work has shown that an ATR-dependent checkpoint can also separate Dbf4 and Cdc7 [41], we investigated the possible role of checkpoint kinases in the binding of XDbf4 to chromatin in the presence of geminin. ATR can activate Xenopus Chk1 by phosphorylating it on Ser344 (equivalent to Ser345 in human Chk1) [42,43]. In extracts treated with geminin, Chk1 becomes phosphorylated (Fig 4D), though the extent of phosphorylation seems to vary somewhat between experiments. Further, Fig 4D shows that the residual binding of XDbf4 to chromatin that occurs in the presence of geminin could be significantly reduced by addition of caffeine. Since caffeine is an efficient inhibitor of the ATM/ATR checkpoint kinases, this suggest a role for these kinases in allowing XDbf4 to bind to chromatin under conditions where XCdc7 does not. Taken together, these results suggest that XDbf4 chromatin binding is independent of any of the proteins involved in the assembly of a pre-replication complex, and indicates that XDbf4 and XCdc7 chromatin association is differentially regulated.
XCdc7 chromatin interaction requires XDbf4
Previously it was described that XCdc7 chromatin binding is completely dependent on licensing [22,30]. We next investigated whether the loading of XCdc7 onto chromatin also requires XDbf4. Sperm nuclei were incubated in non-immune-depleted or XDbf4-depleted extract plus or minus geminin. Chromatin was then isolated and immunoblotted for XOrc1, XCdc6, XMcm7 and XCdc7. Fig 5A shows that, as expected, the addition of geminin to the non-immune-depleted extract blocked the loading of XMcm7 and XCdc7. However, XDbf4-depleted extract did not support the assembly of XCdc7 onto chromatin, even though XDbf4-depleted extract contains a significant quantity of XCdc7 (Fig 2B). The XCdc7 remaining in XDbf4-depleted extract could be induced to bind to chromatin by the addition of recombinant XDbf4; this induced binding appeared physiological as it could be inhibited by geminin. These results suggest that XCdc7 loading is dependent on both licensing and the presence of XDbf4.
Figure 5.
XCdc7 chromatin interaction requires XDbf4. (A) Sperm nuclei were incubated for 30 min in Xenopus interphase extract previously depleted with antibodies specific to XDbd4 (XDbf4-) or with non-immune serum (NI-), minus or plus added recombinant GST-XDbf4 protein, minus or plus geminin. Chromatin was then isolated and immunoblotted with antibodies specific for XCdc7, XMcm7, XOrc1 and XCdc6. (B) Sperm nuclei were incubated for 30 min in XCdc7-depleted extracts (XCdc7-) or non-immune depleted extracts (NI-) containing recombinant GST-XCdc7, recombinant XDbf4 previously treated with thrombin to remove the GST tag (XDbf4), recombinant geminin and/or microcystin. Incubation of recombinant proteins in non-immune depleted extract lacking sperm are shown as negative controls. Chromatin was then isolated and immunoblotted for XDbf4, XCdc7, XMcm7 and XOrc1.
To confirm the requirement of XDbf4 for XCdc7 chromatin loading and to test the possibility that XDbf4 loading was dependent on XCdc7, we investigated the loading of recombinant XCdc7 and XDbf4 proteins in a XCdc7-depleted extract (Fig 5B). In order to avoid a possible dimerization effect between the recombinant proteins due to the GST tag, we removed it from the recombinant XDbf4 protein used in this experiment. A non-immune-depleted extract was incubated with the recombinant proteins in the absence of sperm nuclei as negative controls. The addition of recombinant XCdc7 to XCdc7-depleted extract did not rescue the loading of XCdc7 onto chromatin. However, consistent with the results obtained in a XDbf4-depleted extract and with the observed DNA replication rescue in a XCdc7-depleted (Fig 2), the addition of recombinant XCdc7 and XDbf4 allow the assembly of recombinant XCdc7 onto chromatin. Evidence that this association corresponded to a physiological process was reinforced by the fact that recombinant XCdc7 chromatin binding was blocked by geminin addition, leaving recombinant XDbf4 bound to chromatin, similar to what is observed for endogenous XDbf4 in geminin-treated extract (Fig 4). The recombinant XDbf4 detected on chromatin after reconstitution of XCdc7/Dbf4 chromatin loading showed a retarded mobility that might suggest a XCdc7-dependent XDbf4 phosphorylation, since it was not observed in the absence of recombinant XCdc7. Cdc7-dependent Dbf4 phosphorylation has been described previously in mammalian cells [44,45]. On the other hand, recombinant Dbf4 was loaded onto chromatin in a XCdc7-depleted extract. Consistent with our previous results, this association was licensing independent but was inhibited by microcystin. These results suggest that XCdc7 chromatin association is not only dependent on licensing, but also requires the presence of its regulatory subunit XDbf4. However, XDbf4 chromatin binding is independent of the presence of pre-RC proteins and XCdc7.
Discussion
In this study we have cloned, using a degenerate PCR approach, a Xenopus homologue of Dbf4 (XDbf4) and have characterized its role in DNA replication using the Xenopus cell-free system. Our results suggest that XDbf4 is the regulatory subunit of XCdc7 and is required for DNA replication. We have shown that XDbf4 associates with chromatin during the cell cycle and that this association is independent of licensing and can also occur in the absence of XCdc6 and XORC. We have shown that the binding of XCdc7 to chromatin depends on the presence of XDbf4, whilst the binding of XDbf4 to chromatin does not depend on the presence of Cdc7.
Dbf4 homologues had been described in several organisms but a Xenopus homolog had not been identified when we began this work. The presence in the Dbf4 family of three relative well conserved motifs allowed us to design degenerate primers in order to clone by PCR a Dbf4 homolog in Xenopus. XDbf4 shares low identity (33%) with human Dbf4, but a higher similarity is observed over the three conserved domains. During the preparation of this manuscript two Dbf4-related proteins were described in Xenopus [24,26]. Both proteins are able to activate XCdc7 and phosphorylate Mcm2. However, it was not known which of them corresponded to the regulatory subunit of XCdc7 that is essential for DNA replication. Based on the sequence reported for these two genes, the protein described in our study corresponds to the one described by Furukohri et al. [24], although both proteins showed a few differences in the sequence that could represent genetic variants.
We have addressed the role of XDbf4 in DNA replication by using Xenopus cell free extracts depleted of XCdc7, since we could not prepare extracts depleted of XDbf4 that were suitable for DNA replication assays. XCdc7-depleted extracts, which lack both XCdc7 and XDbf4 (and the XDrf1 homologue of XDbf4 [26]), are deficient for DNA replication as we previously described [22]. XCdc7-depleted extract supplemented with recombinant XCdc7 alone does not support efficient DNA replication, suggesting that one or more cofactors were co-depleted with XCdc7. The efficient rescue of DNA replication when both recombinant XCdc7 and XDbf4 were added to the XCdc7-depleted extract support the idea that the protein cloned by us is a functional Xenopus homologue of Dbf4. Immunodepletion of Drf1 from extracts does not prevent XCdc45 chromatin loading or DNA replication [26]; however it still remains possible that both XCdc7/XDbf4 and XCdc7/Drf1 could have a role in DNA replication. The rescue of DNA replication in a XCdc7 depleted extract by the addition of recombinant XCdc7/XDrf1 could provide a definitive answer.
The assembly of pre-RCs is a highly regulated process. How the XCdc7/XDbf4 kinase complex is targeted to replication origins seems to differ between different organisms. In S. cerevisiae, Dbf4 binds to chromatin at the G1/S transition and remains attached during S phase, whilst Cdc7 appears to remain chromatin-associated throughout the cell cycle [13]. The association of Dbf4 with chromatin depends on the presence of ORC, but does not require Cdc6 or Mcm2-7 [21]. Recently, it has been described that the localization and chromatin binding of endogenous huCdc7 and ASK (human Dbf4 homolog) are independently regulated, and huCdc7 binds to chromatin before ASK is loaded onto chromatin after nuclear re-location [18]. We previously described that in Xenopus the association of Cdc7 with chromatin depends on the presence of Mcm2-7, but does not require the continued presence of XORC or XCdc6 [22]. Our present results suggest that in Xenopus, XCdc7 and XDbf4 are targeted differently to replication origins. XDbf4 seems to bind to chromatin very early during cell cycle, clearly before than XCdc7, and this association occurs in the presence of the licensing inhibitor geminin, or in the absence of functional XCdc6 or XORC. This suggests that XDbf4 chromatin binding, unlike XCdc7, requires neither licensing nor other pre-RC proteins. A similar XDbf4 chromatin binding in the absence of XORC has been described [24].
Recently it has been shown that Cdc7/Dbf4 could be the target of DNA replication checkpoints, activation of which results in the release of XDbf4 from XCdc7 and from chromatin, leaving XCdc7 associated with chromatin [41]. We show here that the binding of XDbf4 to chromatin that occurs when licensing is blocked by geminin is sensitive to inhibition by caffeine, an inhibitor of the ATM/ATR checkpoint kinases. The physiological consequence of this XDbf4 binding is currently unknown. However, geminin induces the phosphorylation of Chk1 in a caffeine-sensitive manner, consistent with a role for checkpoint kinases in this process. It is unclear why geminin should cause checkpoint activation, though it may occur as a consequence of S phase initiating under circumstances where there are only a very small number of licensed origins present. Consistent with this idea, peak levels of chromatin-bound XDbf4 in geminin-treated extract was seen when the extract would normally be in S phase. Checkpoint activation has also been observed in human cancer cells entering S phase whilst forced to overexpress geminin [46]. Taken together, these results suggest that checkpoint kinases may be involved in the separation of XDbf4 and XCdc7 and their differential recruitment to chromatin. The observation that during cell cycle progression XDbf4 shows a peak of chromatin association after Mcm2-7 are loaded and at the time of XCdc7 binding suggests the possibility that XDbf4 binds with a higher affinity to chromatin via Mcm2-7 after licensing has occurred.
Whilst XDbf4 can bind to chromatin independently of XCdc7, the converse does not seem to be true. The lack of XCdc7 chromatin binding when sperm DNA was incubated in a XDbf4-depleted extract and the fact that the addition of recombinant XDbf4 rescues XCdc7 chromatin loading in a licensing-dependent manner, strongly suggest that XCdc7 loading is dependent on two events: the presence of XDbf4 and licensing. This was confirmed by the observation that a XCdc7-depleted extract can only support the binding of recombinant XCdc7 protein on chromatin in the presence of recombinant XDbf4 and licensing. This result provides an explanation for why the rescue of DNA replication in a XCdc7-depleted extracts depends on both addition of recombinant XCdc7 and XDbf4. This is the first time that a requirement for Dbf4 has been demonstrated for the chromatin loading of Cdc7. This conclusion is consistent with studies showing that particular Dbf4 domains can target the kinase complex to DNA, as has been described for yeast and human [17,18,23,47]. These results are in apparent disagreement with the results of Yanow et al [26] who showed that in Drf1-depleted extracts, XCdc7 is not recruited to chromatin. One formal possibility to explain this discrepancy would be that XCdc7 chromatin association is dependent on both regulatory subunits, XDbf4 and XDrf1, and that the recombinant XDbf4 bypasses the requirement of XDrf1 when it is added to a XCdc7 depleted extract. However, this explanation would be difficult to reconcile with the absence of DNA replication defects reported in a Drf1-depleted extracts [26]. An alternative explanation is that XDbf4 and XDrf1 recruit XCdc7 to chromatin in distinct ways: XDbf4 is required to recruit XCdc7 prior to initiation, a function which is complete early in the cell cycle [22]; XDrf1 is involved in checkpoint-dependent recruitment of XCdc7, which is typically observed later in the cell cycle when the observations of Yanow et al [26] were made.
Conclusions
Our work provides evidence that XDbf4 corresponds to the regulatory subunit of XCdc7 required for DNA replication. The different subunits of the XCdc7/XDbf4 kinase complex seem to be differently regulated during cell cycle progression, XDbf4 being recruited to chromatin independently of pre-replication complex formation or the presence of XCdc7. Importantly, however, we show that XCdc7 chromatin binding depends not only on licensing but also on XDbf4.
Methods
Xenopus Dbf4 cloning
Sequence analysis of Dbf4 homologues allowed us to design two degenerate primers containing deoxyinosine that were positioned at conserved regions of the N-domain and C-domain. The forward primer was GAYATIAARIIIYTIGGIGG and the reverse primer was RCAIIIYTCRCARTAICC (Life Technologies). As a template we used a cDNA library from unfertilised Xenopus eggs (Clontech). A PCR fragment of 710 pb was amplified only in the presence of both degenerate primers. This PCR product was cloned and sequenced, showing homology to human Dbf4. Specific XDbf4 primers were designed and combined with primers present in the vector to amplify the N and C-terminus. The PCR fragments were cloned using a pGem easy vector and sequenced. After identification of the initiation codon and the stop codon, specific primers, containing restriction sites for cloning in pet21a or pGEX2T, were designed to amplify full length XDbf4 cDNA using Pfu Taq. Also we amplified a full length XCdc7 cDNA. The PCR was repeated two times and both strands of the cloned product were sequenced.
Recombinant protein production
BL21DE3 cells were transformed with the XDbf4-His construct. Cells were allowed to grow to an OD600 of 0.7 and then were induced with 0.4 mM IPTG for 3 hours. After 30'of induction rifampicin (100 μg/ml) was added to the medium. Inclusion bodies were isolated using Bug Buster reagent (Novagen). XDbf4-His was purified using Ni-agarose (Qiagen) following the manufacturer's instructions. The protein was used to produce a rabbit polyclonal antibody against XDbf4-His. GST-XDbf4 or GST-XCdc7 generated by specific PCR were used to transform BL21DE3 cells. Cells were allowed to grow to an OD600 of 0.7 and then protein expression was induced by addition of IPTG and incubated for 3 hr. In order to get soluble protein we used a modification of a previously reported protocol that recovers soluble recombinant protein in the presence of Sarkosyl [48]. Pelleted cells were resuspended in STE (10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA) plus lysozyme (100 μg/ml), left on ice for 15 min, then supplemented with dithiothreitol (DTT; 25 μM), 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin and PMSF (100 μM). Cells were lysed via two freeze thaw cycles using liquid nitrogen. After incubation on ice for 10 min, benzonase was added (25–30 U/ml, Novagen) and further incubated for 15 min on a rotating mixer at room temperature. Sarkosyl was added to a final concentration of 0.75–1.25 %, mixed gently and spun at 10,000 g at 4°C for 12 min. To the supernatant Triton X-100 was added to a final concentration of 1.5–2%. Recombinant protein was purified using Glutathione Sepharose 4B beads (Amersham) following the manufacturer's instructions, except that the glutathione elution buffer contained a final concentration of 0.2% sarkosyl. Eluted protein solutions were concentrated approximately 5 fold using a Centricon 30 (Schleicher & Schuell). In order to eliminate sarkosyl from the sample, a detergent clean-up matrix was used (Pharmacia) following the manufacturer's instructions. A fraction of GST-XDbf4 protein was digested with thrombin (Pharmacia) in order to get a recombinant XDbf4 protein free of GST. GST-p21Cip/Waf1 [49] and 6His-geminin [33] were prepared as described.
Preparation and use of egg extracts
Activated, metaphase-arrested, and high speed-Xenopus egg extracts were prepared as described [50]. For replication assays, the extracts were supplemented with 100 μg/ml cycloheximide, 25 mM phosphocreatine, 15 μg/ml creatine phosphokinase and 0.3 mM CaCl2. As negative control some incubations were performed in the presence of microcystin (1 μM). Immunodepletion of interphase extracts with specific antibodies or with antibodies from non-immune rabbit serum was performed as described [50]. Immediately after immunodepletion, extracts were snap-frozen in liquid nitrogen in 10 μl aliquots for future use. Demembranated Xenopus sperm nuclei were prepared as described [50]. They were assembled into chromatin by incubation at 5–20 ng/μl for appropriate times in treated extracts. In the case of immunodepleted extracts, sperm nuclei were incubated at 7 ng/μl, to compensate for the threefold dilution that occurs during depletion. GST-XDbf4 and GST-XCdc7 were added to egg extracts to give final concentrations of 40–50 nM; this is slightly higher than the 10 nM reported for the concentration of endogenous XDbf4 [24]. DNA synthesis was measured by incorporation of [α-32P]dATP into acid insoluble material as described [50]. All incubations were performed at 23°C.
Antibodies
Antibodies raised against XOrc1 [36], XCdc6 [51], XMcm3 [39], and XCdc7 [22] were as described previously. Anti-human phospho-Chk1Ser345 was from Cell Signaling Technology. Rabbit polyclonal antibodies were raised against recombinant His-tagged full-length Xenopus Dbf4. The XDbf4 antibody specifically immunoprecipitated a single protein. This protein also was recognised on immunoblots of chromatin assembled in the Xenopus extracts. The predicted molecular weight of full length XDbf4 was about 75 kDa. However, the apparent molecular mass of XDbf4, as determined from its electrophoretic mobility by SDS-PAGE, was higher. Phosphatase treatment of this protein reduced the apparent molecular weight of the protein to that of recombinant XDbf4-His. Although the antibody also cross-reacted in immunoblots of whole extracts with other polypeptides, these polypeptides were not immunoprecipitated by the antibody.
Chromatin and nuclear isolation
For isolation of chromatin, extract was diluted 10 to 20-fold in Nuclear Isolation Buffer [50] (NIB 50 mM KCl, 50 mM HEPES-KOH at pH 7.6, 5 mM MgCl2, 2 mM DTT, 0.5 mM spermine-3HCl, 0.25 mM spermidine-4HCl, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) supplemented with 0.25% Triton X-100 and then underlayered with the same buffer containing 15% sucrose. The chromatin was pelleted at 2100 g in a swinging bucket centrifuge for 5 min at 4°C. The diluted extract and the top part of the cushion were carefully removed, and the chromatin pellet was resuspended in loading buffer and subjected to immunoblotting by standard techniques using 10% SDS-PAGE and ECL detection (Amersham).
For isolation of intact nuclei [52], extract containing nuclei was resuspended 10-fold in buffer containing 40% (w/v) sucrose, 50 mM Hepes-KOH pH 7.5, 100 mM KCl and 2.5 mM MgCl2 and pelleted at 5,000 × g in swing-out rotor for 5 min at 4°C. The pellet was resuspended in 1 ml of the same buffer and re-centrifuged under the same conditions. The supernatant was removed and nuclear samples were resuspended in loading buffer and analysed by immunoblotting.
Authors' contributions
PJ performed most of the experiments and wrote the first draft of the manuscript. MGL prepared some of the recombinant proteins and antibodies, and did experiments involving caffeine, Chk1 and phosphatase treatment. JJB co-ordinated the work.
Acknowledgments
Acknowledgements
Thanks to Anatoliy Li and Margret Michalski for comments on the manuscript. This work was supported by Cancer Research UK grants SP2385/0101 and C303/A3135.
Contributor Information
Pedro Jares, Email: pjares@clinic.ub.es.
M Gloria Luciani, Email: m.g.luciani@dundee.ac.uk.
J Julian Blow, Email: j.j.blow@dundee.ac.uk.
References
- Blow JJ, Hodgson B. Replication licensing - defining the proliferative state? Trends in Cell Biology. 2002;12:72–78. doi: 10.1016/S0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes to Cells. 2002;7:523–534. doi: 10.1046/j.1365-2443.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- Jares P, Donaldson A, Blow JJ. The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint? EMBO Rep. 2000;1:319–322. doi: 10.1093/embo-reports/kvd076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Masai H, Sugino A. A Cdc7p-Dbf4p protein kinase activity is conserved from yeast to humans. Prog Cell Cycle Res. 2000;4:61–69. doi: 10.1007/978-1-4615-4253-7_6. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- Sclafani RA. Cdc7p-Dbf4p becomes famous in the cell cycle - Commentary. J Cell Sci. 2000;113:2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Molecular and Cellular Biology. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HJ, Loo S, Campbell JL. Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Molecular Biology of the Cell. 1993;4:195–208. doi: 10.1091/mbc.4.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proceedings of the National Academy of Sciences of the USA. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro G, Owens JC, Shellman Y, Sclafani RA, Li JJ. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Molecular and Cellular Biology. 1999;19:4888–4896. doi: 10.1128/mcb.19.7.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H. A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Molecular and Cellular Biology. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Collyer T, Hardy CF. Cell cycle regulation of DNA replication initiator factor Dbf4p. Molecular and Cellular Biology. 1999;19:4270–4278. doi: 10.1128/mcb.19.6.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO Journal. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MF, Santocanale C, Drury LS, Diffley JF. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Molecular and Cellular Biology. 2000;20:242–248. doi: 10.1128/mcb.20.1.242-248.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Masai H, Sugino A. First the CDKs, now the DDKs. Trends in Cell Biology. 1999;9:249–252. doi: 10.1016/S0962-8924(99)01586-X. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem Biophys Res Commun. 2000;275:228–232. doi: 10.1006/bbrc.2000.3281. [DOI] [PubMed] [Google Scholar]
- Ogino K, Takeda T, Matsui E, Iiyama H, Taniyama C, Arai K, Masai H. Bipartite binding of a kinase activator activates Cdc7-related kinase essential for S phase. J Biol Chem. 2001;276:31376–31387. doi: 10.1074/jbc.M102197200. [DOI] [PubMed] [Google Scholar]
- Sato N, Sato M, Nakayama M, Saitoh R, Arai K, Masai H. Cell cycle regulation of chromatin binding and nuclear localization of human Cdc7-ASK kinase complex. Genes Cells. 2003;8:451–463. doi: 10.1046/j.1365-2443.2003.00647.x. [DOI] [PubMed] [Google Scholar]
- Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes and Development. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes and Development. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Duncker BP, Schwob E, Gasser SM. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes and Development. 1999;13:2159–2176. doi: 10.1101/gad.13.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Blow JJ. Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6 or CDK activity and is required for XCdc45 loading. Genes and Development. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JF. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- Furukohri A, Sato N, Masai H, Arai K, Sugino A, Waga S. Identification and characterization of a Xenopus homolog of Dbf4, a regulatory subunit of the Cdc7 protein kinase required for the initiation of DNA replication. J Biochem (Tokyo) 2003;134:447–457. doi: 10.1093/jb/mvg163. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Bosotti R, Villa F, Rialland M, Brotherton D, Mercurio C, Berthelsen J, Santocanale C. Drf1, a novel regulatory subunit for human Cdc7 kinase. Embo J. 2002;21:3171–3181. doi: 10.1093/emboj/cdf290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanow SK, Gold DA, Yoo HY, Dunphy WG. Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J Biol Chem. 2003;278:41083–41092. doi: 10.1074/jbc.M307144200. [DOI] [PubMed] [Google Scholar]
- Blow JJ. Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J. 2001;20:3293–3297. doi: 10.1093/emboj/20.13.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Blow JJ. The regulation of replication origin activation. Current Opinion in Genetics and Development. 1999;9:62–68. doi: 10.1016/S0959-437X(99)80009-4. [DOI] [PubMed] [Google Scholar]
- Walter JC. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem. 2000;275:39773–39778. doi: 10.1074/jbc.M008107200. [DOI] [PubMed] [Google Scholar]
- Duncker BP, Shimada K, Tsai-Pflugfelder M, Pasero P, Gasser SM. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc Natl Acad Sci U S A. 2002;99:16087–16092. doi: 10.1073/pnas.252093999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/S1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/S0092-8674(00)81209-X. [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/S0092-8674(00)81346-X. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. European Molecular Biology Organization Journal. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thömmes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. European Molecular Biology Organization Journal. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova TA, Blow JJ. Sequential MCM/P1 subcomplex assembly is required to form a hetero-hexamer with replication licensing activity. Journal of Biological Chemistry. 2000;275:2491–2504. doi: 10.1074/jbc.275.4.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/S1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Sato N, Yamada M, Mahony D, Seghezzi W, Lees E, Arai K, Masai H. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Molecular and Cellular Biology. 1999;19:5083–5095. doi: 10.1128/mcb.19.7.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung AD, Ou J, Bueler S, Brown GW. A conserved domain of Schizosaccharomyces pombe dfp1(+) is uniquely required for chromosome stability following alkylation damage during S phase. Mol Cell Biol. 2002;22:4477–4490. doi: 10.1128/MCB.22.13.4477-4490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Current Biology. 1994;4:876–883. doi: 10.1016/S0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Chong JP, Thömmes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Methods in Enzymology. 1997;283:549–564. doi: 10.1016/S0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- Tada S, Chong JPJ, Mahbubani HM, Blow JJ. The RLF-B component of the replication licensing system is distinct from Cdc6 and functions after Cdc6 binds to chromatin. Current Biology. 1999;9:211–214. doi: 10.1016/S0960-9822(99)80092-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]