Abstract
Prion diseases are fatal neurodegenerative disorders characterized by the accumulation of prion protein (PrPC). To date, there is no effective treatment for the disease. The accumulated PrP, termed PrPSc, forms amyloid fibrils and could be infectious. It has been suggested that PrPSc is abnormally folded and resistant to proteolytic degradation, and also inhibits proteasomal functions in infected cells, thereby inducing neuronal death. Recent work indicates that the ubiquitin-proteasome system is involved in quality control of PrPC. To reveal the significance of prion protein ubiqitination, we focused on ubiquitin-specific protease 14 (USP14), a deubiqutinating enzyme that catalyzes trimming of polyubiquitin chains and plays a role in regulation of proteasomal processes. Results from the present study showed that treatment with a selective inhibitor of USP14 reduced PrPC, as well as PrPSc, levels in prion-infected neuronal cells. Overexpression of the dominant negative mutant form of USP14 reduced PrPSc, whereas wildtype USP14 increased PrPSc in prion-infected cells. These results suggest that USP14 prevents degradation of both normal and abnormal PrP. Collectively, a better understanding about the regulation of PrPSc clearance caused by USP14 might contribute greatly to the development of therapeutic strategies for prion diseases.
Prion diseases are fatal neurodegenerative disorders associated with the conformational conversion of cellular prion protein (PrPC) to β-sheet-rich abnormal prion protein (PrPSc)1. PrPC builds up in the rough endoplasmic reticulum (ER), is transported to the Golgi, and subsequently sorted to the cell surface as a glycosylphosphatidylinositol (GPI)-anchored protein. During its biosynthetic maturation in the ER, PrPC is subject to several posttranslational modifications, including cleavage of the N-terminal signal peptide and C-terminal hydrophobic peptide, addition of N-linked oligosaccharide chains at two sites, formation of a single disulphide bond, and attachment of the GPI anchor2,3,4.
Misfolded proteins are removed from the ER by retrograde transportation to the cytosol and degraded by the ubiquitin–proteasome system, which is referred to as endoplasmic reticulum-associated degradation (ERAD). A recent study revealed that ubiquitin ligase gp78 is involved in unglycosylated prion protein degradation5. However, to the best of our knowledge, there is as yet no convincing evidence demonstrating direct ubiquitination of prion protein. Although both nascent wildtype6,7 and misfolded PrP8,9,10 are actually degraded by ERAD prior to release into the secretory pathway, the precise mechanism by which proteasome degrades prion protein has not been established.
Ubiquitin-specific protease 14 (USP14), one of the deubiquitinating enzymes, is localized at the proteasome11. USP14 plays a special role in rescuing proteins from degradation by trimming ubiquitin chains from its substrate-distal tip of the ubiquitinated protein12,13,14. In yeast and mammals, loss of USP14/Ubp6 (yeast homolog of USP14) results in increased degradation of ubiquitinated proteins and reduced levels of free ubiquitin, suggesting that USP14 is required for ubiquitin recycling at the proteasome to maintain ubiquitin pools15,16,17,18. Thus, the proteasomal function is limited by USP14-dependent chain trimming. It has been shown that endogenous USP14 negatively regulates ERAD19. USP14 overexpression inhibits degradation of the null Hong Kong mutant α1-antitrypsin, an ER luminal unfolded protein of ERAD substrate20, whereas USP14 depletion by small interfering RNA effectively accelerates its degradation. Intriguingly, Lee et al. demonstrated that IU1, a selective small-molecule inhibitor of USP14, accelerated proteasomal degradation of tau and TDP-43, which have been implicated in neurodegenerative diseases21. Thus, we focused on the USP14 system to reveal PrPC degradation in the proteasome, and investigated whether USP14 is related to regulation of prion protein degradation. Results demonstrated that an inhibitor of USP14 reduced PrPC in mouse neuroblastoma cells, as well as PrPSc, indicating that USP14 negatively regulates degradation of prion protein.
Results
Effects of IU1 treatment on prion protein degradation
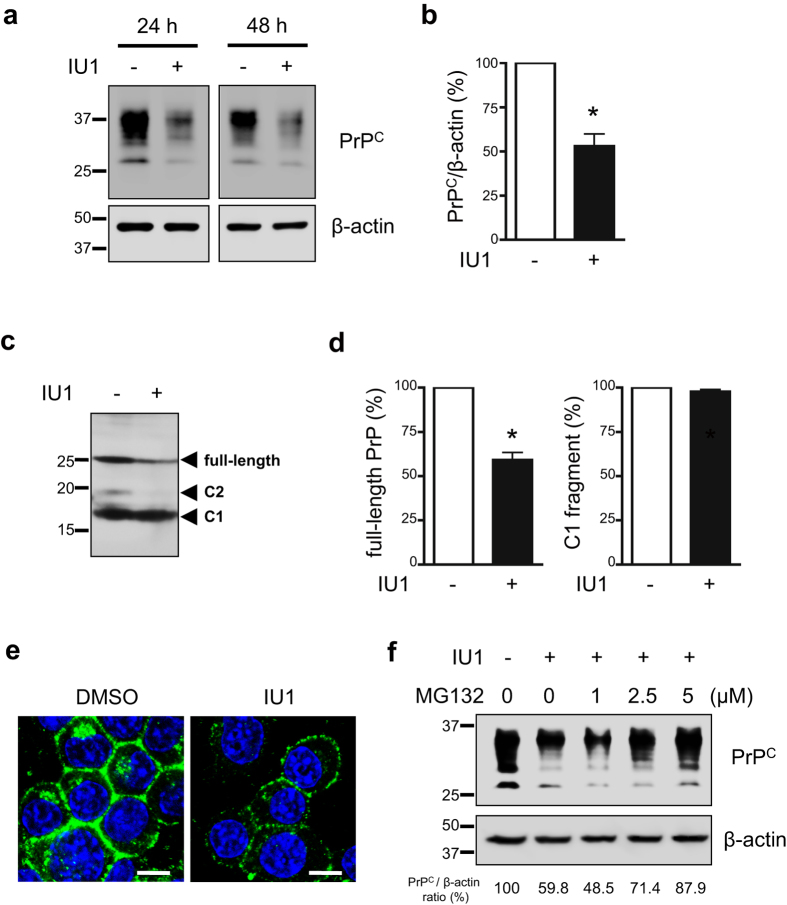
To examine the effect of IU1 on PrPC degradation, mouse neuroblastoma N2a cells expressing endogenous PrP (N2a58 cells) were treated with 100 μM IU1, an agent that inhibits the function of USP1421, for 24 or 48 h. Upon lysis, we observed that protein levels of PrPC were reduced in IU1-treated N2a58 cells compared with DMSO-treated N2a58 cells (Fig. 1a,b). The mRNA levels of PrPC were not altered after the treatment for 48 h (Supplementary Fig. S1a). Reduction of protein levels of PrPC was also observed in mouse hypothalamic GT1-7 cells (Supplementary Fig. S2), indicating that the effect was not restricted to N2a58 cells.
Figure 1. IU1 treatment reduces PrPC levels in N2a58 cells.
(a) N2a58 cells were treated with DMSO or 100 μM IU1 for 24 or 48 h. Lysates were analyzed by immunoblotting with anti-PrP (SAF32) and anti-β-actin antibodies. (b) The graph shows the PrPC levels in N2a58 cells after treatment with DMSO or 100 μM IU1 for 48 h from at least three independent experiments. Asterisk indicates significant difference (*P < 0.05). Mean ± SD. (c) N2a58 cell lysates were deglycosylated with PNGase F followed by immunoblotting with anti-PrP (SAF83) antibody. (d) Quantification of deglycosylated PrP from at least three independent experiments was performed as described in (c). Asterisk indicates significant difference (*P < 0.05). Mean ± SD. (e) N2a58 cells were treated with DMSO or 100 μM IU1 for 48 h. PrPC (SAF32; green) and nuclei (blue) were visualized. Bars: 10 μm. (f) During 48 h of IU1 treatment and 12 h prior to harvest, N2a58 cells were treated with DMSO or MG132 (1, 2.5, 5 μM). Lysates were analyzed by immunoblotting with anti-PrP (SAF32) and anti-β-actin antibodies. Numbers below the gel indicate relative expression of PrPC normalized to β-actin.
For better signal comparison, proteins were deglycosylated with PNGase F. After deglycosylation, we observed full-length PrP, the highest level of C1 fragments, and barely detectable levels of C2 fragments (Fig. 1c). Of note, IU1 treatment significantly reduced full-length PrP levels, but had little effect on C1 fragment levels (Fig. 1c,d). We next analyzed PrPC in cells using immunofluorescence. N2a58 cells were treated with 100 μM IU1 for 48 h and stained with anti-PrP antibody for PrPC detection. As expected, we observed reduced PrPC levels after IU1 treatment (Fig. 1e). To confirm whether PrPC degradation was proteasome-dependent, we utilized the proteasome inhibitor MG132. During 48 h of IU1 treatment and 12 h prior to harvest, vehicle DMSO or MG132 was added to the cultures. As expected, in the presence of MG132, PrPC degradation was prevented in a dose-dependent manner (Fig. 1f).
Autophagy is one of the major cellular degradation pathways involved in protein and organelle turnover, in addition to proteasomal degradation. Therefore, we examined the effect of IU1 on autophagic flux by measuring protein levels of an autophagosomal membrane protein LC3-II, which is involved in autophagosome formation. IU1 treatment had little effect on protein levels of LC3-II, while the lysosome inhibitor (NH4Cl) treatment increased LC3-II levels in N2a58 cells (Supplementary Fig. S3). Furthermore, during 48 h of IU1 treatment and 24 h prior to harvest, NH4Cl was added to the cultures. The combination of IU1/NH4Cl resulted in no further increase of LC3-II, suggesting that PrPC reduction by IU1 treatment was not due to autophagy activation. Taken together, these results demonstrate that IU1 treatment reduces PrPC via proteasome-dependent degradation.
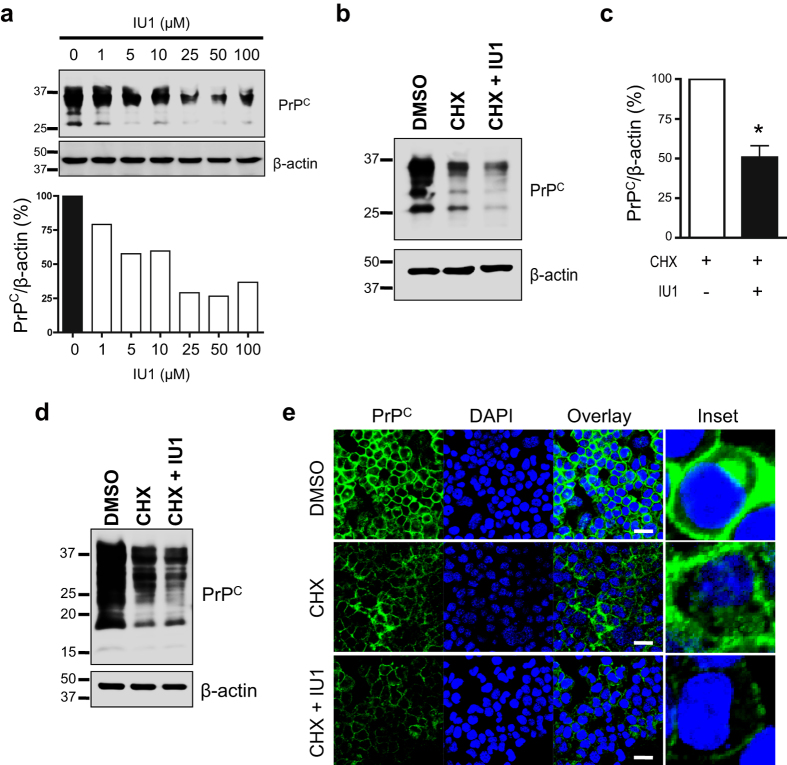
To analyze PrPC protein stability, N2a58 cells were treated with the protein synthesis inhibitor cycloheximide (CHX) combined with various concentrations of IU1 (1–100 μM) for 6 h and subsequently analyzed by immunoblotting with anti-PrP antibody. The reduction of PrPC levels by IU1 treatment was dose-dependent and detectable at IU1 concentrations as low as 1 μM (Fig. 2a). To test whether IU1 treatment could accelerate proteasomal degradation of PrPC, N2a58 cells were treated with CHX alone or in combination with 100 μM IU1 for 6 h, and the lysates were subsequently subjected to immunoblotting with anti-PrP antibody. CHX treatment reduced PrPC level in N2a58 cells (Fig. 2b). Of note, CHX/IU1 combination reduced PrPC more significantly than CHX treatment alone (Fig. 2b,c). Similar results were obtained in PrP-deficient HpL mouse hippocampal cells transiently transfected with PrP, according to immunoblot results (Fig. 2d). Immunofluorescence analysis revealed that CHX/IU1 combination reduced PrPC level more significantly than CHX treatment alone in N2a58 cells (Fig. 2e). These findings indicated that proteasomal degradation of PrPC was indeed dramatically stimulated by IU1 treatment.
Figure 2. IU1 treatment accelerates degradation of PrPC in N2a58 cells.
(a) N2a58 cells were treated with 50 μg/ml CHX combined with various concentrations of IU1 (1–100 μM) for 6 h. Lysates were analyzed by immunoblotting with anti-PrP (SAF32) and anti-β-actin antibodies. (b) N2a58 cells were treated with DMSO, 50 μg/ml CHX alone, or a combination of CHX and 100 μM IU1 for 6 h. Lysates were analyzed by immunoblotting with anti-PrP (SAF32) and anti-β-actin antibodies. (c) Quantification of PrPC from at least three independent experiments was performed as described in (b). Asterisk indicates significant difference (*P < 0.05). Mean ± SD. (d) HpL cells were transfected with pcDNA3.1 mouse PrP expression plasmid. After 48 h, cells were treated with DMSO, 50 μg/ml CHX alone, or a combination of CHX and 100 μM IU1 for 6 h. Lysates were analyzed by immunoblotting with anti-PrP (SAF83) and anti-β-actin antibodies. (e) N2a58 cells were treated with DMSO, 50 μg/ml CHX alone, or a combination of CHX and 100 μM IU1 for 6 h. PrPC (SAF32; green) and nuclei (blue) were visualized. Bars: 20 μm.
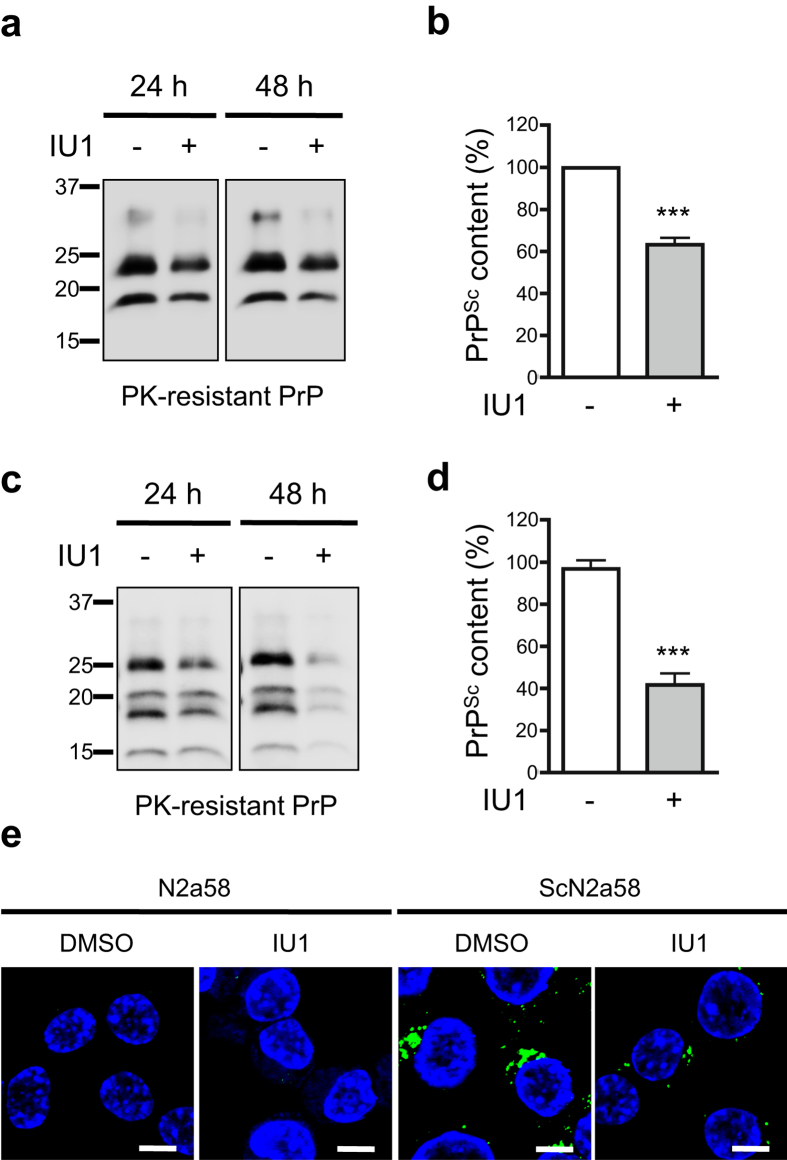
For further experiments, we utilized ScN2a58 cells or FK-N2a58 cells, and persistently scrapie-derived 22 L or Gerstmann-Sträussler-Scheinker (GSS) syndrome-derived Fukuoka-1 strain infected N2a58 cells, respectively. Immunoblotting revealed that IU1 treatment reduced PrPSc levels in ScN2a58 cells (Fig. 3a,b). IU1 treatment also reduced PrPSc levels in FK-N2a58 cells (Fig. 3c,d). To support this, we visualized PrPSc using mAb132, which is a specific monoclonal antibody that reacts with PrPSc in immunofluorescence analysis22. We confirmed reduced PrPSc levels in ScN2a58 cells after IU1 treatment (Fig. 3e). The mRNA levels of PrPC in ScN2a58 cells were not altered after IU1 treatment for 48 h (Supplementary Fig. S1b). Taken together, these results demonstrate that IU1 treatment reduced PrPSc levels in prion-infected cells.
Figure 3. IU1 treatment reduces PrPSc levels in prion-infected cells.
(a) ScN2a58 cells were treated with DMSO or 100 μM IU1 for 24 or 48 h. Lysates were PK-digested followed by immunoblotting with anti-PrP (M20) antibody. (b) Quantification of PrPSc after IU1 treatment for 48 h from at least three independent experiments performed as described in (a). Asterisk indicates significant difference (***P < 0.0001). Mean ± SD. (c) N2a58-FK cells were treated with DMSO or 100 μM IU1 for 24 or 48 h. Lysates were PK-digested followed by immunoblotting with anti-PrP (M20) antibody. (d) Quantification of PrPSc after IU1 treatment for 48 h from at least three independent experiments performed as described in (c). Asterisk indicates significant difference (***P < 0.0001). Mean ± SD. (e) N2a58 or ScN2a58 cells were treated with DMSO or 100 μM IU1 for 48 h. PrPSc (mAb132; green) and nuclei (blue) were visualized. Bars: 10 μm.
USP14 negatively regulates prion protein degradation
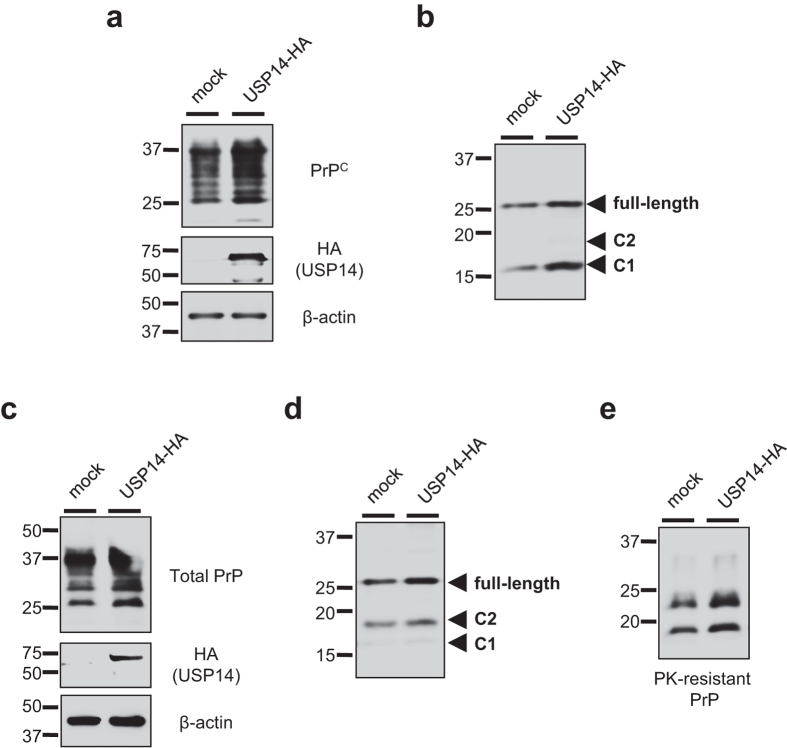
To investigate whether USP14 overexpression could influence prion protein degradation, N2a58 and ScN2a58 cells were transiently transfected with a HA-tagged USP14 expression plasmid for 48 h and subjected to immunoblotting. As expected, we confirmed that total PrPC levels (Fig. 4a), deglycosylated full-length PrP (Fig. 4b), and C1 fragment (Fig. 4b) levels increased in USP14-overexpressed cells compared to mock transfected N2a58 cells. The mRNA levels of PrPC were not altered in USP14-overexpressed cells compared to mock transfected N2a58 cells (Supplementary Fig. S1c). Of note, we observed that total PrP (Fig. 4c), deglycosylated full-length PrP, and C2 fragment (Fig. 4d), as well as PrPSc (Fig. 4e), levels increased in USP14-transfected ScN2a58 cells.
Figure 4. Overexpression of USP14 increases PrPSc levels in prion-infected cells.
(a) N2a58 cells were transfected with an empty plasmid (mock) or a HA-tagged USP14 expression plasmid for 48 h. Lysates were analyzed by immunoblotting with anti-PrP (SAF32), anti-HA, and anti-β-actin antibodies. (b) Mock or USP14-overexpressed N2a58 cell lysates were deglycosylated with PNGase F followed by immunoblotting with anti-PrP (SAF83) antibody. (c) Mock or USP14-overexpressed ScN2a58 cells were analyzed by immunoblotting with anti-PrP (SAF32), anti-HA, and anti-β-actin antibodies. (d) Mock or USP14-overexpressed ScN2a58 cell lysates were deglycosylated with PNGase F followed by immunoblotting with anti-PrP (SAF83) antibody. (e) Mock or USP14-overexpressed ScN2a58 cell lysates were PK-digested followed by immunoblotting with anti-PrP (M20) antibody.
Conversely, to examine whether dominant negative USP14 could counteract PrPSc accumulation in prion-infected cells, a HA-tagged dominant negative USP14 (USP14DN), which was mutated by the substitution of cysteine with alanine at position 114 of USP region in mouse USP14 (Fig. 5a), was prepared. As a control study, we verified an effect of USP14DN against expression of p53 protein, a well-characterized proteasomal substrate, in cells. As expected, p53 proteins were also reduced in N2a58 and ScN2a58 cells transducing the USP14DN compared to mock groups (Fig. 5b). The mRNA levels of PrPC were not altered in USP14DN-overexpressed cells compared to mock transfected N2a58 cells (Supplementary Fig. S1c). Of note, we detected significantly reduced PrPSc in the dominant negative-transfected cells compared to the mock-transfected control cells (Fig. 5c,d). We also confirmed that deglycosylated full-length PrP and C2 fragment levels were reduced in the dominant negative-transfected cells (Fig. 5e).
Figure 5. Overexpression of dominant negative USP14 reduces PrPSc levels in prion-infected cells.
(a) Schematic representation of the dominant negative USP14 (USP14DN). USP14 contains the ubiquitin-like (UBL) domain, which is important for localization at the proteasome, as well as the ubiquitin-specific protease (USP) domain. Asterisk indicates substitution of mouse USP14 cysteine 114 for alanine in the USP domain. (b) N2a58 or ScN2a58 cells were transfected with an empty plasmid (mock) or a HA-tagged dominant negative USP14 (USP14DN) expression plasmid for 48 h. Lysates were analyzed by immunoblotting with anti-p53 and anti-β-actin antibodies. Numbers below the gel indicate relative expression of p53 normalized to β-actin. (c) Mock or USP14DN-overexpressed ScN2a58 cell lysates were analyzed by immunoblotting with anti-HA and anti-β-actin antibodies. Alternatively, lysates were PK-digested followed by immunoblotting with anti-PrP (M20) antibody. (d) Quantification of PrPSc from at least three independent experiments performed as in (c). Asterisk indicates significant difference (**P < 0.01). Mean ± SD. (e) Mock or USP14DN-overexpressed ScN2a58 cell lysates were deglycosylated with PNGase F followed by immunoblotting with anti-PrP (SAF83) antibody.
Finally, we analyzed endogenous USP14 protein expression by immunoblotting. No differences in USP14 expression were detectable between N2a58 and ScN2a58 cells (Supplementary Fig. S4). It is plausible that prion infection did not influence USP14 expression, because no variations in proteasome content or in its subunit composition were detected between scrapie-positive and control samples23, although prion infection inhibits the function24,25.
Discussion
Previous studies have revealed that a series of prion proteins is degraded by ERAD (Step 1 in Supplementary Fig. S5). Wildtype PrPC undergoes ERAD and is degraded by the proteasome6,7. Additionally, GSS-associated PrP mutant (Y145stop, Q217R and Q212P) and Creutzfeldt-Jakob disease-associated PrP mutant (V203, E211Q) are also subject to ERAD and degraded by the proteasome8,9,10. Overexpression of EDEM-3, a protein that recognizes N-linked glycans on aberrantly folded proteins and sorts them for ERAD, resulted in reduced PrPSc accumulation26. Alternatively, the proteasome also cotranslationally degraded nascent peptide chains. Other studies, however, have indicated that PrP is not subject to retrotranslocation from the ER into the cytoplasm prior to degradation by the proteasome27,28. During PrP biosynthesis, some PrP molecules fail to translocate into the ER lumen and are degraded by the proteasome due to the intrinsic inefficiency of signal peptides27,28.
Based on our results, we propose a model that shows that USP14 rescues prion protein from proteasomal degradation. When it comes to degradation, a certain population of prion proteins might escape from proteasomal degradation due to the presence of USP14 (Step 2). Our results demonstrated that USP14 overexpression increased total PrP levels (Fig. 4). In addition, we provide data that IU1 treatment accelerated prion protein degradation (Fig. 2), implicating the existence of the surplus prion protein. PrP cleavage is thought to occur either in endocytic vesicles29 or in the late secretory pathway30,31. In the current study, IU1 treatment reduced full-length PrP levels, but not C1 fragment levels (Fig. 1c,d), indicating that the affected PrP molecules appear to be within an earlier compartment of the secretory pathway. Although it is not abundant, the C2 fragment is apparently present in normal human brains32,33. It has been reported that C2 cleavage can be stimulated by reactive oxygen species34. We also barely detected C2 fragment, and it seems to be affected by IU1 treatment through unknown mechanisms (Fig. 1c).
PrPSc has been shown to be difficult to unfold and inhibits the proteasome24,25. Under circumstances in which the proteasome is inhibited, it facilitates conversion of cytosolic PrPC to an abnormal, PrPSc-like form, with partial protease resistance and detergent insolubility35. Hence, it is likely that IU1 treatment reduced PrPC levels (Fig. 1), and this subsequently reduced further conversion of PrPC to PrPSc in prion-infected cells (Fig. 3) (Step 3).
In summary, our findings demonstrated a novel mechanism by which USP14 negatively regulated prion protein degradation via the proteasome, although the detailed mechanism remains unclear. It has a possiblility that USP14 may regulate PrP degradation via an indirect mechanism because it remains controversial whether PrP is ubiquitinated and USP14 can affect its ubiquitination level. At the current moment, we speculate that it might be important for prion protein degradation that some ubiquitinated cargo proteins bind to PrP, thereby transporting PrP to the proteasome. However, further investigations are required for the elucidation of its opinion.
IU1 has been shown to accelerate proteasomal degradation of aggregation-prone proteins, including proteins associated with neurodegenerative diseases, such as tau, TDP-43, polyglutamine-expanded ataxin-321, and abnormal prion protein in this study. These results suggest USP14 inhibition as a new therapeutic approach for the treatment of protein aggregation diseases through proteasomal degradation of disease-associated proteins.
Materials and methods
Antibodies
Anti-USP14 (Cell Signaling Technology, #11931), anti-β-actin (MBL, 177-3), anti-HA-tag (MBL, 561), anti-LC3B (Cell Signaling Technology, #2775), anti-p53 (Cell Signaling Technology, #2524), and anti-PrP (Santa Cruz Biotechnology, M20; SPI-Bio, SAF32 and SAF83) antibodies were purchased from the indicated vendors. The anti-PrP mAb132 was a kind gift from Prof. Motohiro Horiuchi (Hokkaido University). Horseradish peroxidase-conjugated anti-goat (Jackson ImmunoResearch), anti-mouse, and anti-rabbit (GE Healthcare Life Sciences) IgG antibodies were used for immunoblotting.
Cell cultures
The mouse neuroblastoma Neuro 2a cells were obtained from the American Type Culture Collection (CCL 131). N2a58 cells are mouse PrPC-overexpressing Neuro 2a cells. ScN2a58 cells originated from N2a58 cells infected with a mouse-adapted scrapie strain, 22 L, as previously described36,37,38. FK-N2a58 cells originated from N2a58 cells infected with a mouse-adapted Gerstmann-Sträussler-Scheinker (GSS) strain, Fukuoka-1, as previously described39. HpL2-3 cells are immortalized hippocampal cells derived from PrP-deficient mice, a kind gift from Prof. Takashi Onodera (The University of Tokyo, Japan). The above cells were grown at 37 °C in 5% CO2 in Dulbecco’s-modified Eagle’s medium (DMEM; Sigma-Aldrich) containing 10% heat-inactivated fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Immortalized mouse hypothalamic GT1-7 cells were maintained in DMEM containing 5% heat-inactivated FBS, 5% heat-inactivated horse serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, as previously described36.
Plasmids
The mouse PrP expression plasmid has been previously described40. The mouse USP14 open reading frame was amplified from N2a58 cDNA with primers: 5′-ATT GGG CCC CTC GCC ATG CCA CTC TAC TCT GTT ACA-3′ and 5′-CCG CTC GAG TCA TTA AGC GTA ATC TGG AAC ATC GTA TGG GTA CTG TTC ACT TTC TTC TTC-3′. To generate a dominant-negative mutant of USP14, the substitution of mouse USP14 cysteine 114 for alanine in the UBL domain was produced by PCR with primers: 5′-CTT GGT AAC ACT GCT TAC ATG AAT-3′ and 5′-ATT CAT GTA AGC AGT GTT ACC AAG-3′. Amplified PCR fragments were inserted into the ApaI and XhoI sites of expression plasmid pcDNA3.1 (Invitrogen) and confirmed by sequential analysis.
Immunoblotting
Immunoblotting was performed as previously described41. For PrPSc detection, lysates were digested with 40 μg/ml of proteinase K (PK; Sigma-Aldrich) at 37 °C for 30 min. Bands were visualized using the ECL Western Blotting Detection Kit (GE Healthcare Life Sciences). Band intensities were quantified using ImageJ software (National Institutes of Health).
Plasmid transfection
Transfection with plasmids was performed with PEI “Max” (Polysciences, Warrington). Four micrograms of PEI was used per microgram of transfected DNA. PEI and DNA were each added to 250 μl serum-free Opti-MEM, and the two solutions were combined and mixed by vortex. After a 15-min incubation at room temperature, the mixture was added to 2 ml DMEM in the well. After 48 h, lysates were prepared and subjected to immunoblotting.
IU1 treatment
IU1, a selective inhibitor of USP14, was purchased from Focus Biomolecules. Cells were treated with dimethyl sulfoxide (DMSO) or 100 μM IU1 for 48 h and cell lysates were prepared, unless stated otherwise. Alternatively, cells were treated with 50 μg/ml cycloheximide (CHX; Sigma-Aldrich), combined with 100 μM IU1 for 6 h, and subjected to immunoblotting.
Deglycosylation of proteins
Deglycosylation of proteins was performed according to manufacturer’s protocol (New England Biolabs). Briefly, proteins were denatured by boiling for 10 min in 0.5% SDS and 1% 2-mercaptoethanol. A 1/10-volume of 10% Nonidet P-40, 1/10-volume of 0.5 M sodium phosphate (pH 7.5), and 2 μl of PNGase F (500 units/μl) were added to each sample. After incubation for 2 h at 37 °C, the samples were subjected to immunoblotting.
Real-time PCR
Total RNA was isolated from cells using a TRIZOL® Reagent (Invitrogen) or GenElute™ Mammalian Total RNA Kit® (Sigma). After the extracted total RNA was purified by RNeasy MinElute Cleanup Kit (QIAGEN) with RNase-free DNase set (QIAGEN), the first-strand cDNA was synthesized from 5 μg of total RNA with ThermoScript Reverse Transcriptase® (Invitrogen). The primers were: 5′-GAT CCA TTT TGG CAA CGA CT-3′ and 5′-GTG TGC TGC TTG ATG GTG AT-3′ for mouse prnp gene and 5′-AAA TCG TGC GTG ACA TCA AA-3′ and 5′-AAG GAA GGC TGG AAA AGA GC-3′ for mouse β-actin gene. The level of β-actin mRNA was determined as a positive control and used to normalize as the mRNA expression level as an internal control. For real-time PCR, the synthesized cDNA was reacted with LightCycler® 480 SYBR Green I Master (Roche Applied Science) and measured by Light Cycler 480 instrument (Roche Applied Science).
Immunofluorescence analysis
Immunofluorescence staining was performed as previously described41. Cells were treated with DMSO or 100 μM IU1 for 48 h and washed twice in PBS followed by fixation using 4% formaldehyde for 30 min at room temperature. The cells were permeabilized in 0.5% Triton X-100 for 5 min. The slides were then incubated in blocking solution (5% nonfat dry milk) for 30 min at 37 °C. Incubation with primary antibodies (SAF32 for PrPC; mAb132 for PrPSc) was performed overnight at 4 °C22. For PrPSc detection, the slides were treated with 3 M guanidine thiocyanate for 5 min at room temperature before staining. Alexa Fluor® 488-conjugated anti–mouse IgG antibody (Invitrogen) served as the secondary antibody. Nuclei were stained with DAPI. All images were obtained using a confocal laser-scanning microscope LSM 700 (Carl Zeiss).
Statistical analysis
Results in the graph represent mean ± standard deviation (SD) of at least three independent experiments. Statistical analysis was performed using GraphPad Prism 4 software. P-values were calculated using the Student’s t-test.
Additional Information
How to cite this article: Homma, T. et al. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Sci. Rep. 5, 11028; doi: 10.1038/srep11028 (2015).
Supplementary Material
Acknowledgments
We are most grateful to Prof. Takashi Onodera (The University of Tokyo, Japan) for providing the HpL2-3 cell line and to Prof. Motohiro Horiuchi and Dr. Takeshi Yamasaki (Hokkaido University, Japan) for providing mAb132. This work was supported by JSPS KAKENHI Grant Nos. 23300127 and 24591482 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; a grant-in-aid of the Research Committee of Prion disease and Slow Virus Infection, from the Ministry of Health, Labour and Welfare of Japan; a grant from Takeda Science Foundation; a grant from the Japan Intractable Disease Research Foundation; a grant-in-aid from the Tokyo Biochemical Research Foundation; and a grant provided by The Ichiro Kanehara Fundation.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.H. and D.I. coordinated the entire project. T.H. performed all experiments except Supplementary Fig. 1 (D.I.). T.F., T.N., T.M., K.S., R.A. and N.N. supervised and discussed the data. T.H. wrote the manuscript. D.I. and N.N. revised the manuscript.
References
- Prusiner S. B. Prions. Proc Natl Acad Sci USA 95, 13363–13383 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N., Borchelt D. R., Hsiao K. & Prusiner S. B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240 (1987). [DOI] [PubMed] [Google Scholar]
- Turk E., Teplow D. B., Hood L. E. & Prusiner S. B. Purification and properties of the cellular and scrapie hamster prion proteins. European journal of biochemistry/FEBS 176, 21–30 (1988). [DOI] [PubMed] [Google Scholar]
- Haraguchi T. et al. Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch Biochem Biophys 274, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- Shao J. et al. Ubiquitin ligase gp78 targets unglycosylated prion protein PrP for ubiquitylation and degradation. PLoS One 9, e92290, 10.1371/journal.pone.0092290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. & Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci USA 98, 14955–14960, 10.1073/pnas.011578098 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia Y., Horonchik L., Tzaban S., Yanai A. & Taraboulos A. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. Embo J 20, 5383–5391, 10.1093/emboj/20.19.5383 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanusso G. et al. Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J Biol Chem 274, 23396–23404 (1999). [DOI] [PubMed] [Google Scholar]
- Jin T. et al. The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J Biol Chem 275, 38699–38704, 10.1074/jbc.M005543200 (2000). [DOI] [PubMed] [Google Scholar]
- Mishra R. S., Bose S., Gu Y., Li R. & Singh N. Aggresome formation by mutant prion proteins: the unfolding role of proteasomes in familial prion disorders. J Alzheimers Dis 5, 15–23 (2003). [DOI] [PubMed] [Google Scholar]
- Borodovsky A. et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J 20, 5187–5196, 10.1093/emboj/20.18.5187 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M. et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J 24, 3747–3756, 10.1038/sj.emboj.7600832 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A., Besche H. C. & Goldberg A. L. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36, 794–804, 10.1016/j.molcel.2009.11.015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. J., Lee B. H., Hanna J., King R. W. & Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics 10, R110 003871, 10.1074/mcp.R110.003871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. et al. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem 95, 724–731, 10.1111/j.1471-4159.2005.03409.x (2005). [DOI] [PubMed] [Google Scholar]
- Leggett D. S. et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell 10, 495–507 (2002). [DOI] [PubMed] [Google Scholar]
- Chernova T. A. et al. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J Biol Chem 278, 52102–52115, 10.1074/jbc.M310283200 (2003). [DOI] [PubMed] [Google Scholar]
- Chen P. C. et al. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci 29, 10909–10919, 10.1523/JNEUROSCI.2635-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai A. et al. USP14 inhibits ER-associated degradation via interaction with IRE1alpha. Biochem Biophys Res Commun 379, 995–1000, 10.1016/j.bbrc.2008.12.182 (2009). [DOI] [PubMed] [Google Scholar]
- Hosokawa N. et al. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep 2, 415–422, 10.1093/embo-reports/kve084 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184, 10.1038/nature09299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T. et al. Characterization of intracellular localization of PrP(Sc) in prion-infected cells using a mAb that recognizes the region consisting of aa 119–127 of mouse PrP. J Gen Virol 93, 668–680, 10.1099/vir.0.037101-0 (2012). [DOI] [PubMed] [Google Scholar]
- Amici M. et al. Interplay between 20S proteasomes and prion proteins in scrapie disease. J Neurosci Res 88, 191–201, 10.1002/jnr.22186 (2010). [DOI] [PubMed] [Google Scholar]
- Kristiansen M. et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell 26, 175–188, 10.1016/j.molcel.2007.04.001 (2007). [DOI] [PubMed] [Google Scholar]
- Deriziotis P. et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. Embo J 30, 3065–3077, 10.1038/emboj.2011.224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunziante M. et al. Proteasomal dysfunction and endoplasmic reticulum stress enhance trafficking of prion protein aggregates through the secretory pathway and increase accumulation of pathologic prion protein. J Biol Chem 286, 33942–33953, 10.1074/jbc.M111.272617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisaldi B. et al. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J Biol Chem 278, 21732–21743, 10.1074/jbc.M213247200 (2003). [DOI] [PubMed] [Google Scholar]
- Fioriti L. et al. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J Biol Chem 280, 11320–11328, 10.1074/jbc.M412441200 (2005). [DOI] [PubMed] [Google Scholar]
- Shyng S. L., Huber M. T. & Harris D. A. A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J Biol Chem 268, 15922–15928 (1993). [PubMed] [Google Scholar]
- Walmsley A. R., Watt N. T., Taylor D. R., Perera W. S. & Hooper N. M. alpha-cleavage of the prion protein occurs in a late compartment of the secretory pathway and is independent of lipid rafts. Mol Cell Neurosci 40, 242–248, 10.1016/j.mcn.2008.10.012 (2009). [DOI] [PubMed] [Google Scholar]
- Hachiya N., Komata Y., Harguem S., Nishijima K. & Kaneko K. Possible involvement of calpain-like activity in normal processing of cellular prion protein. Neurosci Lett 490, 150–155, 10.1016/j.neulet.2010.12.046 (2011). [DOI] [PubMed] [Google Scholar]
- Chen S. G. et al. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem 270, 19173–19180 (1995). [DOI] [PubMed] [Google Scholar]
- Jimenez-Huete A. et al. Endogenous proteolytic cleavage of normal and disease-associated isoforms of the human prion protein in neural and non-neural tissues. Am J Pathol 153, 1561–1572, 10.1016/S0002-9440(10)65744-6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. E. et al. Cleavage of the amino terminus of the prion protein by reactive oxygen species. J Biol Chem 276, 2286–2291, 10.1074/jbc.M007243200 (2001). [DOI] [PubMed] [Google Scholar]
- Ma J. & Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science 298, 1785–1788, 10.1126/science.1073619 (2002). [DOI] [PubMed] [Google Scholar]
- Nishida N. et al. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J Virol 74, 320–325 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi D. et al. Protective role of interferon regulatory factor 3-mediated signaling against prion infection. J Virol 86, 4947–4955, 10.1128/JVI.06326-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma T. et al. Persistent prion infection disturbs the function of Oct-1, resulting in the down-regulation of murine interferon regulatory factor-3. Sci Rep 4, 6006, 10.1038/srep06006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi D. et al. Antigenic mimicry-mediated anti-prion effects induced by bacterial enzyme succinylarginine dihydrolase in mice. Vaccine 29, 9321–9328, 10.1016/j.vaccine.2011.10.017 (2011). [DOI] [PubMed] [Google Scholar]
- Atarashi R., Sim V. L., Nishida N., Caughey B. & Katamine S. Prion strain-dependent differences in conversion of mutant prion proteins in cell culture. J Virol 80, 7854–7862, 10.1128/JVI.00424-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma T. et al. Increased expression of p62/SQSTM1 in prion diseases and its association with pathogenic prion protein. Sci Rep 4, 4504, 10.1038/srep04504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.