Abstract
Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum are the most frequently identified protozoan parasites causing waterborne disease outbreaks. The morbidity and mortality associated with these intestinal parasitic infections warrant the development of rapid and accurate detection and genotyping methods to aid public health efforts aimed at preventing and controlling outbreaks. In this study, we describe the development of an oligonucleotide microarray capable of detecting and discriminating between E. histolytica, Entamoeba dispar, G. lamblia assemblages A and B, and C. parvum types 1 and 2 in a single assay. Unique hybridization patterns for each selected protozoan were generated by amplifying six to eight diagnostic sequences/organism by multiplex PCR; fluorescent labeling of the amplicons via primer extension; and subsequent hybridization to a set of genus-, species-, and subtype-specific covalently immobilized oligonucleotide probes. The profile-based specificity of this methodology not only permitted for the unequivocal identification of the six targeted species and subtypes, but also demonstrated its potential in identifying related species such as Cryptosporidium meleagridis and Cryptosporidium muris. In addition, sensitivity assays demonstrated lower detection limits of five trophozoites of G. lamblia. Taken together, the specificity and sensitivity of the microarray-based approach suggest that this methodology may provide a promising tool to detect and genotype protozoa from clinical and environmental samples.
Entamoeba histolytica, Giardia lamblia (syn. intestinalis or duodenalis) and Cryptosporidium parvum are three of the most common intestinal protozoan parasites infecting humans worldwide (25). The disease manifestation of E. histolytica infections, invasive intestinal amoebiasis, causes up to 100,000 deaths per year globally (31). Even more staggering are the 600 million estimated intestinal infections caused by the diplomonad G. lamblia (giardiasis) and the apicomplexan C. parvum (cryptosporidiosis). Both are recognized as common causes of diarrheal disease worldwide, and although usually self-limiting in immunocompetent individuals, G. lamblia and C. parvum infections can become chronic and deadly in immunocompromised patients and malnourished children (3, 37). In addition to posing a continuing health threat to civilian populations, the disabling gastrointestinal disease caused by these organisms also presents a serious threat to the overall mission readiness of deployed military personnel as outbreaks of all three diseases usually occur through fecal-oral transmission and are most often associated with the consumption of untreated water or contaminated food (18).
The global distribution and increasingly frequent presence of E. histolytica, G. lamblia, and C. parvum in ambient and source waters suggest that rapid and accurate identification methods are important for public health efforts to prevent and control outbreaks. Traditionally, laboratory detection of these three parasites has mostly relied on the microscopic examination of stool samples and water concentrates (25, 41), which is laborious, insensitive, and requires professional training. The paramount limitation of this method is its inability to differentiate closely related species and heterogeneity within species, as it is often difficult to differentiate these pathogenic organisms from the cysts of nonpathogenic intestinal protozoa via microscopic examination. For example, the nonpathogenic protist Entamoeba dispar is morphologically indistinguishable from its sister species, pathogenic E. histolytica (11), but is responsible for approximately 10 times as many infections as E. histolytica that do not require treatment (15). In addition to genus-level identification, species differentiation and subtype differentiation also provide important epidemiological, surveillance, and host range information. G. lamblia isolates capable of causing infectious disease in humans are morphologically indistinguishable, yet can genetically be differentiated into two major groups, designated assemblages A and B (42). Similarly, morphologically indistinguishable C. parvum isolates can be differentiated into two distinct genotypes, 1 and 2: genotype 1 members exclusively infect humans and nonhuman primates, while genotype 2 members have a wider range of hosts, including humans and livestock (27). In this regard, antibody-based diagnostic methods are useful in that they can discriminate between different species or genotypes, but are far from optimal due to problems of nonspecific binding, variability among clinical isolates, and interference from sample debris (28).
Molecular methods, such as PCR, have aided in alleviating some of the sensitivity and specificity issues traditionally associated with the detection of protozoan pathogens. A number of PCR-based assays, including gene amplification with specific primers (17, 24, 33), multiplex PCR (12, 32), restriction fragment length polymorphism (3, 5, 10, 46), and real-time PCR (2, 4, 16, 23, 40), have been developed for the identification of protozoan infections. However, the shortcomings of PCR-based assays become apparent during practical applications. The generation of nonspecific DNA fragments from environmental and clinical samples poses a significant problem that often results in false-positive results. Conversely, the failure to amplify a single diagnostic sequence due to inhibitors in the sample or possible mutations in the primer binding region may result in false-negative results. Furthermore, although real-time PCR assays are sensitive enough to detect a single cell (4), the limited number of probes that can be applied in one reaction hinders its utility for confident multitarget detection and genotyping analyses.
Recently, oligonucleotide microarrays have been used successfully for the detection of bacterial and viral pathogens (7, 8, 13, 43, 44). The distinct advantage of this detection approach is that it combines powerful DNA amplification strategies with subsequent hybridization to oligonucleotide probes specific for multiple target sequences. This method allows for the simultaneous analysis of a larger number of genetic features in a single experiment (8). Thus, the amplification and hybridization approach produces a highly sensitive and specific platform with high-throughput capacity for pathogen detection and genotyping. Due to the increasing reliance upon genetic tests for identification and differentiation, the low concentration or number of organisms required to cause disease, and the often-found presence of multiple protozoan species in a single environmental or clinical sample, such methods would be ideally suited for the detection of waterborne protozoan parasites. In the present study, we demonstrate the first oligonucleotide microarray capable of simultaneously detecting and differentiating the primary waterborne protozoa pathogenic for humans, E. histolytica, G. lamblia, and C. parvum.
MATERIALS AND METHODS
Parasite isolates and DNA.
Purified genomic DNA extracted from cultures of E. histolytica strain HM-1:IMSS clone 9, E. dispar SAW760 and Entamoeba moshkovskii strain Laredo were kindly provided by C. Graham Clark (London School of Hygiene and Tropical Medicine). Trophozoites of G. lamblia WB, GS-H7, and CM strains were generous gifts from Theodore E. Nash (National Institutes of Health, Bethesda, Md.). Genomic DNA and oocysts of C. parvum TU502, UG502, GCH1, C. meleagridis, and C. muris were obtained from Donna Akiyoshi (Tufts University, Medford, Mass.). Formalin-fixed oocysts of C. parvum Iowa isolates were purchased from Waterborne, Inc. (New Orleans, La.). Genomic DNA from G. lamblia trophozoites was isolated using the DNA STAT-60 kit (Tel-Test, Inc., Friendswood, Tex.) or simply released by suspension in distilled water. C. parvum DNA from purified oocysts was extracted following three cycles of freezing and thawing (35).
PCR primers.
The primers used in the multiplex PCRs to amplify fragments from different target genes are listed in Table 1. These primers were either derived from publications or designed by PCR primer design software: Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) and Oligos (http://www.biocenter.helsinki.fi/bi/bare-1_html/oligos.htm).
TABLE 1.
Protozoan gene-targeted primers used in this study
| Targeta | Name | Sequence (5′→3′) | Length (nt) | Tm (°C)b | GC (%) | PCR product size (nt) | Source or reference |
|---|---|---|---|---|---|---|---|
| Entamoeba | |||||||
| Locus1-2 | R1 | CTGGTTAGTATCTTCGCCTGT | 21 | 50.6 | 48 | ||
| R2 | CTTACACCCCCATTAACAAT | 20 | 49.6 | 40 | 402/495 | 48 | |
| Locus5-6 | R5A | CTAAAGCCCCCTTCTTCTATAATT | 24 | 57.5 | 38 | ||
| R6A | CTCAGTCGGTAGAGCATGGT | 20 | 49.6 | 55 | 485/510 | 48 | |
| ITS | 18SP1 | AGGTGAACCTGCGGAAGGATCATTA | 25 | 62.6 | 48 | ||
| 28SP2 | TCATTCGCCATTACTTAAGAAATCATTGTT | 30 | 65.2 | 30 | 433/423 | 34 | |
| sod-act | SAF2 | GAGCTGCTTACTTAGAACATTGGTGG | 26 | 59.1 | 46 | 14 | |
| SAR2 | CCAGATCCATTATCTACAACAAGTGC | 26 | 57.0 | 42 | 497 | This study | |
| prrdx | prrdxF | GTCAAGAGAAAGAATGTTGTAAAGA | 25 | 51.6 | 32 | 39 | |
| prrdxR1 | TTGATTTCTTTCAATTGTCCTGCA | 24 | 56.3 | 33 | 274/304 | This study | |
| cpl | EntaCP1-F | GCAGCACTTGAAGGAAGATTATT | 23 | 54.3 | 39 | ||
| EntaCP1-R | CCATCAACAACACCATATCCAA | 22 | 52.2 | 41 | 441 | This study | |
| G. lamblia | |||||||
| hsp | ABB97F | AGGGCTCCGGCATAACTTTCC | 21 | 60.5 | 57 | ||
| ABB220R | GTATCTGTGACCCGTCCGAG | 20 | 51.5 | 60 | 163 | 32 | |
| tpiA | TPIAF | CGAGACAAGTGTTGAGATGC | 20 | 47.7 | 50 | ||
| TPIAR | GGTCAAGAGCTTACAACACG | 20 | 48.5 | 50 | 476 | 3 | |
| tpiB | TPIBF | GTTGCTCCCTCCTTTGTGCA | 20 | 54.1 | 55 | ||
| TPIBR2 | AGGCAATTACAACGTTCTCCCA | 22 | 56.3 | 46 | 384 | This study | |
| giardin | GIA40F | CCGACGACCTCACCCGCAG | 19 | 58.5 | 74 | 5 | |
| GIA773R | GAGAGGCCGCCCTGGATC | 18 | 56.3 | 72 | 749 | This study | |
| gdh | GDHF | CCGCTTCCACCCCTCTGTCAA | 21 | 59.6 | 62 | 26 | |
| GDHR | CCTTGCACATCTCCTCCAGGAA | 21 | 57.0 | 55 | 389 | This study | |
| c4-orf | C4-F | AGCTCATCTTCGTCCTCTA | 19 | 46.4 | 48 | 47 | |
| C4-R | CAATCTTGTTTGCATACGA | 19 | 46.2 | 37 | 445 | ||
| C. parvum | |||||||
| cowp1 | CpR1-F2 | AAAGAAGCACCTCCTGTTTCAG | 22 | 53.5 | 46 | 21 | |
| CpR1-R2 | GCAGCTGCTAATCTTCTTAGTGC | 23 | 54.6 | 48 | 485 | ||
| SSUrRNA | CpB-DIAGF | AAGCTCGTAGTTGGATTTCTG | 21 | 50.8 | 43 | 30 | |
| CpB-DIAGR | TAAGGTGCTGAAGGAGTAAGG | 21 | 50.9 | 48 | 435 | ||
| dhf | DHFR1 | TTGTTGTGGCAGCTTCTGTTTTGA | 24 | 57.7 | 42 | 30 | |
| DHFR4 | AAAGTTATCCTTTAAAGCATCCCT | 24 | 56.8 | 33 | 359 | This study | |
| ptg | PolyTF1 | TCCCAGTTCAAACTCACAAGAGTA | 24 | 52.9 | 42 | 30 | |
| PolyTR1 | GGAGGAATAATACCACCATCTTCA | 24 | 55.2 | 42 | 496 | This study | |
| RAPD | SB012F | CTCCGTTCGATGATGCAGATG | 21 | 51.2 | 52 | 45 | |
| SB012R | CGGCCCCTGTAGAAATAAGTCA | 22 | 57.0 | 50 | 433 | ||
| TRAP-C2 | TRAP-C2F | CATATTCCCTGTCCCTTGAG | 20 | 50.4 | 50 | 38 | |
| TRAP-C2R | TGGACAACCCAAATGCAGAC | 20 | 51.9 | 50 | 369 | ||
| hsp | CPHSPT2F | TCCTCTGCCGTACAGGATCTCTTA | 24 | 57.2 | 50 | ||
| CPHSPT2R | TGCTGCTCTTACCAGTACTCTTATCA | 26 | 55.5 | 42 | 346 | 22 | |
| p23 | P23-45 | ATTATTTTTACGTTCCTTCCACTTG | 25 | 57.0 | 32 | ||
| P23-569 | AACCTTAATAAAAAACACTCTATTG | 25 | 51.9 | 24 | 537 | 37 |
Abbreviations of target genes or gene products: ITS, intergenic sequence between rDNA; sod-act, intergenic region between superoxide dismutase and actin 3 genes; prrdx, perosiredoxin; cp1, cysteine protease 1; hsp, heat shock protein; gdh, glutamate dehydrogenase; tpi, triose phosphate isomerase; cowp, Cryptosporidium oocyst wall protein; ssu, small subunit; dhf, dihydrofolate reductase-thymidylate synthase; ptg, polythreonine-rich glycoprotein; TRAP-C2, thrombospondin-related anonymous protein 2.
Basic melting temperature (Tm) was calculated with Oligos software downloaded from (http://www.biocenter.helsinki.fi/bi/bare-1_html/oligos.htm).
Microarray design and fabrication.
Target sequences were downloaded from either GenBank, the TIGR parasite database (The Institute for Genomic Research, Rockville, Md.), or the Giardia lamblia Genome Project (Marine Biology Laboratory, Woods Hole, Mass.). Orthologous sequences between E. histolytica and E. dispar and polymorphic sequences within G. lamblia and C. parvum were aligned by using ClustalW (http://www.ebi.ac.uk/clustalw/). Oligonucleotide probes were designed with Array Designer 2.02 (Premier Biosoft, Palo Alto, Calif.) and the Oligonucleotide Properties Calculator (http://www.basic.nwu.edu/biotools/OligoCalc.html) by using the following criteria: (i) probe length range between 23 and 30 nucleotides (nt) and (ii) melting temperature range of 55 to 65°C. At least two oligonucleotide probes were designed for each target sequence to identify and differentiate species of Entamoeba, G. lamblia assemblages, and C. parvum genotypes. For example, probes ssrDNA and locus1-2.3 are common to both E. histolytica and E. dispar (genus-specific probes), whereas the remainder of the Entamoeba probes were species specific. For G. lamblia, both species-specific and assemblage-specific probes were selected from the gene coding for giardin and gdh, whereas the two hsp70 probes were only species specific and the tpi and c4 probes were assemblage specific. As the available C. parvum sequences with polymorphic characteristics were limited, probes from only three genes (dhf, ptg, and p23) were genotype specific, while the remaining probes were species specific. The sequences of all 92 probes and one additional internal positive control probe used in this study are presented in Table 2. Based on the design specificities, the expected hybridization patterns are shown in Fig. 2B (internal positive hybridization spots at left and right ends of each array not included). The probes were synthesized with a 5′ amino modifier and 12-carbon spacer (QIAGEN Operon, Alameda, Calif.) and were resuspended in a carbonate-bicarbonate buffer (100 mM, pH 9.0) at a final concentration of 50 μM. The probes were printed onto 3-aminopropyltriethoxysilane (silanization)-plus-1,4-phenylene diisothiocyanate (cross-linker)-modified glass slides for covalent probe immobilization (6), using a Virtek ChipWriter Pro contact printer at KamTek, Inc. (Gaithersburg, Md.). The printed slides were stored desiccated at room temperature.
TABLE 2.
Oligonucleotide probe set used in this study
| Name | Sequencea | Positionb | Length (nt) | Tm (°C) | GC (%) | Coordinate(s)c |
|---|---|---|---|---|---|---|
| Entamoeba | ||||||
| Ehlocus1-2.3 | CTGGTTAGTATCTTCGCCTGTCACG | 1 | 25 | 59.3 | 52 | A3, C3 |
| EssrDNA.1 | AATGAATTGCGATAAGTGATAGGAAC | 171 | 26 | 55.0 | 35 | B3, D3 |
| E. histolytica | ||||||
| Ehlocus5-6.1 | TCTTTGAGACTTATTTCTACTTTATTTCTT | 37 | 30 | 53.7 | 23 | A1 |
| Ehlocus5-6.2 | TGTACGTCTTTAACTTTAAAAACAA | 415 | 25 | 54.7 | 24 | B1 |
| Ehlocus1-2.1 | ATATTCTTATCACTTCCTACTACTCTTATT | 116 | 30 | 53.6 | 27 | A2 |
| Ehlocus1-2.2 | TACTACTCTT*(6)CTTACTATACCT*(6)CTTACTAC | 331 | 30 | 54.5 | 33 | B2 |
| EhlTS.1 | AATCTACAAAGAAAATAATAATAAGTAAGA | 54 | 30 | 50.4 | 17 | A4 |
| EhlTS.2 | AATATCAATAGACAGACCAGACCAATA | 368 | 27 | 54.8 | 33 | B4 |
| EhlTS.3 | GCAAGTACAACAGAGAAGAAGTAGC | 304 | 25 | 53.1 | 44 | A5 |
| EhSA.1 | ACTTTTCCCCTCAATTATTTCGTTTT | 76 | 26 | 55.1 | 31 | A6 |
| EhSA.2 | AAGACAGGTATTTAAAGATCATAATAAACT | 401 | 30 | 53.5 | 23 | B6 |
| Ehprrdx.1 | GAAAGAATGTTGTAAAGAATGTTGTTGTCC | 9 | 30 | 61.6 | 33 | A7 |
| Ehprrdx.2 | AACAGAAATGATTGGATATAGTGAACTT | 224 | 28 | 54.8 | 29 | B7 |
| Ehcp1.1 | TATGGTTCAATGTACTAGGGAAGATG | 73 | 26 | 55.1 | 39 | A8 |
| Ehcp1.2 | AGTTATACAAGAGTGGAGCATATACAG | 342 | 27 | 55.4 | 37 | B8 |
| E. dispar | ||||||
| Edlocus5-6.1 | CTATATTCTTTTTATGTACTTCCCTTA | 33 | 27 | 54.3 | 26 | C1 |
| Edlocus5-6.2 | TTCCTTTTATACAAATACTCTCATG | 356 | 25 | 51.4 | 28 | D1 |
| Edlocus1-2.1 | GTCACTTATACTATTAACTTTATCTATTCC | 102 | 30 | 52.2 | 27 | C2 |
| Edlocus1-2.2 | CCTACTATACCTACTACTCTTACTACTCCT | 428 | 30 | 54.1 | 40 | D2 |
| EdITS.1 | AATCTACAAAGAAAATAATAA*AAGTAAGA | 54 | 29 | 55.9 | 17 | C4 |
| EdITS.2 | TTAACCAGATATCTATAAGTGAGTTAATA | 356 | 29 | 53.7 | 24 | D4 |
| EdITS.3 | TTAGTAGAAGTGAGAAGTAGCTAGTG | 303 | 26 | 50.1 | 39 | C5 |
| EdSA.1 | GTTTTTGTTAAGTTTTTGGTATTACTG | 110 | 27 | 55.5 | 26 | C6 |
| EdSA.2 | GAGCCAGGTATTTAAAGATCATATCAA | 401 | 27 | 58.2 | 33 | D6 |
| Edprrdx.1 | GAGAAAGAATGTTGTAAAGAGTATTGTTGT | 6 | 30 | 58.2 | 30 | C7 |
| Edprrdx.2 | AACAGAAATGATTGGATATAGTGAAGTT | 194 | 28 | 56.8 | 29 | D7 |
| Edcp1.1 | GTACTAGAGATAATGGAAACAATGG | 84 | 25 | 52.2 | 36 | C8 |
| Edcp1.2 | GTGGTGCATACAGTGATACTAAATG | 353 | 25 | 51.8 | 40 | D8 |
| G. lamblia | ||||||
| Glhsp.1 | AATTGATTAGTATTAGTAGGATGCCT | 46 | 26 | 52.6 | 31 | E1, G1 |
| Glhsp.2 | CTTGGCGTTCCCGAAGTCTGTC | 113 | 22 | 59.8 | 59 | F1, H1 |
| girAB.1 | TCAAGCTCAGCAACATGAACCAGC | 36 | 24 | 58.8 | 50 | E5, G5 |
| girAB.2 | ATGGAGAACGAGATCGAGGTCCG | 84 | 23 | 60.4 | 57 | F5, H5 |
| gdhAB.1 | GAGCAGATCCTGAAGAACTCCCT | 49 | 23 | 57.0 | 52 | E8, G8 |
| gdhAB.2 | GACAACGAGGTCATGCGCTT | 134 | 20 | 54.1 | 55 | F8, H8 |
| Assemblage A | ||||||
| tpiA.1 | ATGGGTTTGAAGCATGTGATAGTAG | 29 | 25 | 55.8 | 40 | E2 |
| tpiA.2 | TATGACGATGATCGACATTCTTACG | 505 | 25 | 55.7 | 40 | F2 |
| girA.1 | ATGTACCTAACGATCAAGGAGGAGA | 246 | 25 | 57.3 | 44 | E3 |
| girA.2 | AGAACGCAGAAAGGAAGAAGATGTA | 442 | 25 | 56.9 | 40 | F3 |
| girA.3 | ATCGCACACCTCGACAGGCTCAT | 150 | 23 | 60.1 | 57 | E4 |
| girA.4 | CCACGACAGAAGCGCTCACAAACA | 561 | 24 | 60.0 | 54 | F4 |
| gdhA.1 | AATCTTTCGATTCTCAAGTTCCT | 20 | 23 | 57.6 | 35 | E6 |
| gdhA.2 | GGTACCTGTACGGACAGTACAAG | 246 | 23 | 57.8 | 52 | F6 |
| gdhA.3 | TTGACCCAAAGGGCAAGTCCGA | 113 | 22 | 61.8 | 55 | E7 |
| c4A.1 | GCCATGATGAAAGGGAGTGCTGTA | 37 | 24 | 58.9 | 50 | E9 |
| c4A.2 | TGGACCCCACTGTTCAAAAGAAGA | 158 | 24 | 58.5 | 46 | F9 |
| c4A.3 | CATCCAAAGATATGAATGAGCTTCC | 218 | 25 | 57.4 | 40 | E10 |
| c4A.4 | CAGATGGAATCAAGCTTCTTAGCG | 362 | 24 | 58.4 | 46 | F10 |
| Assemblage B | ||||||
| tpiB.1 | CTGAGCCATGTAATAATAGGACACT | 145 | 25 | 54.9 | 40 | G2 |
| tpiB.2 | GCCAATAACACTATGGAGGTGAATA | 289 | 25 | 55.4 | 40 | H2 |
| girB.1 | ATGTACCTGACGATCAAGGAGGAGA | 246 | 25 | 57.5 | 48 | G3 |
| girB.2 | AGAACGCCGAGAGGAAGAAGATGTA | 442 | 25 | 60.6 | 48 | H3 |
| girB.3 | ATCGCGCACCTCGACAGACTCAT | 150 | 23 | 60.4 | 57 | G4 |
| girB.4 | CCACGACAGAGGCCCTCACAAACA | 561 | 24 | 61.9 | 58 | H4 |
| gdhB.1 | AACCTCTCGATCCTTAAGTTCCT | 20 | 23 | 55.0 | 44 | G6 |
| gdhB.2 | GTTATCTGTTTGGACAGTATAAGCG | 246 | 25 | 54.5 | 40 | H6 |
| gdhB.3 | TCGATCCTAAGGGCAAGTCGGA | 113 | 22 | 60.2 | 55 | G7 |
| c4B.1 | GGCTATGACGAAAGAAGATGCTGTA | 36 | 25 | 57.8 | 44 | G9 |
| c4B.2 | AGCCGGATGCTCATTGTTGCG | 68 | 21 | 60.8 | 57 | H9 |
| c4B.3 | TATCTGAGAAGGACATGAAGGACCT | 215 | 25 | 55.7 | 44 | G10 |
| c4B.4 | ATCCAGCTTCTCAGTGCTTCTCTG | 370 | 24 | 56.0 | 50 | H10 |
| C. parvum | ||||||
| dhf.1 | GGATAGGAATTAACGGACAATTACC | 45 | 25 | 58.3 | 40 | I1 |
| dhf.2 | TCAATAGAGAATCTTATGAATGATGACT | 281 | 28 | 54.8 | 29 | J1 |
| dhf.3 | AATGTGACTCGAATAAGAAGAATGC | 120 | 25 | 54.7 | 36 | K1 |
| dhf.4 | AAGCCGATCCTAATGTTGTTGTATT | 240 | 25 | 55.8 | 36 | L1 |
| ptg.2 | GAGTGATGATTCCAGGTTCTTTAGG | 415 | 25 | 55.9 | 44 | J4 |
| Cphsp.1 | TGCCGTACAGGATCTCTTATTATTG | 6 | 25 | 55.2 | 40 | K4 |
| cowp.1 | TCTCCTGCTTCAACTGTATGTCCTA | 133 | 25 | 57.7 | 44 | I6 |
| cowp.2 | TGTGTTAGAAGAAGCCAATATGACT | 391 | 25 | 54.9 | 36 | J6 |
| rDNA.1 | TTTACTTTGAGAAAATTAGAGTGCTTAAA | 106 | 29 | 54.3 | 24 | K6 |
| rDNA.2 | TAACAGTCAGAGGTGAAATTCTTAGAT | 249 | 27 | 55.2 | 33 | L6 |
| RAPD.1 | CATACAGGTGAGGAAGTTGATCTAT | 75 | 25 | 52.7 | 40 | I7 |
| RAPD.2 | ATCGCAATAATCCTTGTAACTTGTG | 372 | 25 | 56.5 | 36 | J7 |
| TRAPC2.1 | CCTTGAGTTGTACTGTCTCTGAATG | 14 | 25 | 56.1 | 44 | K7 |
| TRAPC2.2 | GGAGGAAAGACCTTCAGATTGTTTA | 324 | 25 | 55.4 | 40 | L7 |
| Type 1 | ||||||
| dhfI.1 | CTGTTTTGAGCAGTGGGATAGGAAT | 16 | 25 | 58.8 | 44 | I2 |
| dhfI.2 | CAAAGATAACTAATAATAAATGTGACTCG | 89 | 29 | 57.2 | 28 | J2 |
| ptgI.1 | TTAGATCCAGTTAGTTTGATTCCATTC | 161 | 27 | 57.3 | 33 | I3 |
| ptgI.2 | ATCCAATCTCAGATGAGATCATGAAT | 214 | 26 | 55.3 | 35 | J3 |
| Cphsp.2 | TGGTATCTTGAATGTATCTGCTGTT | 297 | 25 | 53.5 | 36 | L4 |
| p23I.1 | CCAATTAGCAACCAAGCCCAACAAA | 188 | 25 | 64.4 | 44 | I5 |
| p23I.2 | TACACTC*(3)GAGTACTTAACATGGGTT | 427 | 25 | 53.5 | 40 | J5 |
| Type 2 | ||||||
| dhfII.1 | CTGTTTTGAGTAGAGGAATAGGAAT | 16 | 25 | 53.6 | 36 | K2 |
| dhfII.2 | CAAAGATAACTAGTAATAATTGTGACTCG | 89 | 29 | 56.5 | 31 | L2 |
| ptgII.1 | TTGGATCCAGTCAGTTTGATTCCGTT | 161 | 26 | 61.8 | 42 | K3 |
| ptgII.2 | ATCCAATATCAGATGAGATAATGAAT | 214 | 26 | 53.4 | 27 | L3 |
| ptg.1 | TTATCGGTATCATATCTTGCTGCTAA | 117 | 26 | 55.0 | 35 | I4 |
| p23II.1 | CCAATCAGCAACCAAGCTCAACAAA | 188 | 25 | 61.4 | 44 | K5 |
| p23II.2 | CTCAGAGAGTACTTAACATGGGTT | 431 | 24 | 51.0 | 42 | L5 |
| Internal positive control | ||||||
| fliC.1 | AACGGTTAGCAATCGCCTGACCTGCGGCGTCATCCTTCGCGCTGTTAATACGCAAGCCAGAAGACAGACG | 70 | 80.6 | 56 |
Mismatched nucleotides between homologous target sequences are underlined, and deleted nucleotides are designated by an asterisk (with the number in parentheses).
The first nucleotide of the probe corresponding to the position within an amplicon starting from the 5′ end.
The position of each probe on the microarray is represented by coordinate number corresponding to Fig. 2B.
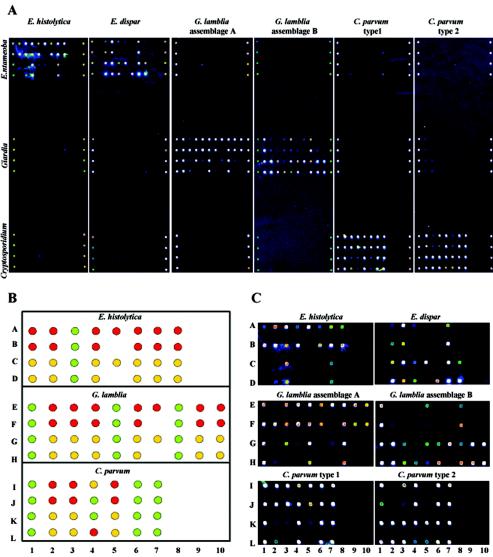
FIG. 2.
Genus-, species-, and subtype-level microarray specificity. (A) Identical protozoan microarrays were individually hybridized with the labeled multiplex PCR products from the six protozoan strains shown in Fig. 1. The portions of the array dedicated to a particular genus are indicated on the left, whereas the original hybridization sample source is indicated above each array. The hybridization spots at the left and right ends of each array represent internal positive controls. (B) Microarray template showing the coordinates and identity of each probe (see Table 2) and the expected subarray hybridization pattern for each species, assemblage, or genotype. In each template, red spots represent E. histolytica, G. lamblia assemblage A, or C. parvum type 1-specific probes; yellow spots represent E. dispar, G. lamblia assemblage B, or C. parvum type 2-specific probes; and green spots represent probes common for both E. histolytica and E. dispar, G. lamblia assemblages A and B, or C. parvum types 1 and 2. (C) Observed subarray hybridization patterns.
Multiplex PCR and synthesis of Cy5-labeled targets.
Multiplex PCR was conducted with the HotStarTaq Multiplex PCR kit according to the standard protocol (QIAGEN, Valencia, Calif.). The amplification reaction mixture (25 μl) consisted of 12.5 μl of 2× Master buffer, a mixture of 200 nM (each) forward and reverse primers, and 1 to 2 μl of template DNA. PCR was carried out in a Peltier Thermal Cycler PTC225 (MJ Research, Inc., Reno, Nev.) with an activation step at 95°C for 15 min; followed by 45 cycles of 94°C for 30 s, 54°C for 90 s, and 72°C for 90 s; and a final extension at 72°C for 10 min. PCR products (5 μl) were analyzed by electrophoresis with 2% agarose gels, and the remaining reaction volumes were purified with the DNA Clean & Concentrator-25 kit (Zymo Research, Orange, Calif.) and eluted in 35 μl of water or directly transferred to the following labeling step. Cy5-labeled ssDNA for microarray hybridization was synthesized from the multiplex PCR products by the primer extension method (20, 43). The labeling reaction was performed in a volume of 25 μl containing 2.5 U of Taq polymerase (QIAGEN); 1× PCR buffer with 2 mM MgCl2; 200 nM (each) reverse primers; 200 nM dATP, dGTP, and dTTP; 40 nM dCTP; 40 nM Cy5-dCTP (Amersham Bioscience UK Ltd., Amersham, Buckinghamshire, United Kingdom); and 2.5 μl of unpurified double-stranded DNA (dsDNA) or 16 μl of purified dsDNA from the previous multiplex PCRs. The primer extension protocol included preliminary denaturing at 95°C for 1 min followed by 35 cycles at 95°C for 20 s, 52°C for 20 s, and 72°C for 2 min, with a final extension step at 72°C for 10 min. The Cy5-labeled ssDNA products were then purified and dried.
Microarray hybridization and processing.
Oligonucleotide-printed slides were blocked with a 3% bovine serum albumin-casein solution (pH 7.4) for 15 min at room temperature, rinsed with distilled water, air dried, and placed in a MAUI hybridization dual chamber lid (BioMicro, Salt Lake City, Utah) immediately prior to hybridization. The fluorescently labeled ssDNA was resuspended in 20 μl of 1× hybridization buffer (5× Denhardt's solution, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 0.2% sodium dodecyl sulfate) containing 0.1 μM Cy5-labeled internal positive control probe (Table 2). The target hybridization sample was denatured at 95°C for 5 min, chilled on ice for 2 min, and applied to the microarray. Hybridization was performed on MAUI Hybridization System (BioMicro) for 1 h at 58°C. After hybridization, the slides were washed once with 4× SSC plus 0.2% sodium dodecyl sulfate at 58°C for 5 min and twice with 1× SSC at room temperature for 1 min. Slides were dried under a nitrogen stream and subsequently scanned with a GSI Lumonics ScanArray Lite confocal laser-scanning system (Perkin-Elmer, Torrance, Calif.). Unless otherwise noted, the microarray images were captured at laser power 80/PMT gain 80.
RESULTS
Target selection and multiplex PCR.
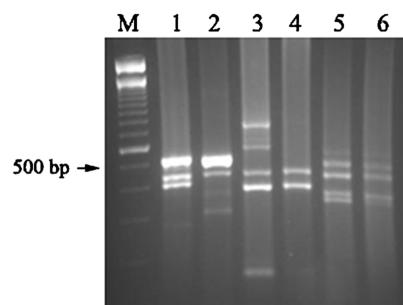
In addition to conserved genes, such as rRNA and hsp, which have been widely used as diagnostic markers, many genus- and species-specific genes were selected as amplification targets so as to avoid potential coamplification and cross-hybridization issues. The selected targets and their respective primer sets are shown in Table 1. Each of the Entamoeba targets was selected from previously reported variable regions: extrachromosomal rRNA, the intergenic region between the superoxide dismutase (sod) and actin genes, and the cysteine protease gene (cp1). Each pair of primers was adopted or designed to be able to amplify homologous genes from different species (E. histolytica and E. dispar) and genotypes (G. lamblia and C. parvum). Only the two sets of tpi primers were Giardia assemblage A and B specific. Individual PCRs with each primer set were conducted to confirm primer specificity and verify expected amplicon sizes (data not shown). Once confirmed, all genus-specific primers were pooled for Entamoeba-, Giardia-, and Cryptosporidium-specific multiplex PCRs (Fig. 1). Differences in the electrophoretic profiles between E. histolytica (lane 1) and E. dispar (lane 2) are due to homologous gene-length size variation as shown in Table 1. A 749-bp fragment of the giardin gene appeared to be missing from G. lamblia GS-H7 (lane 4), suggesting that this gene was not efficiently amplified in the multiplex PCR. C. parvum type 1 (lane 5) and type 2 (lane 6) isolates showed nearly identical electrophoretic profiles. In addition to the expected amplified products, there were also a few unexpected bands on the gel due to nonspecific amplification.
FIG. 1.
Amplification of protozoan target genes by multiplex PCR. A combination of seven Entamoeba-specific primer pairs were used to amplify the genomic DNA of E. histolytica HM-1:IMSS (lane 1) and E. dispar SAW760 (lane 2). Similarly, six pairs of G. lamblia primers were used to amplify G. lamblia WB (lane 3) and G. lamblia GS-H7 (lane 4), and eight pairs of C. parvum primers were used to amplify the genomic DNA of C. parvum TU502 (lane 5) and C. parvum GCH1 (lane 6).
Genus-level hybridization specificity.
Genomic DNA from each of the six protozoan species displayed in Fig. 1 was subjected to multiplex PCR amplification, primer extension fluorescent labeling, and hybridization to the protozoan microarrays. The unique hybridization pattern for each of the six representative protozoan isolates is shown in Fig. 2A. Importantly, the target DNA from each genus did not cross-hybridize with extra-genus probes, indicating the genus specificity of each protozoan subarray.
Entamoeba subarray.
Hybridization of E. histolytica strain HM-1:IMSS clone 9 and E. dispar SAW760 amplified products to the protozoan microarray (Entameoba subarray) resulted in the generation of two distinct profiles (Fig. 2C). As expected, the locus1-2.3 and ssrDNA probes, which are common for both Entamoeba species and served as Entamoeba-specific markers, showed strong signals in both the E. histolytica and E. dispar hybridizations. The prrdx and cp1 genes, which are unique to the genus Entamoeba, were the only gene coding sequences used to differentiate the two species tested. However, the E. histolytica prrdx probes cross-hybridized with E. dispar target DNA and vice versa, albeit with lower average hybridization signal intensities when compared to the isogenic probe/target hybridizations. This result is not surprising based on the number and location of nucleotide mismatches. A single mismatch between Ehprrdx.2 and Edprrdx.2 and two mismatches between Ehprrdx.1 and Edprrdx.1 were all located at the 3′ ends of the probes (Table 2). E. dispar target DNA also cross-hybridized with the E. histolytica locus1-2.1 and locus1-2.2 probes, although there are more than 10 nt differences in these regions between the two species. Again, the cross-hybridization signals were of lower average signal intensities when compared to the isogenic probe/target hybridizations. False-negative results were observed for the Edlocus5-6.1 and Edlocus5-6.2 probes when tested with E. dispar target DNA, despite the presence of a 510-bp locus 5-6 amplicon (Fig. 1, lane 2) and appropriately designed probes. However, the few instances of cross-hybridization and false-negative results within the Entamoeba subarray did not interfere with the interpretation of array results: the observed hybridization patterns clearly permitted the differentiation of both species.
Another apparently noninvasive but prevalent Entamoeba species, E. moshkovskii, was tested on the microarray as it is morphologically indistinguishable from E. histolytica and E. dispar (1). When genomic DNA from E. moshkovskii strain Laredo was amplified by the same set of multiplex primers and hybridized to the Entamoeba subarray, only two probes, Ehlocus1-2.3 and EssrDNA.1, which are common to both E. histolytica and E. dispar, showed positive hybridization signals (data not shown).
G. lamblia subarray.
The hybridization of G. lamblia WB (assemblage A isolate) and G. lamblia GS-H7 (assemblage B isolate) amplified targets to the Giardia subarray resulted in two distinct profiles (Fig. 2C). Probes targeting the hsp70, giardin, and gdh genes were used as species-specific markers and were positive for both assemblage members. All other probes were shown to be specific for either assemblage A (isolate WB, top two rows) or assemblage B (isolate GS-H7, bottom two rows). The profile results indicated a single false-positive result (girB.1) and false-negative result (tpiA.1) for the assemblage A representative and two false-negative results (girB.3 and girAB.2) for the assemblage B representative. A confirmatory profile generated by the hybridization of another assemblage B isolate, G. lamblia CM, was identical to that of G. lamblia GS-H7 (data not shown). Despite the observed absence of the 749-bp giardin amplicon from the G. lamblia GS-H7 multiplex PCR (Fig. 1, lane 4), six of the eight probes targeting the giardin amplicon were hybridization positive, thus highlighting microarray-based detection sensitivity.
C. parvum subarray.
Distinct profiles were also generated when the amplicons from representative C. parvum type 1 and type 2 strains, TU502 and GCH1, respectively, were hybridized to the C. parvum subarray (Fig. 2C). The observed type 1 hybridization profile revealed unexpected false-positive signals arising from three probes: dhfII.2, ptg1, and (low positive) p23II.1. The observed type 2 hybridization profile matched the expected profile (Fig. 2B). Confirmatory profiles generated by the hybridization of two other C. parvum isolates, UG502 (type 1) and Iowa (type 2), to the C. parvum subarray were identical to the presented TU502 (type 1) and GCH1 (type 2) profiles, respectively (data not shown). Although the C. parvum subarray contained the smallest number of genotype-specific probes, the probes designed to target three genes, dhf, ptg, and p23, were clearly enough to ably discriminate between the two major C. parvum genotypes.
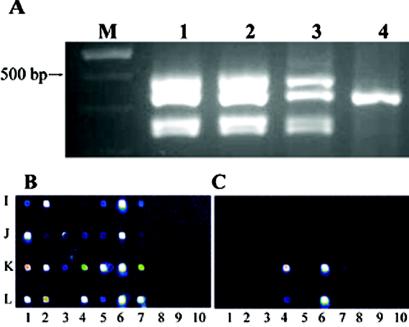
Although the probes and primers used for C. parvum subarray-based detection were specifically designed to differentiate the two major human infectious genotypes, we sought to potentially expand the utility of the current assay by testing two nonhuman-pathogenic Cryptosporidium species, C. meleagridis (primarily a bird pathogen but which can infect humans) and C. muris (a rodent pathogen). The electrophoretic profiles demonstrated in Fig. 3A suggested that the C. parvum multiplex primers amplified C. meleagridis DNA (lane 3) almost as efficiently as C. parvum DNA (lanes 1 and 2), but only appeared to amplify a single product from the C. muris isolate (lane 4). Application of the C. meleagridis amplicons to the C. parvum subarray resulted in hybridization-positive signals from 22 of the possible 28 probes, including the dhf, hsp, cowp, rRNA, randomly amplified polymorphic DNA (RAPD; one of two), and TRAP-C2 probes that were common to both cryptosporidial genotypes (Fig. 3B). Hybridization of the C. muris amplified material to the C. parvum subarray revealed two cryptosporidial targets were amplified: hsp70 and rRNA (Fig. 3C). The results suggested that (i) as expected, the hsp70 and rRNA sequences are the most conserved sequences among Cryptosporidum species; (ii) based solely on the number of hybridization-positive probes (and hence, primary sequence conservation), C. meleagridis appears to be more closely related to C. parvum than is C. muris (a result consistent with Cryptosporidium gene phylogenies) (38, 46); and (iii) although specifically designed for the detection of C. parvum, this subarray may also be used for the detection of other Cryptosporidium spp. once characteristic profiles using known templates have been established.
FIG. 3.
Electrophoretic and microarray hybridization profiles of three species of Cryptosporidium. (A) Comparative electrophoretic profiles of C. parvum TU502 (lane 1), C. parvum GCH1 (lane 2), C. meleagridis (lane 3), and C. muris (lane 4) multiplex PCR amplicons. (B and C) Hybridization pattern of C. meleagridis (B) and C. muris (C) according to the Cryptosporidium subarray.
Sensitivity of protozoan microarray.
One of the main advantages of microarray-based protozoan pathogen detection was assay sensitivity. As shown in Table 3, the combination of multiplex PCR, primer extension, and microarray hybridization resulted in the detection of as few as five G. lamblia WB and GS-H7 trophozoites. A comparison with individual PCR amplicon visualization via gel electrophoresis revealed that microarray-based detection sensitivity was either as sensitive or more sensitive in every instance, except for the detection of the GS-H7 giardin target amplicon.
TABLE 3.
Microarray versus PCR sensitivity with G. lamblia trophozoites
| Gene (no. of trophozoites) | Detection bya:
|
|||
|---|---|---|---|---|
| PCR
|
Microarray (no. positive/ no. of probes)
|
|||
| WB | GS-H7 | WB | GS-H7 | |
| hsp | ||||
| 200 | + | + | 2/2 | 2/2 |
| 5 | + | − | 2/2 | 2/2 |
| tpi | ||||
| 200 | + | + | 1/2 | 2/2 |
| 5 | − | + | 1/2 | 2/2 |
| giardin | ||||
| 200 | + | + | 6/6 | 6/6 |
| 5 | ? | ? | 6/6 | 0/6 |
| gdh | ||||
| 200 | + | + | 5/5 | 5/5 |
| 5 | − | − | 5/5 | 4/5 |
| c4 | ||||
| 200 | + | + | 4/4 | 4/4 |
| 5 | − | − | 4/4 | 4/4 |
Detection of each gene by PCR is designated as positive (+), ambiguous (?), or negative (−). Detection of each gene by microarray is represented by the number of positive spots out of the number of probes per gene.
As protozoan coinfections are not uncommon, we also sought to test whether the simultaneous amplification of two protozoan species would result in lower detection sensitivity for the less-abundant protozoan in the mixture. Two 10:1 G. lamblia mixtures (500:50 WB/GS-H7 and 50:5 WB/GS-H7 trophozoite ratios) were amplified and hybridized to the Giardia subarray. In both cases, all of the probes within the Giardia subarray showed hybridization-positive signals revealing Giardia assemblage A and B profiles (data not shown). These results indicated that the microarray was able to simultaneously detect members of both assemblages and that the detection of both coinfectants (at a 50:5 ratio) did not adversely effect the lower detection threshold of 5 trophozoites seen when detecting a single species (Table 3).
DISCUSSION
This study represents the first report documenting the use of oligonucleotide microarrays for the detection and genotyping of multiple protozoan parasites. The developed experimental protocol combining multiplex PCR, primer extension-based labeling, and microarray hybridization was shown to be a successful strategy for obtaining genus-level specificity capable of unequivocally detecting and differentiating members of the genera Entamoeba, Giardia, and Cryptosporidium. Furthermore, the hybridization of diagnostic targets to species- and genotype-specific probes generated unique profiles enabling the identification and differentiation of E. dispar from E. histolytica, G. lamblia assemblages A and B, and C. parvum genotypes 1 and 2 by using a single assay. In addition to distinguishing between the targeted principal genotypes, this assay may also have utility in detecting other related isolates, such as C. meleagridis and C. muris, and differentiating innocuous species (E. moshkovskii) from pathogenic species (E. histolytica).
The current accepted methods for the environmental detection of Cryptosporidium and Giardia spp. are labor and resource intensive. For example, Environmental Protection Agency method 1623 requires a series of filtration, immunomagnetic separation, and immunofluorescence assays for detection and determination of pathogen concentrations followed by vital dye staining and microscopy confirmatory assays. Although effective, the observed limits in sensitivity, specificity, and reproducibility have warranted the development of alternate detection strategies. Molecular detection methods, primarily PCR based, have become increasingly common for the identification of viral and bacterial pathogens and appear to be especially attractive for the detection of protozoan parasites that are difficult to culture, morphologically similar, and capable of causing disease in low infectious doses. Thus, the sensitivity and specificity afforded by molecular detection methods such as PCR, restriction fragment length polymorphism, and real-time PCR have resulted in the development of rapid approaches for the detection and genotyping of protozoa in clinical and environmental samples (4, 16, 40).
However, as most PCR-based detection methods are reliant upon amplicon size analyses or generic dsDNA intercalating dye fluorescence, subsequent sequencing or hybridization assays are necessary to confirm the identity of the amplified target. In comparison to a single PCR-based detection method, the combined amplification and microarray hybridization strategy employed in this study to detect and genotype protozoa had the following advantages. (i) Assay sensitivity was achieved not only by amplification of target sequences, but also by DNA-DNA hybridization, which was visualized by fluorescent labeling. Our data demonstrated that as few as five trophozoites of G. lamblia could be accurately detected by this method, whereas the use of PCR analysis alone at identical concentrations generated false-negative results (Table 3). (ii) Pathogen identification was not reliant upon amplicon size analyses or nonspecific fluorescent dye intercalation, but rather two sequential hybridization events—primer hybridization for target generation and target/probe hybridization. The combination of primer and probe specificity enhanced assay specificity as genetic variants were unambiguously identified and genotyped despite the existence of nonspecific multiplex PCR products that tend to confound electrophoretic analyses. (iii) Multiplex PCR amplification and microarray hybridization allowed for the simultaneous detection of multiple genetic markers. The redundancy of species- and genotype-specific targets and probes not only increased the confidence in the results but also reduced the vulnerability to spontaneous mutations that may occur in circulating clinical or environmental isolates. This experimental redundancy was found to be necessary for accurate and reliable data interpretation. (iv) Assay throughput was increased as the microarray format enabled the simultaneous analysis of multiple organisms with a large number of genetic markers in one experiment.
The sensitivity and specificity afforded by amplification and hybridization schemes has been highlighted by a number of microarray-based pathogen detection studies that have utilized a single highly conserved gene as the amplification and hybridization target (9, 13, 19, 29, 36). As demonstrated in these studies, the amplification of a single conserved target sequence with a pair of specific or degenerate primers is often more efficient and convenient than multiplex or random amplification, but there are two salient caveats. (i) Often the selected gene, such as 16S rRNA, is so highly conserved that sequence variability among species and strains is small or nonexistent, making it difficult to confidently differentiate between closely related species or subtypes. (ii) The reliance upon a primer pair to amplify a single target region provides little margin for error. A mutation in the primer binding site could potentially reduce or prevent target amplification and subsequently produce false-negative results. The potential pitfalls associated with the use of a single target led us to select multiple diagnostic sequences, both conserved and highly variable, as amplification and hybridization targets. Highly conserved target genes, such as rRNA and hsp70, were selected for general identification of the genus or species. Identification down to the species, assemblage, or genotype level was accomplished by targeting highly variable genes or genes that were unique to Entamoeba (cp1), Giardia (the gene encoding giardin and c4), and Cryptosporidium (cowp, ptg, RAPD, the gene encoding TRAP-C2, and p23). As demonstrated in this study, the use of multiple genetic markers, both conserved and variable, increased the certainty of detection and discrimination.
The use of short probes also aided in the differentiation of species as 20- to 30-mer oligonucleotides are ideal for distinguishing closely related species and monitoring intraspecies genetic variability. For example, Straub et al. recently reported genotyping C. parvum with an hsp70 single-nucleotide polymorphism (SNP) microarray using short probes (36). In this study, we also chose to target the Cryptosporidium hsp70 gene and designed short probes based on primary sequence from a C. parvum genotype 1 human isolate (accession no. AF221535). As hybridization to the C. parvum subarray with C. parvum genotype 1 (TU502) and genotype 2 (GCH1) demonstrated, genotype 2 strain GCH1 target hybridization to probe Cphsp.2 generated a markedly lower fluorescent signal (14-fold less) than that observed with the genotype 1 strain TU502 (Fig. 2C, C. parvum subarray, coordinate L-4). A comparison of the Cphsp.2 probe sequence to C. parvum genotype 2 hsp70 genes in GenBank (accession no. U71181) revealed an SNP at position 1404 of hsp70. This SNP finding turned out to be the same as described by Straub et al. when using the hsp70 SNP microarray (36). Thus, in addition to only using the presence or absence of fluorescent signals to determine genotype, in certain instances, the variations in hybridization signal intensities due to SNPs were also useful in differentiating closely related strains.
Although they did not hinder the interpretation of results, each of the subarrays generated a few false-positive and false-negative hybridization results. We suspect that these erroneous results were due to the amount of available sequence information, the probe location within the target, and/or probe design. First, in comparison to the amount of primary sequence information available for viral and bacterial enteric pathogens, the amount of sequence data available for protozoan enteric pathogens is limited. Thus, limited or incorrect sequence information in public databases may have resulted in the design of PCR primers and oligonucleotide probes that were not representative of the targeted genes. For example, the numbers of E. dispar sequences deposited in GenBank are limited, and often there was only a single sequence available for each gene target. This limitation was clearly highlighted by the number of false-positive and false-negative results obtained within the E. dispar SAW760 hybridization profile. The lack of sequence redundancy and sampling may have resulted in a potentially high design error rate for both primers and probes. Secondly, the positions of oligonucleotide probes (as they relate to the target sequence) and chosen method of labeling may have hindered results. Usually, two probes were designed for each target, with one probe located near the 5′ end and the other near 3′ end of the amplicon. In most cases, false-negative or low-intensity hybridization signals were found associated with probes located on the 5′ end of the target amplicon (such as Ehlocus5-6.1, EhITS.1, EhSA.1, and tpiA.1). As the amplicons were labeled by primer extension incorporation of Cy5-dCTP from the 3′ end, it is possible that premature termination of polymerization and dye incorporation resulted in fewer labeled full-length amplicons, thus decreasing the number of labeled targets for hybridization to the 5′-localized probes. In addition, as the probes were covalently immobilized via a 5′ reactive amine group, hybridization to the 5′ target terminus may have been hindered by increased steric and spatial constraints at the probe-target interface.
In summary, we have developed a microarray for the parallel detection and genotyping of E. histolytica, G. lamblia, and C. parvum. The amplification and hybridization of multiple diagnostic regions to short genus-, species-, and subtype-specific probes allowed for the unequivocal detection and discrimination of E. histolytica, E. dispar, G. lamblia assemblages A and B, and C. parvum genotypes 1 and 2 in a single assay. Thus, this method may aid in confidently expediting the detection of these three major waterborne parasites while simultaneously providing valuable epidemiological information. The relatively rapid and accurate nature of this platform has great potential for use as a diagnostic tool, and efforts are currently under way to further test the utility of this microarray on environmental water and clinical human fecal samples.
Acknowledgments
We kindly thank Carolyn Meador for excellent technical assistance and KamTek, Inc., for their microarray printing support. We also are grateful to C. Graham Clark (London School of Hygiene and Tropical Medicine), Theodore E. Nash (National Institutes of Health), and Donna Akiyoshi (Tufts University) for their generous provision of the protozoan isolates and to Vladimir Chizhikov (Food and Drug Administration) for his scientific advice and help.
This work was supported by the Office of Naval Research. Z.W. is an American Society for Engineering Education Postdoctoral Fellow, and G.J.V. is a National Research Council Postdoctoral Fellow.
The opinions and assertions contained herein are those of the authors and are not to be construed as those of the U.S. Navy or military service at large.
REFERENCES
- 1.Ali, I. K., M. B. Hossain, S. Roy, P. F. Ayeh-Kumi, W. A. Petri, Jr., R. Haque, and C. G. Clark. 2003. Entamoeba moshkovskii infections in children, Bangladesh. Emerg. Infect. Dis. 9:580-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amar, C. F., P. H. Dear, and J. McLauchlin. 2003. Detection and genotyping by real-time PCR/RFLP analyses of Giardia duodenalis from human faeces. J. Med. Microbiol. 52:681-683. [DOI] [PubMed] [Google Scholar]
- 3.Amar, C. F. L., P. H. Dear, S. Pedraza-Diaz, N. Looker, E. Linnane, and J. McLauchlin. 2002. Sensitive PCR-restriction fragment length polymorphism assay for detection and genotyping of Giardia duodenalis in human feces. J. Clin. Microbiol. 40:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blessmann, J., H. Buss, P. A. Nu, B. T. Dinh, Q. T. V. Ngo, A. L. Van, M. D. A. Alla, T. F. H. G. Jackson, J. I. Ravdin, and E. Tannich. 2002. Real-time PCR for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in fecal samples. J. Clin. Microbiol. 40:4413-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caccio, S. M., M. De Giacomo, and E. Pozio. 2002. Sequence analysis of the beta-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int. J. Parasitol. 32:1023-1030. [DOI] [PubMed] [Google Scholar]
- 6.Charles, P. T., G. J. Vora, J. D. Andreadis, A. J. Fortney, C. E. Meador, C. S. Dulcey, and D. A. Stenger. 2003. Fabrication and surface characterization of DNA microarrays using amine- and thiol-terminated oligonucleotide probes. Langmuir 19:1586-1591. [Google Scholar]
- 7.Cherkasova, E., M. Laassri, V. Chizhikov, E. Korotkova, E. Dragunsky, V. I. Agol, and K. Chumakov. 2003. Microarray analysis of evolution of RNA viruses: evidence of circulation of virulent highly divergent vaccine-derived polioviruses. Proc. Natl. Acad. Sci. USA 100:9398-9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, C. G. 1997. Riboprinting: a tool for the study of genetic diversity in microorganisms. J. Eukaryot. Microbiol. 44:277-283. [DOI] [PubMed] [Google Scholar]
- 11.Diamond, L. S., and C. G. Clark. 1993. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 40:340-344. [DOI] [PubMed] [Google Scholar]
- 12.Evangelopoulos, A., G. Spanakos, E. Patsoula, N. Vakalis, and N. Legakis. 2000. A nested, multiplex, PCR assay for the simultaneous detection and differentiation of Entamoeba histolytica and Entamoeba dispar in faeces. Ann. Trop. Med. Parasitol. 94:233-240. [DOI] [PubMed] [Google Scholar]
- 13.Fukushima, M., K. Kakinuma, H. Hayashi, H. Nagai, K. Ito, and R. Kawaguchi. 2003. Detection and identification of Mycobacterium species isolates by DNA microarray. J. Clin. Microbiol. 41:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, S., A. Debnath, A. Sil, S. De, D. J. Chattopadhyay, and P. Das. 2000. PCR detection of Giardia lamblia in stool: targeting intergenic spacer region of multicopy rRNA gene. Mol. Cell Probes 14:181-189. [DOI] [PubMed] [Google Scholar]
- 15.Gonin, P., and L. Trudel. 2003. Detection and differentiation of Entamoeba histolytica and Entamoeba dispar isolates in clinical samples by PCR and enzyme-linked immunosorbent assay. J. Clin. Microbiol. 41:237-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston, S. P., M. M. Ballard, M. J. Beach, L. Causer, and P. P. Wilkins. 2003. Evaluation of three commercial assays for detection of Giardia and Cryptosporidium organisms in fecal specimens. J. Clin. Microbiol. 41:623-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keramas, G., D. D. Bang, M. Lund, M. Madsen, S. E. Rasmussen, H. Bunkenborg, P. Telleman, and C. B. Christensen. 2003. Development of a sensitive DNA microarray suitable for rapid detection of Campylobacter spp. Mol. Cell Probes 17:187-196. [DOI] [PubMed] [Google Scholar]
- 20.Laassri, M., V. Chizhikov, M. Mikheev, S. Shchelkunov, and K. Chumakov. 2003. Detection and discrimination of orthopoxviruses using microarrays of immobilized oligonucleotides. J. Virol. Methods 112:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laberge, I., A. Ibrahim, J. R. Barta, and M. W. Griffiths. 1996. Detection of Cryptosporidium parvum in raw milk by PCR and oligonucleotide probe hybridization. Appl. Environ. Microbiol. 62:3259-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeChevallier, M. W., G. D. Di Giovanni, J. L. Clancy, Z. Bukhari, S. Bukhari, J. S. Rosen, J. Sobrinho, and M. M. Frey. 2003. Comparison of method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Appl. Environ. Microbiol. 69:971-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limor, J. R., A. A. Lal, and L. Xiao. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahbubani, M. H., A. K. Bej, M. H. Perlin, F. W. Schaefer III, W. Jakubowski, and R. M. Atlas. 1992. Differentiation of Giardia duodenalis from other Giardia spp. by using polymerase chain reaction and gene probes. J. Clin. Microbiol. 30:74-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall, M. M., D. Naumovitz, Y. Ortega, and C. R. Sterling. 1997. Waterborne protozoan pathogens. Clin. Microbiol. Rev. 10:67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monis, P. T., R. H. Andrews, G. Mayrhofer, and P. L. Ey. 1999. Molecular systematics of the parasitic protozoan Giardia intestinalis. Mol. Biol. E vol. 16:1135-1144. [DOI] [PubMed] [Google Scholar]
- 27.Morgan, U. M., P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, K. D. Sargent, A. Elliot, and R. C. Thompson. 1999. Sequence and PCR-RFLP analysis of the internal transcribed spacers of the rDNA repeat unit in isolates of Cryptosporidium from different hosts. Parasitology 118:49-58. [DOI] [PubMed] [Google Scholar]
- 28.Morgan, U. M., K. D. Sargent, P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, A. Elliot, and R. C. Thompson. 1998. Molecular characterization of Cryptosporidium from various hosts. Parasitology 117:31-37. [DOI] [PubMed] [Google Scholar]
- 29.Peplies, J., F. O. Glöckner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petri, W. A., Jr., and U. Singh. 1999. Diagnosis and management of amebiasis. Clin. Infect. Dis. 29:1117-1125. [DOI] [PubMed] [Google Scholar]
- 32.Rochelle, P. A., R. De Leon, M. H. Stewart, and R. L. Wolfe. 1997. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl. Environ. Microbiol. 63:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanuki, J., T. Asai, E. Okuzawa, S. Kobayashi, and T. Takeuchi. 1997. Identification of Entamoeba histolytica and E. dispar cysts in stool by polymerase chain reaction. Parasitol. Res. 83:96-98. [DOI] [PubMed] [Google Scholar]
- 34.Som, I., A. Azam, A. Bhattacharya, and S. Bhattacharya. 2000. Inter- and intra-strain variation in the 5.8S ribosomal RNA and internal transcribed spacer sequences of Entamoeba histolytica and comparison with Entamoeba dispar, Entamoeba moshkovskii and Entamoeba invadens. Int. J. Parasitol. 30:723-728. [DOI] [PubMed] [Google Scholar]
- 35.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straub, T. M., D. S. Daly, S. Wunshel, P. A. Rochelle, R. DeLeon, and D. P. Chandler. 2002. Genotyping Cryptosporidium parvum with an hsp70 single-nucleotide polymorphism microarray. Appl. Environ. Microbiol. 68:1817-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturbaum, G. D., B. H. Jost, and C. R. Sterling. 2003. Nucleotide changes within three Cryptosporidium parvum surface protein encoding genes differentiate genotype I from genotype II isolates. Mol. Biochem. Parasitol. 128:87-90. [DOI] [PubMed] [Google Scholar]
- 38.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tachibana, H., and X. J. Cheng. 2000. Entamoeba dispar: cloning and characterization of peroxiredoxin genes. Exp. Parasitol. 94:51-55. [DOI] [PubMed] [Google Scholar]
- 40.Tanriverdi, S., A. Tanyeli, F. Başlamişli, F. Köksal, Y. Kilinçc, X. Feng, G. Batzer, S. Tzipori, and G. Widmer. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanyuksel, M., and W. A. Petri, Jr. 2003. Laboratory diagnosis of amebiasis. Clin. Microbiol. Rev. 16:713-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, R. C., R. M. Hopkins, and W. L. Homan. 2000. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol. Today 16:210-213. [DOI] [PubMed] [Google Scholar]
- 43.Volokhov, D., A. Rasooly, K. Chumakov, and V. Chizhikov. 2002. Identification of Listeria species by microarray-based assay. J. Clin. Microbiol. 40:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, Z., I. Nagano, A. Matsuo, S. Uga, I. Kimata, M. Iseki, and Y. Takahashi. 2000. Specific PCR primers for Cryptosporidium parvum with extra high sensitivity. Mol. Cell Probes 14:33-39. [DOI] [PubMed] [Google Scholar]
- 46.Xiao, L., J. Limor, U. M. Morgan, I. M. Sulaiman, R. C. A. Thompson, and A. A. Lal. 2000. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl. Environ. Microbiol. 66:5499-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yong, T., K. Han, H. Yang, and S. Park. 2002. PCR-RFLP analysis of Giardia intestinalis using a Giardia-specific gene, GLORF-C4. Parasite 9:65-70. [DOI] [PubMed] [Google Scholar]
- 48.Zaki, M., P. Meelu, W. Sun, and C. G. Clark. 2002. Simultaneous differentiation and typing of Entamoeba histolytica and Entamoeba dispar. J. Clin. Microbiol. 40:1271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]