Abstract
Brachyspira pilosicoli, the causative agent of porcine intestinal spirochetosis, usually has hippurate-cleaving capacity. We have regularly isolated hippurate-negative B. pilosicoli from cases of porcine diarrhea. In this study, we show that these biochemically atypical B. pilosicoli isolates can be classified as B. pilosicoli. 16S ribosomal DNA was partially sequenced from eight hippurate-negative and two hippurate-positive B. pilosicoli-like isolates from seven herds. The differences in nucleotide sequence with B. pilosicoli P43/6/78 type strain were not associated with hippurate cleavage. In 877 bp, the hippurate-negative isolates had a similarity of 98.63 to 100% to the type strain, with the corresponding figures for the two hippurate-positive isolates being 98.86 and 100%. The nucleotide sequences of hippurate-positive isolates were identical to the respective sequences of hippurate-negative isolates from one herd. The DNA macrorestriction patterns of a total of 20 hippurate-negative and -positive B. pilosicoli isolates were diverse, and no clustering in conjunction with the hippurate reaction was found. In two herds, hippurate-positive and -negative B. pilosicoli isolates had a common macrorestriction pattern. The ultrastructure of hippurate-negative isolates was similar to the type strain. In conclusion, B. pilosicoli can be either hippurate positive or negative and, thus, the scheme for biochemical differentiation of porcine Brachyspira should be revised to include identification of hippurate-negative B. pilosicoli.
Brachyspira pilosicoli is a weakly beta-hemolytic, anaerobic intestinal spirochete that occurs worldwide in pigs. B. pilosicoli has also been detected in several other farmed and wild animal species, as well as in humans (3, 9, 15, 17, 23, 31, 38). In growing pigs, B. pilosicoli causes diarrhea and affects the production and welfare of animals (6, 32, 33).
According to biochemical and phylogenetic studies, Brachyspira spp. can be divided into biochemical groups I, II, IIIa, IIIb, IIIc, and IV (8). B. pilosicoli belongs to group IV. The species of porcine Brachyspira can be determined by evaluating the degree of hemolysis and by biochemical tests based on the classification scheme of Fellström et al. (10). According to the species description, B. pilosicoli hydrolyzes hippurate, does not produce indole from tryptophan, and does not have β-glucosidase activity (37) (Table 1). Several species-specific PCR applications are used to detect B. pilosicoli in cultures and feces (1, 7, 19, 20, 24). PCR speeds up the identification of Brachyspira species and is economical for use in high throughput laboratories. DNA macrorestriction profiling by pulsed-field gel electrophoresis (PFGE) or amplified fragment length polymorphism methods has been used for subspecies genotyping of B. pilosicoli strains (2, 11, 21).
TABLE 1.
| Species | Group | Hemolysis | Indole production | Hippurate hydrolysis | α-Galactosidase activity | β-Glucosidase activity |
|---|---|---|---|---|---|---|
| B. hyodysenteriae | I | Strong | ± | − | − | + |
| B. intermedia | II | Weak | + | − | − | + |
| B. murdochii | IIIa | Weak | − | − | − | + |
| B. innocens | IIIbc | Weak | − | − | + | + |
| B. pilosicoli | IV | Weak | − | ± | ± | − |
According to Fellström et al. (10).
B. pilosicoli can be hippurate positive or negative according to the present study.
Brachyspira hyodysenteriae, a cause of swine dysentery, was considered to be indole positive until indole-negative strains were identified in Germany, Belgium, and Canada (10). Confusion with other porcine Brachyspira species was avoided because of the strong beta-hemolysis of these strains. Characterization of hippurate-negative B. pilosicoli strains has not been reported to date. Such strains could easily be confused with nonpathogenic Brachyspira species of biochemical groups IIIa to c when a simplified classification scheme (7) with no glucosidase tests is applied.
Thomson et al. (34, 35) reported that some weakly beta-hemolytic porcine spirochetes could not be classified biochemically according to the current scheme. These authors also obtained inconsistent results between biochemical classification and molecular typing of some porcine spirochetes (35). Thus, phylogenetically new groups of Brachyspira may be characterized in the future.
The cell of B. pilosicoli is smaller than that of the other porcine Brachyspira species, and this difference can be seen by using light microscopy (32). However, Brachyspira aalborgi, which has been recognized in humans and nonhuman primates (13, 22), is also smaller than, for example, Brachyspira hyodysenteriae. Thus, a comparative study using light microscopy is not appropriate for distinguishing between different Brachyspira species.
B. pilosicoli has been frequently isolated from Finnish pigs (12). Since 1997, we have isolated from diarrheic pigs weakly beta-hemolytic spirochetes that are hippurate negative and indole negative and lack β-glucosidase activity. Under light microscopy, the hippurate-negative isolates resemble B. pilosicoli in size. They are positive in a B. pilosicoli-specific PCR designed for 16S ribosomal DNA (rDNA) (7) and 23S rDNA (19). We have presumed these hippurate-negative isolates to be biochemical variants of B. pilosicoli and designated them B. pilosicoli hipp−. We have isolated B. pilosicoli hipp− from two to seven unrelated Finnish herds annually. For comparison, the mean annual herd diagnosis of typical hippurate-positive B. pilosicoli is 24 cases.
Our objective here was to verify the inclusion of hippurate-negative, B. pilosicoli-like porcine spirochetes in the species B. pilosicoli and, if verified, to amend the classification scheme of B. pilosicoli with regard to hippurate hydrolysis (Table 1). We studied the genetic relationship between Finnish B. pilosicoli hipp− isolates, Finnish B. pilosicoli isolates, and Brachyspira reference strains by using 16S rDNA nucleotide sequence analysis and PFGE. We further examined the ultrastucture of B. pilosicoli hipp− by transmission electron microscopy (TEM).
MATERIALS AND METHODS
Eleven B. pilosicoli hipp− isolates from seven unrelated pig herds were studied. Diarrhea was present among weaners and/or fatteners of all of the herds. A typical B. pilosicoli was concomitantly isolated from four of these herds. Altogether, nine B. pilosicoli isolates from three herds were included in the study. B. pilosicoli hipp− was found alone in two herds, and both Lawsonia intracellularis and B. pilosicoli hipp− were detected in one herd. The Brachyspira isolates studied and the reference strains used are summarized in Table 2.
TABLE 2.
Brachyspira isolates and strains used in the study
| Species (biotype) | Designation | Origin | Investigationsa | Accession no.b | Source or reference |
|---|---|---|---|---|---|
| B. pilosicoli hipp− | Br710 | Herd A | PFGE; SEQ | AY514025 | This study |
| Br860 | Herd B | PFGE; seq | ID to U14927 | This study | |
| Br944 | Herd B | PFGE; seq | ID to U14927 | This study | |
| Br972 | Herd C | PFGE | This study | ||
| Br980 | Herd D | PFGE; seq; TEM | ID to AY514026 | This study | |
| Br983 | Herd D | PFGE | This study | ||
| Br986 | Herd D | PFGE | This study | ||
| Br1048 | Herd E | PFGE; seq; TEM | AY514026 | This study | |
| Br1620 | Herd F | PFGE; SEQ | ID to AY514024 | This study | |
| Br1622 | Herd F | PFGE; SEQ | AY514024 | This study | |
| Br1940 | Herd G | PFGE; seq | ID to AY514026 | This study | |
| B. pilosicoli | Br858 | Herd B | PFGE | This study | |
| Br868 | Herd B | PFGE | This study | ||
| Br869 | Herd B | PFGE; seq | ID to U14927 | This study | |
| Br973 | Herd C | PFGE | This study | ||
| Br964 | Herd D | PFGE; seq | ID to AY514026 | This study | |
| Br977 | Herd D | PFGE | This study | ||
| Br981 | Herd D | PFGE | This study | ||
| Br985 | Herd D | PFGE | This study | ||
| P43/6/78T | ATCCT 51139 | PFGE; TEM | U14927 | 27, 32 | |
| B. hyodysenteriae | B-78T | ATCCT 27164 | TEM | 30 | |
| B204 | ATCC 31212 | PFGE | 16, 30 |
SEQ, sequencing of 1,435 bp from 16S rDNA; seq, sequencing of 877 bp from 16S rDNA.
Italics indicate accession numbers of sequences determined in this study. ID, identical.
Hippurate hydrolysis.
At the time of the primary isolation, the B. pilosicoli hipp− isolates had been tested for hippurate hydrolysis two or three times before being stored at −70°C in beef broth (Merck, Darmstadt, Germany) supplemented with 12% horse serum and 15% glycerol. In our study, the permanence of the lack of hippurate hydrolysis of the isolates was studied. Altogether, seven B. pilosicoli hipp− isolates, one from each herd, were thawed and grown anaerobically on Fastidious Anaerobe agar (LabM, Lancashire, United Kingdom) supplemented with 5% bovine blood. The isolates were cultured and passaged as doublets: one culture at 38°C and another at 42°C for all of the passages. The passage interval was 3 or 4 days. At least 7 and a maximum of 10 consecutive passages of each isolate were tested for hippurate hydrolysis by using the method described by Rübsamen and Rübsamen (28), with a minor modification concerning incubation temperature: bacteria were suspended in 0.5 ml of 1% sodium hippurate in water and then incubated for 4 h at 38 or 42°C, depending on the incubation temperature used for the passage. Next, 0.2 ml of 3.5% ninhydrin was added, followed by incubation for 10 min at 38°C. Nontransparent, deep blue color was judged to be a positive, and no color was considered a negative reaction. B. pilosicoli P43/6/78 type strain (B. pilosicoli P43) and B. hyodysenteriae reference strain B204 (B. hyodysenteriae B204) were used as positive and negative controls, respectively.
Nucleotide sequence analysis.
Eight B. pilosicoli hipp− and two B. pilosicoli isolates were subjected to sequence analysis. A partial 16S rDNA sequence was obtained from the amplicon produced by B. pilosicoli-specific PCR (7). The 877-bp sequence ranged from nucleotide positions 183 to 1059, the numbering following the Brachyspira consensus sequence (27). The amplicon was sequenced directly by using the PCR primers or cloned with a PCR-Script Amp cloning kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. The sequencing was purchased from the AIV Institute, Kuopio, Finland. In addition, 16S rDNA of three B. pilosicoli hipp− isolates was almost completely sequenced, as previously described (18). The 1,435-bp sequence obtained covered the Brachyspira consensus sequence from position −8 to position 1431.
The sequences were aligned with the 16S rDNA sequence of B. pilosicoli P43 (accession no. U14927) by using the MultAlin program (http://prodes.tolouse.inra.fr/multalin/) (4). The sequences were also screened via the internet for homology with database sequences from the National Center for Biotechnology (Washington, D.C. [http://www.ncbi.nlm.nih.gov/BLAST]).
The obtained 16S rDNA sequences of the isolates Br1622, Br710, and Br1048 have been deposited in GenBank under accession numbers AY514024 to AY514026.
PFGE.
All of the B. pilosicoli hipp− and B. pilosicoli field isolates, B. pilosicoli P43, and B. hyodysenteriae B204 were subjected to PFGE and subsequently to clustering analysis of macrorestriction profiles (MRPs). Rare-cutting enzyme MluI (New England Biolabs, Inc., Beverly, Mass.) was used for DNA digestion. The macrorestriction and PFGE were done as previously described (11), with minor modifications as follows: 40 U of MluI per sample was used for the DNA digestion, and the digestion lasted 16 h at 37°C. In the electrophoresis, a pulse ramp from 2 to 45 s was used.
The MRPs were analyzed by using the GelCompar II program (version 1.01; Applied Maths, Kortrijk, Belgium). Clustering analysis was based on the unweighted pair-group method with arithmetic averages.
TEM.
B. pilosicoli hipp− isolate Br980 from herd D, B. pilosicoli hipp− isolate Br1048 from herd E, B. hyodysenteriae type strain B-78 (B. hyodysenteriae B78), and B. pilosicoli P43 were subjected to ultrastructural study by TEM. The bacteria were suspended in 0.1 M phosphate buffer (pH 7.4) at a concentration of 1.0 × 107 to 1.0 × 108 cells/ml. Negative stainings were performed by using 1% phosphotungstic acid, as previously described (39). The mean values for bacteria dimensions were calculated from the measurements of 20 single bacteria per isolate or strain.
RESULTS
Hippurate hydrolysis.
The hippurate hydrolysis test results for each culture passage of all seven B. pilosicoli hipp− isolates at incubation temperatures of 38°C and 42°C were negative.
Nucleotide sequence analysis.
The search for homologous nucleotide sequences showed 16S rDNA of B. pilosicoli P43 to have the lowest number of nucleotide differences with the Finnish isolates. Within the nucleotide positions 183 to 1059, the 16S rDNA sequences of B. pilosicoli hipp− isolates had no differences or 10-, 11-, or 12-nucleotide differences with B. pilosicoli P43 (Table 3). The sequences of one B. pilosicoli and two B. pilosicoli hipp− isolates from herd B were identical to B. pilosicoli P43. One B. pilosicoli and one B. pilosicoli hipp− isolate from herd D had the same differences in 10 nucleotides. The residues and positions for the first 10 nucleotide differences with B. pilosicoli P43 were identical between the isolates that had at least 10 nucleotide differences. Within the partial sequence of 877 bp, the homology between B. pilosicoli hipp− isolates and B. pilosicoli P43 was 98.63 to 100%. Between the two B. pilosicoli isolates and B. pilosicoli P43, the homologies were 98.86 and 100%.
TABLE 3.
Differences among the 16S rDNA sequences of B. pilosicoli P43/6/78 type strain, Finnish B. pilosicoli, and B. pilosicoli hipp− field isolates
| Herd | Strain/isolate | Biotype | Nucleotide at positiona:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 214 | 215 | 222 | 223 | 229 | 234 | 240 | 243 | 588 | 589 | 996 | 1025 | |||
| NAb | Type strain P43/6/78 | B. pilosicoli | C | T | A | G | T | T | A | T | A | G | G | G |
| B | Br860, Br944 | B. pilosicoli hipp− | - | - | - | - | - | - | - | - | - | - | - | - |
| B | Br869 | B. pilosicoli | - | - | - | - | - | - | - | - | - | - | - | - |
| D | Br980 | B. pilosicoli hipp− | T | A | G | A | A | C | G | A | G | A | - | - |
| D | Br964 | B. pilosicoli | T | A | G | A | A | C | G | A | G | A | - | - |
| E | Br1048 | B. pilosicoli hipp− | T | A | G | A | A | C | G | A | G | A | - | - |
| G | Br1940 | B. pilosicoli hipp− | T | A | G | A | A | C | G | A | G | A | - | - |
| A | Br710 | B. pilosicoli hipp− | T | A | G | A | A | C | G | A | G | A | C | - |
| F | Br1620, Br1622 | B. pilosicoli hipp− | T | A | G | A | A | C | G | A | G | A | C | A |
Nucleotide positions are according to the Brachyspira (Serpulina) consensus sequence (Pettersson et al. [27]). Dashes indicate the same nucleotides as in B. pilosicoli P43/6/78 type strain.
NA, not applicable.
In the three almost completely sequenced B. pilosicoli hipp− isolates, the sequence portions beyond the 877-bp partial sequence were identical to the respective sequence of B. pilosicoli P43. The database search revealed the sequence alignment of 98.95 to 99.93% homology between the Finnish isolates and the group IV Brachyspira comprising two porcine, three human, and six canine strains (accession numbers U14928 and -9, AY187057, U23031, U23034, AY349943 to -6, AF245120, and AF24523).
The first eight nucleotide differences from B. pilosicoli P43 were located quite tightly between nucleotide positions 214 and 243, and the next two nucleotide differences in positions 588 and 589. None of the 10 unique nucleotide positions differing from the sequence of B. pilosicoli P43 matched the sequences of the other strains of group IV Brachyspira.
PFGE.
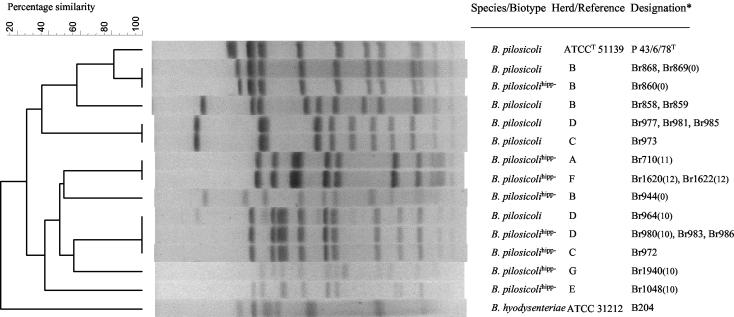
The macrorestriction of bacterial DNA by MluI yielded 9 to 12 bands from B. pilosicoli hipp− isolates and 10 to 12 bands from B. pilosicoli isolates (Fig. 1). The MRPs were fairly diverse, and no clustering based on the hippurate reaction was detected. Two herds had several MRPs; three and two distinctly different MRPs were found in herds B and D, respectively.
FIG. 1.
Dendrogram of PFGE patterns of 11 B. pilosicoli hipp− and 9 B. pilosicoli field isolates, B. pilosicoli P43/6/78 type strain, and B. hyodysenteriae B204 reference strain. ✽, the number of nucleotide differences in the 16S rDNA between field isolates and B. pilosicoli P43/6/78 type strain is in parentheses. B. pilosicolihipp−, B. pilosicoli, hipp−.
B. pilosicoli hipp− isolates were divided into six MRPs, and B. pilosicoli isolates into four MRPs. B. pilosicoli and B. pilosicoli hipp− had a common MRP in herds B and D. In herd B, two B. pilosicoli isolates and one B. pilosicoli hipp− isolate had a common MRP, which was very close to the MRP of B. pilosicoli P43. B. pilosicoli hipp− isolates from herds A and F shared a MRP, as did B. pilosicoli hipp− isolates from herds C and D. A common MRP was also found for B. pilosicoli isolates from herds C and D.
TEM.
The mean diameters of B. pilosicoli hipp− Br980 and Br1048 were 0.28 μm (95% confidence interval [95% CI] = 0.26 to 0.30 μm) and 0.27 μm (95% CI = 0.25 to 0.28 μm), respectively, and those of B. hyodysenteriae B78 and B. pilosicoli P43 were 0.40 μm (95% CI = 0.36 to 0.44 μm) and 0.27 μm (95% CI = 0.25-0.28 μm), respectively. The mean lengths of Br980 and Br1048 were 6.26 μm (95% CI = 6.11-6.41 μm) and 6.53 μm (95% CI = 6.21 to 6.85 μm), respectively, whereas the corresponding figures for B. hyodysenteriae B78 and B. pilosicoli P43 were 9.44 μm (95% CI = 9.00 to 9.88 μm) and 6.83 μm (95% CI = 6.53 to 7.13 μm), respectively. Most Br980, Br1048, and B. pilosicoli P43 cells had pointed ends, and on the ends of a few bacteria cells of Br980, a hazy, lattice-like structure on the surface was present. Six periplasmic flagella could be seen at the end of the Br980 cell. B. pilosicoli P43 had 6 and B. hyodysenteriae B78 had 9 to 10 periplasmic flagella at the end of the cell (not shown).
DISCUSSION
Reports of hippurate-negative B. pilosicoli-like spirochetes to date are few. Thomson et al. (35) isolated hippurate-negative and β-glucosidase-negative, weakly beta-hemolytic spirochetes from diarrheic pigs in the United Kingdom. Further characteristics of these isolates were not described. De Smet et al. (5) studied the partial nucleotide sequences of 16S rDNA from weakly beta-hemolytic human intestinal spirochetes. One of these human strains was hippurate-negative. In 286 bp, this human hippurate-negative isolate had 100% similarity to B. pilosicoli P43.
In the present study, the 16S rDNA sequence of Finnish B. pilosicoli and B. pilosicoli hipp− isolates had a similarity of at least 98.63% to B. pilosicoli P43 in a partial nucleotide sequence of 877 bp. For the entire region of the Brachyspira consensus sequence (1,435 bp), the similarity was upwards of 99.16%. The homology of the 16S rDNA nucleotide sequence between two bacteria species in a genus may range from about 85 to 99% (25, 26). The species in the genus Brachyspira are closely related. The 16S rDNA sequence homology between the species of porcine Brachyspira is at least 98.1% (26, 27, 30). Pettersson et al. (27) found 99.9% similarity between 16S rDNA nucleotide sequences of porcine group IV Brachyspira strains.
In the present study, 16S rDNA nucleotide sequences from seven B. pilosicoli and B. pilosicoli hipp− isolates showed lower similarity to B. pilosicoli P43 than did the Swedish porcine group IV Brachyspira strains studied by Pettersson et al. (27). The similarity between 16S rDNA sequences of two species of the genus Brachyspira can be higher than 99.16% for the Brachyspira consensus sequence of 1,435 bp (27). Thus, the nucleotide similarity percentage of 16S rDNA alone does not prove that the hippurate-negative isolates belong to the species B. pilosicoli. Group IV Brachyspira characteristically has six consecutive uridines in the 16S RNA between nucleotide positions 175 and 182 (27). In the present study, the 1,435-bp sequences from three B. pilosicoli hipp− isolates displayed the typical “TTTTTT” pattern between positions 175 and 182. B. pilosicoli has 18 unique nucleotide positions in the 16S rDNA (27). Six of these in the present study were located in the partially sequenced region. Almost all of the unique nucleotide positions were found in the 16S rDNA of seven Finnish isolates, the only exception being a thymine in position 229, which was replaced by an adenine. The presence of virtually all of the residues in positions characteristic of B. pilosicoli supports the conclusion that hippurate-negative isolates belong to the species B. pilosicoli.
Seven field isolates had ten common nucleotide positions in their 16S rDNA that were different from the sequences of group IV Brachyspira strains deposited in GenBank. Eight of these nucleotides were located within a 295-bp sequence of 16S rDNA, which is known to have one hippurate-positive and one hippurate-negative B. pilosicoli isolate in humans (accession no. Y10314 [5]). However, none of the unique nucleotides seen here were present in the human isolates. The 10 first unique 16S rDNA nucleotide positions were found also in one hippurate-positive B. pilosicoli isolate. However, two B. pilosicoli hipp− isolates had a sequence identical to that of B. pilosicoli P43. In conclusion, the unique nucleotide positions observed in seven Finnish isolates were not associated with hippurate negativity.
The cell diameter of B. pilosicoli is 0.24 to 0.30 μm, and the length is 5.29 to 7.25 μm, whereas the corresponding measurements for B. hyodysenteriae, for example, are 0.33 to 0.37 μm and 7.91 to 11.65 μm (38). The B. pilosicoli cell has 4 to 6 periplasmic flagella, whereas the cells of other species of swine Brachyspira have 7 to 14 periplasmic flagella at each end. The periplasmic flagella overlap in the middle of the bacterium (29, 38). The B. pilosicoli cell also has a unique lattice-like structure at its pointed end, which can be observed by electron microscopy (29). In the present study, the size of B. pilosicoli hipp− was similar to that of B. pilosicoli P43. The lattice-like structure on the end of the cell was observed in one of the two B. pilosicoli hipp− isolates. The number of periplasmic flagella was also similar to that of B. pilosicoli P43. The ultrastructural resemblance between B. pilosicoli hipp− and B. pilosicoli P43 is consistent with the conclusion that the hippurate-negative isolates represent a distinct biotype of B. pilosicoli.
Trott et al. (36) found that PFGE with MluI macrorestriction effectively discriminates between B. pilosicoli strains. Fellström et al. (10) studied rare, indole-negative biotypes of B. hyodysenteriae isolated from Belgium and Germany. All 14 indole-negative B. hyodysenteriae strains showed the same unique PFGE pattern with MluI macrorestriction. In the present study, PFGE with MluI failed to differentiate between hippurate-positive and hippurate-negative B. pilosicoli isolates because of common MRPs. We conclude that PFGE is not suitable for epidemiological study of B. pilosicoli with regard to hippurate negativity.
Thomson et al. (33) observed the pathogenicity of B. pilosicoli to vary between different strains. We cannot draw any conclusions about the pathogenicity of B. pilosicoli hipp− because most of the Finnish pig herds containing these organisms also concomitantly had other microbes with potential pathogenicity. The strong hemolytic capacity of porcine B. hyodysenteriae is related to its virulence (14). The pathogenicity of the biochemically unusual, indole-negative B. hyodysenteriae appeared to be similar to that of indole-positive B. hyodysenteriae (10). The relation of selected biochemical properties of the other porcine Brachyspira species to virulence remains unknown. Based on the close genetic relationship between B. pilosicoli hipp− and the group IV Brachyspira observed here, the potential pathogenicities of porcine B. pilosicoli hipp− and porcine hippurate-positive B. pilosicoli might be expected to be similar. However, the potential pathogenicity of hippurate-negative B. pilosicoli should be verified.
Today, species-specific PCR is widely used as the primary or even the only diagnostic method for detection of B. pilosicoli. However, in small laboratories, classification of Brachyspira may be based solely on biochemical tests and the evaluation of hemolysis intensity. Based on our findings, a weakly beta-hemolytic hippurate-negative and β-glucosidase-negative porcine intestinal spirochete is likely B. pilosicoli. Thus, the glucosidase test should not be overlooked. The evaluation of bacteria size by using a high-quality light microscope with B. hyodysenteriae and B. pilosicoli reference strains on the same slide will aid in the recognition of B. pilosicoli.
Acknowledgments
This study was financially supported by the Finnish Ministry for Agriculture and Forestry and by a personal grant from the Finnish Veterinary Foundation.
REFERENCES
- 1.Atyeo, R. F., S. L. Oxberry, B. G. Combs, and D. J. Hampson. 1998. Development and evaluation of polymerase chain reaction tests as an aid to diagnosis of swine dysentery and intestinal spirochaetosis. Lett. Appl. Microbiol. 26:126-130. [DOI] [PubMed] [Google Scholar]
- 2.Atyeo, R. F., S. L. Oxberry, and D. J. Hampson. 1996. Pulsed-field gel electrophoresis for subspecies differentiation of Serpulina pilosicoli (formerly “Anguillina coli”). FEMS Microbiol. Lett. 141:77-81. [DOI] [PubMed] [Google Scholar]
- 3.Brooke, C. J., A. N. Clair, A. S. J. Mikosza, T. V. Riley, and D. J. Hampson. 2001. Carriage of intestinal spirochaetes by humans: epidemiological data from Western Australia. Epidemiol. Infect. 127:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Smet, K. A. L., D. E. Worth, and S. P. Barrett. 1998. Variation amongst human isolates of Brachyspira (Serpulina) pilosicoli based on biochemical characterization and 16S rRNA gene sequencing. Int. J. Syst. Bacteriol. 48:1257-1263. [DOI] [PubMed] [Google Scholar]
- 6.Duhamel, G. E. 1997. Porcine colonic spirochaetosis associated with Serpulina pilosicoli: experimental reproduction, epidemiology, and control. Proc. Am. Assoc. Swine Pract. 29:487-495. [Google Scholar]
- 7.Fellström, C., B. Pettersson, J. Thomson, A. Gunnarsson, M. Persson, and K.-E. Johansson. 1997. Identification of Serpulina species associated with porcine colitis by biochemical analysis and PCR. J. Clin. Microbiol. 35:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellström, C., B. Pettersson, M. Uhlén, A. Gunnarsson, and K.-E. Johansson. 1995. Phylogeny of Serpulina based on sequence analyses of the 16S rRNA gene and comparison with a scheme involving biochemical classification. Res. Vet. Sci. 59:5-9. [DOI] [PubMed] [Google Scholar]
- 9.Fellström, C., B. Pettersson, U. Zimmerman, A. Gunnarsson, and R. Feinstein. 2001. Classification of Brachyspira spp. isolated from Swedish dogs. Anim. Health Res. Rev. 2:75-82. [PubMed] [Google Scholar]
- 10.Fellström, C., M. Karlsson, B. Pettersson, U. Zimmerman, A. Gunnarsson, and A. Aspan. 1999. Emended descriptions of indole negative and indole positive isolates of Brachyspira (Serpulina) hyodysenteriae. Vet. Microbiol. 70:225-238. [DOI] [PubMed] [Google Scholar]
- 11.Fossi, M., T. Pohjanvirta, and S. Pelkonen. 2003. Molecular epidemiological study of Brachyspira pilosicoli in Finnish sow herds. Epidemiol. Infect. 131:967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinonen, M., M. Fossi, J.-P. Jalli, H. Saloniemi, and V. Tuovinen. 2000. Detectability and prevalence of Brachyspira species in herds rearing health class feeder pigs in Finland. Vet. Rec. 146:343-347. [DOI] [PubMed] [Google Scholar]
- 13.Hovind-Hougen, K., A. Birch-Andersen, R. Henrik-Nielsen, M. Orholm, J. O. Pedersen, P. S. Teglbjærg, and E. H. Thaysen. 1982. Intestinal spirochetosis: morphological characterization and cultivation of the spirochete Brachyspira aalborgi gen. nov., sp. nov. J. Clin. Microbiol. 16:1127-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyatt, D. R., A. A. H. M. ter Huurne, B. A. M. van der Zeijst, and L. A. Joens. 1994. Reduced virulence of Serpulina hyodysenteriae hemolysin-negative mutants in pigs and their potential to protect pigs against challenge with a virulent strain. Infect. Immun. 62:2244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansson, D., C. Bröjer, D. Gavier-Widén, A. Gunnarsson, and C. Fellström. 2001. Brachyspira spp. (Serpulina spp.) in birds: a review and results from a study of Swedish game birds. Anim. Health Res. Rev. 2:93-100. [PubMed] [Google Scholar]
- 16.Kinyon, J. M., and D. L. Harris. 1979. Treponema innocens, a New Species of Intestinal Bacteria, and Emended Description of the type strain of Treponema hyodysenteriae. Int. J. Syst. Bacteriol. 29:102-109. [Google Scholar]
- 17.Kraatz, W., U. Thunberg, B. Pettersson, and C. Fellström. 2001. Human intestinal spirochetosis diagnosed with colonoscopy and analysis of partial 16S rDNA sequences of involved spirochetes. Anim. Health Res. Rev. 2:111-116. [PubMed] [Google Scholar]
- 18.L'Abee-Lund, T. M., R. Heiene, N. F. Friis, P. Ahrens, and H. Sørum. 2003. Mycoplasma canis and urogenital disease in dogs in Norway. Vet. Rec. 153:231-235. [DOI] [PubMed] [Google Scholar]
- 19.Leser, T. D., K. Møller, T. K. Jensen, and S. E. Jorsal. 1997. Specific detection of Serpulina hyodysenteriae and potentially pathogenic weakly β-hemolytic porcine intestinal spirochetes by polymerase chain reaction targeting 23S rDNA. Mol. Cell. Probes 11:363-372. [DOI] [PubMed] [Google Scholar]
- 20.Mikosza, A. S. J., T. La, K. R. Margawani, C. J. Brooke, and D. J. Hampson. 2001. PCR detection of Brachyspira aalborgi and Brachyspira pilosicoli in human faeces. FEMS Microbiol. Lett. 197:167-170. [DOI] [PubMed] [Google Scholar]
- 21.Møller, K., T. K. Jensen, M. Boye, T. D. Leser, and P. Ahrens. 1999. Amplified fragment length polymorphism and pulsed field gel electrophoresis for subspecies differentiation of Serpulina pilosicoli. Anaerobe 5:313-315. [Google Scholar]
- 22.Munshi, M. A., N. M. Taylor, A. S. J. Mikosza, P. B. S. Spencer, and D. J. Hampson. 2003. Detection by PCR and isolation assays of the anaerobic intestinal spirochete Brachyspira aalborgi from the feces of captive nonhuman primates. J. Clin. Microbiol. 41:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oxberry, S. L., and D. J. Hampson. 2003. Colonization of pet shop puppies with Brachyspira pilosicoli. Vet. Microbiol. 93:167-174. [DOI] [PubMed] [Google Scholar]
- 24.Park, N. Y., C. Y. Chung, A. J. McLaren, R. F. Atyeo, and D. J. Hampson. 1995. Polymerase chain reaction for identification of human and porcine spirochaetes recovered from cases of intestinal spirochaetosis. FEMS Microbiol. Lett. 125:225-230. [DOI] [PubMed] [Google Scholar]
- 25.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 26.Paster, B. J., F. E. Dewhirst, W. G. Weisburg, L. A. Tordoff, G. J. Fraser, R. B. Hespell, T. B. Stanton, L. Zablen, L. Mandelco, and C. R. Woese. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersson, B., C. Fellström, A. Andersson, M. Uhlén, A. Gunnarsson, and K.-E. Johansson. 1996. The phylogeny of intestinal porcine spirochetes (Serpulina species) based on sequence analysis of the 16S rRNA gene. J. Bacteriol. 178:4189-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rübsamen, S., and S. Rübsamen. 1986. Hippurat-Hydrolyse: ein Schnelltest zur Unterscheidung von Treponema hyodysenteriae und Treponema innocens. Tierärztl. Umschau. 41:673-677. [Google Scholar]
- 29.Sellwood, R., and A. P. Bland. 1997. Ultrastructure of intestinal spirochaetes, p. 109-149. In D. J. Hampson and T. B. Stanton (ed.), Intestinal spirochaetes in domestic animals and humans. CAB International, Oxon, United Kingdom.
- 30.Stanton, T. B., N. S. Jensen, T. A. Casey, L. A. Tordoff, F. E. Dewhirst, and B. J. Paster. 1991. Reclassification of Treponema hyodysenteriae and Treponema innocens in a new genus, Serpula gen. nov., as Serpula hyodysenteriae comb. nov. and Serpula innocens comb. nov. Int. J. Syst. Bacteriol. 41:50-58. [DOI] [PubMed] [Google Scholar]
- 31.Stephens, C. P., and D. J. Hampson. 2001. Intestinal spirochete infections of chickens: a review of disease associations, epidemiology, and control. Anim. Health Res. Rev. 2:83-91. [PubMed] [Google Scholar]
- 32.Taylor, D. J., J. R. Simmons, and H. M. Laird. 1980. Production of diarrhoea and dysentery in pigs by feeding pure cultures of a spirochaete differing from Treponema hyodysenteriae. Vet. Rec. 106:326-332. [DOI] [PubMed] [Google Scholar]
- 33.Thomson, J. R., W. J. Smith, B. P. Murray, and S. McOrist. 1997. Pathogenicity of three strains of Serpulina pilosicoli in pigs with a naturally acquired intestinal flora. Infect. Immun. 65:3693-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson, J. R., W. J. Smith, and B. P. Murray. 1998. Investigations into field cases of porcine colitis with particular reference to infection with Serpulina pilosicoli. Vet. Rec. 142:235-239. [DOI] [PubMed] [Google Scholar]
- 35.Thomson, J. R., W. J. Smith, B. P. Murray, D. Murray, J. E. Dick, and K. J. Sumption. 2001. Porcine enteric spirochete infections in the UK: surveillance data and preliminary investigations of atypical isolates. Anim. Health Res. Rev. 2:31-36. [PubMed] [Google Scholar]
- 36.Trott, D. J., A. S. J. Mikosza, B. G. Combs, S. L. Oxberry, and D. J. Hampson. 1998. Population genetic analysis of Serpulina pilosicoli and its molecular epidemiology in villages in the Eastern Highlands of Papua New Guinea. Int. J. Syst. Bacteriol. 48:659-668. [DOI] [PubMed] [Google Scholar]
- 37.Trott, D. J., T. B. Stanton, N. S. Jensen, and D. J. Hampson. 1996. Phenotypic characteristics of Serpulina pilosicoli, the agent of intestinal spirochaetosis. FEMS Microbiol. Lett. 142:209-214. [DOI] [PubMed] [Google Scholar]
- 38.Trott, D. J., T. B. Stanton, N. S. Jensen, G. E. Duhamel, and D. J. Hampson. 1996. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int. J. Syst. Bacteriol. 46:206-215. [DOI] [PubMed] [Google Scholar]
- 39.Utriainen, M., K. Jalava, A. Sukura, and M.-L. Hänninen. 1997. Morphological diversity of cultured canine gastric Helicobacter spp. Comp. Immun. Microbiol. Infect. Dis. 20:285-297. [DOI] [PubMed] [Google Scholar]