Abstract
The present study, using robotized DNA isolation and quantitative PCR based on the Neisseria meningitidis-specific capsular transport A gene, demonstrates the ease, rapidity, specificity, and sensitivity of quantifying neisserial DNA in plasma (n = 65) and cerebrospinal fluid (CSF) (n = 12) from patients with systemic meningococcal disease. We found a close correlation between the levels of neisserial DNA and lipopolysaccharides in plasma (r = 0.905) and in CSF (r = 0.964). The median concentration of neisserial DNA in plasma in 23 patients with persistent shock was 2 × 107 copies/ml, versus <103 copies/ml in 42 nonshock patients. Furthermore, quantitative PCR made possible estimates of the total number of meningococci in plasma, as opposed to conventional blood cultures, suggesting about 1,000 dead meningococci for every viable bacterium. Finally, with logistic regression analyses, neisserial DNA may predict a patient's disease severity and outcome at hospital admission. The number of meningococci in plasma and CSF appears to be the main determinant of the lipopolysaccharide levels, clinical presentation, and outcome.
Studies of consecutively hospitalized patients with systemic meningococcal disease have documented a close association between the clinical presentation and their lipopolysaccharide (LPS) levels in plasma and cerebrospinal fluid (CSF) (8, 9). The LPS levels in plasma may be 5 logs higher in patients with fulminant meningococcal septicemia, characterized by circulatory collapse, than in patients with clinically distinct meningitis or a mild meningococcemia without disturbed circulation (8). In patients with distinct symptoms of meningitis, the levels of LPS in CSF are 3 to 5 logs higher than found in patients with fulminant meningococcal septicemia or mild meningococcemia (9). Neisseria meningitidis is known to produce large quantities of outer membrane vesicles, evaginations of the bacterial outer membrane which are subsequently shed during log-phase growth (13). Outer membrane vesicles are potent inducers of proinflammatory cytokines, activate complement, and are released during fulminant meningococcal septicemia (2, 4, 7, 24). It has been suggested that certain strains of N. meningitidis produce more outer membrane vesicles than other strains (1) and that this could explain the differences in clinical presentation.
Patients with the highest levels of LPS in plasma, i.e., patients with rapidly deteriorating circulation, have a shorter time span between recognition of the first disease symptoms and admission to hospital than patients with distinct meningitis (33). This observation favors the hypothesis that the host's ability to contain the growth of N. meningitidis within the circulation is the major determinant of the clinical presentation. Massive bacterial growth within 12 to 24 h generates high levels of LPS in plasma. LPSs are the major inflammation-inducing substances produced by meningococci, although non-LPS components may also contribute to the inflammatory response of the host (3, 5, 6, 22). Using quantitative PCR, Hacket et al. (18) recently showed that the number of N. meningitidis in plasma was positively associated with a clinical severity score and could be as high as 108 meningococci per ml of blood in the most severe cases of septicemia. These observations support the idea that invasive strains of N. meningitidis vary markedly in their ability to proliferate in the blood of nonimmune persons.
The aim of the present study has been to establish a robotized real-time PCR method to quantify N. meningitidis DNA in biological fluids and to compare the relationship between neisserial DNA concentrations and the corresponding levels of bioactive meningococcal LPS in plasma and CSF as quantified by the chromogenic Limulus amoebocyte lysate assay (8, 10). A close correlation between the number of neisserial DNA copies and LPS levels favors the hypothesis that bacterial growth is the primary determinant of the LPS level. We also studied the relationship between the number of viable meningococci, as determined by quantitative blood culture, and the number of neisserial DNA copies in plasma in a subgroup of patients in order to estimate the true bacterial load in these patients. Finally, we wanted to evaluate to what extent the amount of neisserial DNA quantified at admission to the hospital predicts severity and outcome for the individual patient.
MATERIALS AND METHODS
Patients.
A total of 65 patients admitted to Ullevål University Hospital from 1985 to 2003 with systemic meningococcal disease verified bacteriologically, serologically, or by PCR were studied. N. meningitidis was isolated from cultures of blood (41 positive of 65 patients), CSF (15 positive), or nasopharynx/tonsillopharynx (3 positive). The last three patients had a typical clinical picture of fulminant meningococcal septicemia (two patients) or mild meningococcemia (one patient) with fever and hemorrhagic skin lesions. Three patients revealed a significant antibody response to serogroup B meningococcal surface antigens, and three patients had a positive qualitative PCR in CSF or plasma (11, 19). With a previously developed clinical classification system which closely reflects the underlying pathophysiology (6, 8, 9, 14, 33), the patients were assigned to one of the four clinical categories defined as follows: fulminant septicemia, persistent severe septic shock and minimal CSF pleocytosis, i.e., <108 leukocytes/liter of CSF (n = 21; 9 of these patients [43%] died as a consequence of circulatory collapse and multiple organ failure); septicemia and meningitis, ≥108 leukocytes/liter of CSF combined with severe septic shock (n = 2; none died); mild systemic meningococcal disease, patients with a documented systemic N. meningitidis infection but without persistent septic shock or meningitis (n = 14; none died); and distinct meningitis, >108 leukocytes/liter of CSF and the absence of severe septic shock (n = 28; 1 died due to herniation).
The patients' samples were analyzed after informed consent was obtained in accordance with the guidelines of the Regional Medical Ethics Committee of Health Region I in Norway.
Materials.
Blood (n = 65) and CSF (n = 12 for neisserial DNA and 11 for LPS) samples were collected from patients at hospital admission. Sixty-two of the patients had samples drawn before initiation of antibiotic therapy, one patient had samples drawn 10 h after initiation, and two patients had samples drawn 12 h after because they were transferred from other hospitals. In a time course study, neisserial DNA and LPS concentrations were measured in plasma samples collected at irregular intervals from eight of the patients with fulminant meningococcal septicemia. Whole blood (±15 IU of heparin/ml of blood, LPS-free; BD VacutainerSystems, Plymouth, United Kingdom) was centrifuged (1,400 × g, room temperature, 10 min). Plasma, serum, and CSF were aliquoted, with LPS-free equipment, to cryotubes (Nunc A/S, Roskilde, Denmark) and stored at −70°C.
DNA isolation.
DNA from plasma, serum, and CSF was isolated with robotized equipment (BioRobot 48; Qiagen Inc., Valencia, Calif.) containing several safety features to eliminate contamination risks. DNA was isolated based on adsorbance to magnetic silica particles (MagAttract DNA Blood M96 kit 951436; Qiagen), according to the manufacturer's instructions, eluted into 75 μl of H2O, and stored at 4°C until analyzed. Overall robot isolation time for six samples was 15 min.
Real-time PCR.
Quantification of DNA was performed with real-time PCR and SYBR Green fluorescence detection (LightCycler; Roche Diagnostics GmbH, Mannheim, Germany). Optimized, specific PCR primers (5′-GTA-GGT-GGT-TCA-ACG-GCA-AA-3′ and 5′-TCG-CGG-ATT-TGC-AAC-TAA-AT-3′; designed with the primer design program Primer 3 [http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi] by using a sequence from accession number AF315863, strain M4440 [generating a PCR product from nucleotide positions 466 to 567]) for the N. meningitidis capsular transport gene (ctrA) (17) were used for PCR amplification (total volume, 20 μl), including 5 μl of DNA solution, 4 mM MgCl2, 0.5 mM primers, and 2 μl of LC-FastStart DNA Master SYBR Green I. The following cycle conditions were used: 10 min of denaturation at 95°C and 35 amplification cycles at 95°C for 1 s, 60°C for 10 s, and 72°C for 10 s. To ensure specificity, melting curve analyses were performed at 95°C for 0 s, 65°C for 15 s, and 95°C for 0 s. The total amplification time for 32 samples was 45 min.
Specificity of the PCR primers.
PCR primer sequences were tested for homology with other sequences at the NCBI gene BLAST website (http://www.ncbi.nlm.nih.gov/BLAST). The PCR product was sequenced (Beckman Coulter CEQ-2000) (DNA Sequencing Core Facility, Medical Faculty, University of Oslo, Oslo, Norway) and found identical with the genome of N. meningitidis with the NCBI gene BLAST. DNA isolated from humans and bacteria (Streptococcus pneumoniae, Haemophilus influenzae, and serogroup B streptococci; kindly provided by the Department of Microbiology, Ullevål University Hospital) were experimentally tested for specificity in the PCR assay and found not to cross-react.
Determination of DNA content in N. meningitidis preparations used to generate standard curves.
N. meningitidis strain 44/76-SL (B:15:PI.7,16,L3,7,9) (27), stored at −70°C in Greaves' medium, was grown overnight on Columbia horse blood agar plates in 5% CO2 in air at 35°C and then inoculated to a confluent, thin layer on brain heart infusion agar plates (27). These were likewise incubated and then harvested in Hanks' balanced salt solution with 0.1% (wt/vol) bovine serum albumin (HBSS-BSA) after 2, 4, 8, 18, and 24 h. The number of CFU in the suspensions was determined by plating 10-fold HBSS-BSA dilutions in triplicate. The bacteria were heated for 30 min at 56°C and stored at 4°C in HBSS-BSA. These meningococcal suspensions underwent robotized DNA isolation (see above) and DNA quantification by UV spectrophotometry. The number of meningococci in each suspension was calculated from the measured DNA concentrations, setting the meningococcal genome size to 2,272,351 bp (31).
As a meningococcal culture will always include a certain proportion of dead cells, we compared the CFU counted after different lengths of incubation. In suspensions with an optical density of 0.25, we found the number of CFU at 4 h to be about 10 times higher than that at 8 and 18 h. After 24 h, the number of CFU was only about 50% of the values at 8 and 18 h, indicating that the highest proportion of viable cells was present at 4 h. This procedure with growth for 4 h is also the way to secure log-phase growth for use in the serum bactericidal assay for antimeningococcal antibodies (27). In the suspension harvested at 4 h, the calculated number of bacteria and the number of observed CFU were equivalent (1010 CFU/ml, considering that one meningococcal CFU is at least two cells). Consequently, the 4-h suspension was chosen as the stock solution for our standard curves. As each bacterium contains only one capsular transport A gene, the DNA load used further on in the project equates the bacterial cell load.
Standard curves.
Ten microliters of heat-killed N. meningitidis (strain 44/76-SL, stock solution) was added to 190 μl of either plasma, serum, or CSF, and 10-fold dilutions were generated; 100 μl of each dilution was subjected to robotic DNA isolation, and 5 μl of standard curve DNA solutions then underwent PCR quantification. The amount of DNA in each standard curve solution was applied to the software of the LightCycler, and a standard curve was generated after each run (26). Samples containing small quantities of neisserial DNA (that could not be extrapolated from the standard curves, i.e., <10 standard deviation increase in fluorescence) were all set to zero to exclude false-positive samples. All positive samples had a melting point peak at 83.2°C (range, 82.8 to 84.2°C) determined by a melting curve analysis consistent with the specific PCR fragment amplified.
PCR efficiencies.
PCR efficiencies for the standard curves were determined with the calculated slope from the LightCycler software. PCR efficiencies in patient samples (plasma or CSF samples) were determined from PCR amplification of twofold dilutions (n = 4), plotting the recorded crossing point against the log of the initial concentrations of each dilution and calculating the slope (26). Finally, PCR efficiencies were calculated; E = 10−1/slope.
Controls.
The method was evaluated by running a negative control (heparinized plasma or CSF sample from a non-systemic meningococcal disease patient) and a positive control (heparinized plasma from a patient with fulminant meningococcal septicemia diluted 1:100 in normal, heparinized plasma) following the samples in both the DNA isolation and the PCR quantification steps.
Sensitivity of the method.
Known amounts of heat-inactivated N. meningitidis (108 to 102/ml) or neisserial DNA (43.5 μg/ml to 43.5 pg/ml) (kindly provided by Dominique Caugant, Division of Infectious Disease Control, Norwegian Institute of Public Health, Oslo, Norway) were added to plasma (heparinized or EDTA), serum, or CSF. Subsequently, DNA was isolated from a 100-μl specimen, and the neisserial DNA was quantified.
LPS quantification.
LPS quantification was performed by the chromogenic Limulus amoebocyte lysate assay, as previously described (8, 10).
Blood and CSF cultures.
Blood was collected on plain or hypertonic broths, as previously detailed (8).
Quantitative blood culture.
Blood collected from 10 of the 65 patients had previously been cultured quantitatively, as described earlier (8).
Statistical analyses.
Median and range were used for descriptive statistics. When comparing the different patient groups, the Mann-Whitney test was applied. Since the concentrations of both LPS and neisserial DNA are exponential in nature, logarithmic transformation with the 10-logarithm as the base was performed. These variables showed no significant deviation from a normal distribution. The transformed variables (called Lg-LPS and Lg-neisserial DNA) were used when calculating Pearson's correlation coefficient and when applying logistic regression analysis to investigate to what extent LPS and neisserial DNA would be useful in predicting survival of patients with systemic meningococcal disease. The computer program Statistical Package for Social Sciences (SPSS version 11.0) was used for all statistical calculations, and P < 0.05 was considered statistically significant.
RESULTS
Evaluation of the neisserial DNA quantification methodology.
Standard curves were generated by diluting N. meningitidis in the same milieu as the samples to be quantified, DNA was isolated, and quantitative PCR was performed. Diluting the N. meningitidis standards in normal plasma or CSF prior to DNA isolation gave similar results irrespective of donor. N. meningitidis diluted in serum from different donors revealed variable yields, indicating serum to be less suitable as a diluent. Standard curve performances were evaluated based on data collected from plasma (n = 11) and CSF (n = 2) samples over a period of 6 months. Between-day variations in plasma standard curves, calculated from the y intercepts (mean, 35.25) and slopes (mean, −3.396), were 1.9 and 4.3%, respectively. Mean and standard error of the mean on each standard's crossing points were calculated to show the reproducibility (Table 1). The standard curves showed linearity over 5 orders of magnitude, indicating constant PCR efficiency over the concentration range studied.
TABLE 1.
Reproducibility of standard curvesa
| No. of N. meningitidis organisms (copies/ml of plasma) | Mean cycle no. (SEM)
|
|
|---|---|---|
| CSF | Plasma | |
| 108 | 16.43 (0.15) | 16.97 (0.13) |
| 107 | 19.88 (0.15) | 20.38 (0.09) |
| 106 | 23.39 (0.04) | 23.72 (0.10) |
| 105 | 26.68 (0.02) | 27.14 (0.12) |
| 104 | 30.20 (0.08) | 30.15 (0.20) |
| 103 | 33.70 (0.63) | |
Standard curves were generated by isolating neisserial DNA from plasma (n = 11) or CSF (n = 2), and quantitative PCR was performed on different days.
The sensitivity of the method allowed detection of 103 neisserial DNA copies per ml of plasma with the second derivative maximum method. The positive control showed a within-run variation coefficient of variation of 6% (n = 6), whereas the between-run coefficient of variation was 22% (n = 15). The PCR amplification efficiencies of the neisserial DNA in the standards and in patient samples should be similar in order to use the standard curves for conversion of the crossing point values into concentrations. The mean PCR efficiencies were calculated to 1.95 (range, 1.89 to 2.03; n = 10) and 1.93 (range, 1.91 to 2.06; n = 3) for standards in plasma and in CSF, respectively. The PCR efficiencies in patient plasma samples were calculated to be 1.93 (range, 1.81 to 2.06; n = 4) and 2.05 (2.03 to 2.07; n = 2) in patient CSF samples.
Quantification of neisserial DNA in patients with systemic meningococcal disease. (i) Neisserial DNA in plasma samples.
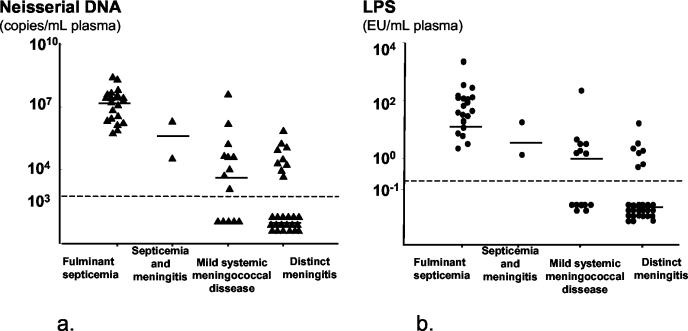
Neisserial DNA was detected in 41 of the 65 plasma samples (63%) from patients with verified systemic meningococcal disease (Fig. 1). The median levels and ranges for each clinical category are given in Table 2. Patients without detectable neisserial DNA were all clinically categorized as having mild systemic meningococcal disease (n = 5) or distinct meningitis (n = 19). Twenty-three patients with persistent septic shock had a median neisserial DNA level of 2 × 107 versus <103 for 42 patients without persistent shock (P < 0.001). Patients with fulminant septicemia had significantly higher levels of neisserial DNA in their plasma samples than patients with distinct meningitis (P < 0.001) and patients with mild systemic meningococcal disease (P < 0.001). The levels of neisserial DNA for nine nonsurvivors with fulminant septicemia (median, 4.6 × 107; range, 1.4 × 105 to 5.4 × 108 neisserial DNA copies/ml) were significantly higher (P = 0.029) than the levels for 12 survivors (median, 1 × 107; range, 5.7 × 105 to 2.9 × 107 neisserial DNA copies/ml).
FIG. 1.
Levels of neisserial DNA (a) and LPS (b) in plasma samples from patients with systemic meningococcal disease presenting with various clinical syndromes. The dotted lines indicate the detection limits, 103 neisserial DNA copies/ml of plasma and 0.25 IU of LPS/ml of plasma. Horizontal bars denote median values.
TABLE 2.
Neisserial DNA and LPS in samples from patients with systemic meningococcal disease presenting with various clinical syndromes
| Clinical presentation (no. of patients) | DNA (copies/ml)
|
LPS (IU/ml)
|
||
|---|---|---|---|---|
| Median | Range | Median | Range | |
| Fulminant septicemia (21) | 2.0 × 107 | 5.7 × 105-5.4 × 108 | 43.0 | 2.1-2,150 |
| Septicemia and meningitis (2) | 1.1 × 107 | 3.5 × 104-2.1 × 106 | 9.4 | 1.3-17.5 |
| Mild systemic meningococcal disease (14) | 7.7 × 103 | <103-4.2 × 107 | <0.5 | <0.5-251 |
| Distinct meningitis (28) | <103 | <103-7.4 × 105 | <0.5 | <0.5-16 |
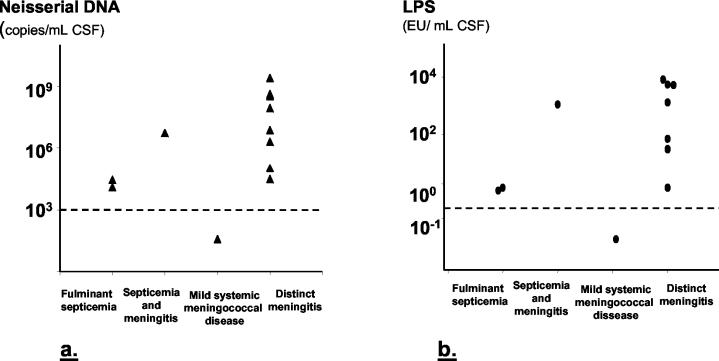
(ii) Neisserial DNA in CSF samples.
Neisserial DNA was detected in 11 of 12 CSF samples (92%) from patients with verified systemic meningococcal disease (Fig. 1a).
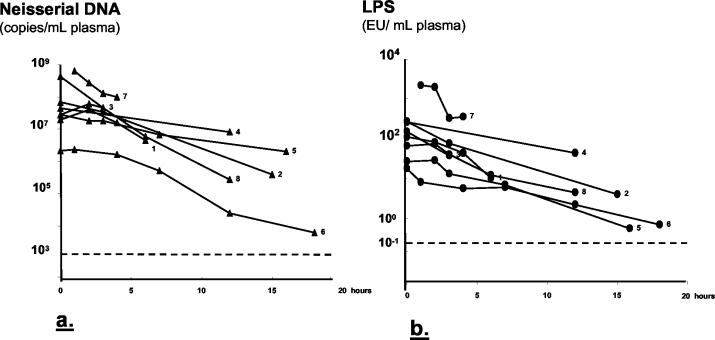
Half-life of neisserial DNA.
Plasma samples serially collected from eight patients with fulminant septicemia (three of whom died) from initiation of antibiotic treatment showed decreasing levels of neisserial DNA. The neisserial DNA half-life ranged from 3 to 4 h. In two patients (2 and 3), the neisserial DNA level increased from the first to the second plasma sample collected after admission and then declined (Fig. 3a).
FIG. 3.
Clearance of neisserial DNA (a) and LPS (b) in plasma samples collected after admission to the hospital from eight patients with fulminant meningococcal septicemia. The first sample (hour 0) was collected immediately before initiation of antibiotic therapy from seven of eight patients. The first sample from patient 7 was collected 1 h after the initiation of antibiotic therapy. Patients 2, 3, and 7 died. The half-life of neisserial DNA (a) was 3 to 4 h and that of LPS (b) was 1 to 3 h. The dotted lines indicate the detection limits, 103 neisserial DNA copies/ml of plasma and 0.25 IU of LPS/ml of plasma.
Quantification of LPS in patients with systemic meningococcal disease. (i) LPS in plasma samples.
LPS quantified by the Limulus amoebocyte lysate assay in the same plasma samples as neisserial DNA were examined resulted in 36 positive samples out of 65 (55%) (Fig. 1b). The median levels and ranges for each clinical category are given in Table 2. Patients with undetectable LPS had a mild systemic meningococcal disease (n = 7) or distinct meningitis (n = 22). Patients with fulminant septicemia had significantly higher levels of LPS in plasma than patients with meningitis (P < 0.001) and patients with mild systemic meningococcal disease (P < 0.001). Levels of LPS in plasma samples from patients with fulminant septicemia (n = 8) collected at admission to hospital and onwards in the course of disease showed decreasing amounts of LPS after initiation of antibiotic therapy. The LPS half-time ranged from 1 to 3 h (Fig. 3b).
(ii) LPS in CSF samples.
LPS was detected in 9 of the 11 CSF samples (82%) (Fig. 2b).
FIG. 2.
Levels of neisserial DNA (a) and LPS (b) in CSF samples from patients with systemic meningococcal disease presenting with various clinical syndromes. The dotted lines indicate the detection limits, 103 neisserial DNA copies/ml of CSF and 0.25 IU of LPS/ml of CSF.
Quantitative blood culture.
The number of live meningococci, i.e., CFU per milliliter of blood, and the number of neisserial DNA copies per milliliter of plasma are given in Table 3 for 10 adult patients with positive blood cultures (8). For every live meningococcus found in the blood cultures, approximately 103 to 104 nonviable meningococci were present, as reflected by the amount of neisserial DNA.
TABLE 3.
Number of N. meningitis organisms from quantitative blood cultures and levels of LPS and neisserial DNA in plasma obtained upon admission to hospital for 10 patients with systemic meningococcal disease
| Patient no. | LPS (IU/ml) | No. of positive blood cultures/totala | Quantitative blood culture (CFU/ml) | DNA (copies/ml of plasma) |
|---|---|---|---|---|
| 1 | 38 | ND | 8.0 × 102 | 1.3 × 107 |
| 2 | 7.5 | 3/9 | 5.0 × 102 | 7.4 × 106 |
| 3 | <0.5 | 7/7 | 2.4 × 102 | 1.8 × 105 |
| 4 | 1.5 | 6/6 | NGb | 4.0 × 104 |
| 5 | 1.1 | 4/4 | NG | 1.0 × 104 |
| 6 | <0.5 | 3/4 | NG | 2.0 × 104 |
| 7 | <0.5 | 1/6 | NG | 1.1 × 104 |
| 8 | <0.5 | 2/6 | NG | 4.6 × 103 |
| 9 | <0.5 | 3/4 | NG | <1 × 103 |
| 10 | <0.5 | 1/6 | NG | <1 × 103 |
3/9 denotes three vials each containing 5 ml of blood grew N. meningitidis out of nine vials collected. ND, not determined.
NG, no growth.
Relation between neisserial DNA, LPS, and survival in patients with systemic meningococcal disease.
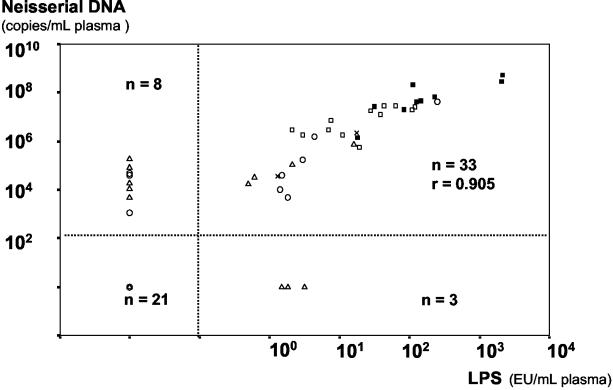
There was a high correlation between Lg-neisserial DNA and Lg-LPS in patients with detectable levels of these substances at admission to hospital (Pearson r = 0.906, P < 0.001, n = 33) (Fig. 4). Plasma samples collected from eight patients after initiation of antibiotic therapy in the time course of disease also showed a high correlation between Lg-neisserial DNA and Lg-LPS (Pearsons r = 0.868, P < 0.001, n = 31), as did Lg-neisserial DNA and Lg-LPS in CSF drawn at admission to hospital (r = 0.975, P < 0.001, n = 9).
FIG. 4.
Relationship between neisserial DNA and LPS in plasma samples from patients with systemic meningococcal disease at admission to hospital. The dotted lines separate samples as either positive or negative for neisserial DNA or LPS. ▪, fulminant septicemia, fatal cases (n = 9); ▪, fulminant septicemia, survivors (n = 12); ×, septicemia and meningitis (n = 2); ○, mild systemic meningococcal disease (n = 14); ▵, distinct meningitis (n = 28).
Relationship between neisserial DNA, LPS, and death from septic shock.
None of the patients with undetectable levels of neisserial DNA or LPS in plasma had a fatal outcome owing to circulatory collapse and multiple organ failure (Fig. 4). Only patients with fulminant septicemia died as a consequence of circulatory collapse. Logistic regression with survival as the dependent variable was performed on patients with positive neisserial DNA and LPS findings (n = 33). Two univariate analyses with either Lg-neisserial DNA or Lg-LPS as the explanatory variable were carried out. Both variables were significant predictors of survival, correctly classifying the outcome for 29 (Lg-neisserial DNA) and 27 (Lg-LPS) of the 33 patients, respectively. Log likelihood values of the models (low values indicating good fit) were practically identical (Table 4). In Table 5, the observed risk of death at different levels of neisserial DNA is compared to predicted risk from the model.
TABLE 4.
Model parameters found when performing logistic regression with dead and alive as dependent variables and either Lg-LPS or Lg-neisserial DNA as the explanatory variable
| Dependent variable | Explanatory variable | Beta | Constant | Significance of beta | 2*log likelihood |
|---|---|---|---|---|---|
| Dead/alive | Lg-LPS | −2.58 | 5.1 | 0.010 | 22.8 |
| Dead/alive | Lg-DNA | −2.29 | 17.2 | 0.017 | 22.7 |
TABLE 5.
Comparison of observed and predicted risk of death at different levels of Lg-neisserial DNA
| Lg-DNA range (copies/ml) | Observed risk of death (%) | Lg-DNA levela (copies/ml) | Predicted risk of death (%) |
|---|---|---|---|
| 2-4 | 0 | 3 | 0 |
| 4-6 | 0 | 5 | 0 |
| 6-8 | 30 | 7 | 24 |
| 8-10 | 100 | 9 | 97 |
Mean of corresponding Lg-DNA range.
DISCUSSION
The main findings in the present study were the ease, rapidity, and sensitivity of quantifying neisserial DNA in plasma and CSF from patients with systemic meningococcal disease. This was accomplished by robotized DNA isolation coupled to quantitative PCR. The levels of neisserial DNA correlated closely with the levels of LPS as determined by the chromogenic Limulus amoebocyte lysate assay. Furthermore, by using logistic regression analyses, we could show that the amount of neisserial DNA in patient specimens was related directly to disease outcome.
Before the development of PCR techniques, microbiological diagnosis of meningococcal infections relied on the cultivation or visualization of N. meningitidis from usually sterile body sites (blood, CSF, or petechiae), detection of capsular polysaccharide in CSF, or significant antibody responses several weeks after the acute infection (15, 19, 25, 29). Quantitative blood culture of meningococci is a cumbersome technique, and few such studies have been published over the years (8, 21, 30, 35). In countries where the health authorities advocate preadmission antibiotic treatment of systemic meningococcal disease, the number of positive blood cultures at hospital admission may now be below 5% (18, 28, 32).
Adding PCR to the diagnostic tools was a major leap forward in diagnosing systemic meningococcal disease (12, 16, 17, 20, 28, 32). In the present study, based on the amplification of the capsular transport A gene, we have demonstrated that robotized DNA extraction followed by quantitative PCR is a feasible, rapid, and sensitive diagnostic tool in specifically demonstrating neisserial DNA in patient plasma and CSF. To obtain reliable results, however, several aspects of the procedure should be stressed. DNA extraction from a complex biological medium such as plasma (or serum) should be as complete as possible. The design and efficiency of PCR primers should address both specificity and sensitivity in the PCR amplification. Several potentially interfering substances, such as components of body fluids and reagents encountered in clinical settings, may occur (34).
In the present study, plasma from different persons (anticoagulated with EDTA or heparin) and CSF spiked with whole N. meningitidis compared well with similar amounts of N. meningitidis isolated from H2O (data not shown). This implies adequate recovery and minimal interference by inhibitory substances in plasma or CSF with the DNA isolation method used. It should be pointed out, however, that serum spiked with N. meningitidis resulted in lower yields than DNA diluted in H2O or plasma. This indicates interference in one of the steps involved, resulting in reduced PCR product formation. PCR efficiencies, on the other hand, showed no significant differences between plasma and serum, indicating that the PCR method operated similarly, indicating that the amount of DNA isolated from serum was lower than that from plasma. Based on our present findings, we advocate the use of plasma for neisserial DNA measurements.
Quantification of DNA in patient samples with real-time PCR and standard curves assumes that the PCR efficiencies for standards and targets are equal (23). To ensure equal PCR efficiencies in our assay, the standard curves were performed under the same conditions as the unknown samples, i.e., plasma or CSF. Under such conditions, our mean PCR efficiencies for standards and patient samples were fairly equal as well as close to the optimal PCR efficiency, E = 2. This indicates a DNA isolation method giving equal recoveries from the two sources and a minimal amount of contaminating substances that could inhibit the PCR.
By applying our method to patient samples, we could document a close association between the levels of neisserial DNA and clinical disease manifestations as well as outcome. Patients who developed a severe, persistent septic shock are characterized by high levels of LPS in plasma and similarly high levels of proinflammatory cytokines, complement activation products, and other inflammatory mediators (6, 8, 33). In the present study, we have shown that this group of patients also had high levels of neisserial DNA in their plasma (Fig. 1a). Furthermore, the neisserial DNA levels were significantly higher among nine patients with fatal septic shock than among 12 patients who survived shock (Fig. 4). On the other hand, patients defined clinically as having either mild systemic meningococcal disease or distinct meningitis have comparatively low levels of LPS (<7 IU/ml) and inflammatory mediators in plasma (6, 8, 33). The present study documents that the plasma levels of neisserial DNA among these patients are several orders of magnitude lower than found in patients with persistent shock. Conversely, patients with symptoms of meningitis had significantly higher levels of neisserial DNA in CSF than patients without meningitis (Fig. 2a).
Measurements of neisserial DNA in serially collected plasma samples from individual patients after the start of antibiotic therapy show that bacterial DNA was cleared with a half-life calculated to be 3 to 4 h. This is slightly longer than the half-life of meningococcal LPS, which was found previously and in this study to be 1 to 3 h (8). Despite the difference in half-lives, the levels of neisserial DNA and LPS correlated closely in samples collected after initiation of antibiotic treatment. In two patients the levels of neisserial DNA increased from the first to the second sample and declined thereafter (Fig. 3a). The levels of neisserial DNA decreased from the first to the second sample in the six other patients studied longitudinally.
One interesting aspect of this study is the relationship between the number of live meningococci, as determined by quantitative blood cultures (8), and the number of neisserial DNA copies, as detected by quantitative PCR (Table 3). The difference between dead and live meningococci appears to be three to four orders of magnitude. This observation is in accordance with our LPS measurements. In a previous publication (8), we speculated whether the number of live meningococci detected by quantitative blood culture could only account for a fraction of the total LPS activity detected in the plasma samples. Our present neisserial DNA results support this conclusion. In several patients, the blood cultures were negative, and yet the patients had detectable LPS by the Limulus amoebocyte lysate assay, and the presence of meningococci was verified by PCR in this study. So far, the Limulus amoebocyte lysate assay has performed remarkably well in giving an estimate of the quantity of circulating N. meningitidis. This has now been verified by measuring the real bacterial load in these patients. However, the Limulus amoebocyte lysate assay cannot specify the source of LPS. By combining the two methods, one can obtain a specific, quantitative, and reliable estimate of meningococcal load in plasma and CSF. It should be noted that samples from several patients were culture and antigen negative but contained high levels of both LPS and neisserial DNA.
Logistic regression analyses with survival as the dependent variable showed that both neisserial DNA and LPS were significant predictors of disease outcome. We found no significant difference between these methods in predicting outcome. Since neisserial DNA probably will be more easily available in routine diagnostics as an emergency test, we chose to concentrate on neisserial DNA as a predictor variable. Our results indicate that neisserial DNA may be used to predict outcome and possibly also to decide to whom experimental treatment should be given. However, the conclusions should be taken with caution, since the number of patients included in the model was relatively small (n = 33).
In conclusion, robotized quantitative PCR of meningococcal DNA extracted from plasma and CSF with primers for the capsular transport A gene is an important step forward in diagnosing systemic meningococcal disease and elucidating the pathophysiology of the disease. The number of meningococci in plasma and CSF appears to be the main determinant of the patient's clinical presentation and chances of survival. Neisserial DNA levels correlate remarkably well with the Limulus amoebocyte lysate measurements and confirm that massive bacterial proliferation takes place in plasma in patients who develop fulminant meningococcal septicemia and in CSF in patients with clinically distinct meningitis. In the future, the estimation of neisserial DNA levels may become as important a predictor of clinical course and outcome as LPS measurements have been for the last 15 years and take its place as a standard procedure in microbiological laboratories, providing a reliable answer within 3 h.
REFERENCES
- 1.Andersen, B. M., O. Solberg, K. Bryn, L. O. Froholm, P. Gaustad, E. A. Hoiby, B. E. Kristiansen, and K. Bovre. 1987. Endotoxin liberation from Neisseria meningitidis isolated from carriers and clinical cases. Scand. J. Infect. Dis. 19:409-419. [DOI] [PubMed] [Google Scholar]
- 2.Bjerre, A., B. Brusletto, T. E. Mollnes, E. Fritzsøonn, E. Rosenqvist, E. Wedege, E. Namork, P. Kierulf, and P. Brandtzaeg. 2002. Complement activation induced by purified Neisseria meningitidis lipopolysaccharides (LPS), outer membrane vesicles, whole bacteria, and an LPS-free mutant. J. Infect. Dis. 185:220-228. [DOI] [PubMed] [Google Scholar]
- 3.Bjerre, A., B. Brusletto, R. Ovstebo, G. B. Joo, P. Kierulf, and P. Brandtzaeg. 2003. Identification of meningococcal LPS as a major monocyte activator in IL-10 depleted shock plasmas and CSF by blocking the CD14-TLR4 receptor complex. J. Endotoxin Res. 9:155-163. [DOI] [PubMed] [Google Scholar]
- 4.Bjerre, A., B. Brusletto, E. Rosenqvist, E. Namork, P. Kierulf, R. Ovstebo, G. B. Joo, and P. Brandtzæg. 2000. Cellular activation properties and morphology of membrane-bound and purified meningococcal lipopolysaccharide. J. Endotoxin Res. 6:437-445. [PubMed] [Google Scholar]
- 5.Brandtzaeg, P. 2003. Host response to Neisseria meningitidis lacking lipopolysaccharides. Expert Rev. Anti-infect. Ther. 1:89-96. [DOI] [PubMed] [Google Scholar]
- 6.Brandtzaeg, P., A. Bjerre, R. Ovstebo, B. Brusletto, G. B Joo, and P. Kierulf. 2001. Neisseria meningitidis lipopolysaccharides in human pathology. J. Endotoxin Res. 7:401-420. [PubMed] [Google Scholar]
- 7.Brandtzaeg, P., K. Bryn, P. Kierulf, R. Ovstebo, E. Namork, B. Aase, and E. Jantzen. 1992. Meningococcal endotoxin in lethal septic shock plasma studied by gas chromatography, mass-spectrometry, ultracentrifugation, and electron microscopy. J. Clin. Investig. 89:816-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandtzaeg, P., P. Kierulf, P. Gaustad, A. Skulberg, J. N. Bruun, S. Halvorsen, and E. Sorensen. 1989. Plasma endotoxin as a predictor of multiple organ failure and death in systemic meningococcal disease. J. Infect. Dis. 159:195-204. [DOI] [PubMed] [Google Scholar]
- 9.Brandtzaeg, P., R. Ovstebo, and P. Kierulf. 1992. Compartmentalization of lipopolysaccharide production correlates with clinical presentation in meningococcal disease. J. Infect. Dis. 166:650-652. [DOI] [PubMed] [Google Scholar]
- 10.Brandtzaeg, P., R. Ovstebo, and P. Kierulf. 2001. Quantitative detection of bacterial lipopolysaccharides in clinical specimens, p 427-439. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease, methods and protocols. The Humana Press, Totowa, N.J. [DOI] [PubMed]
- 11.Caugant, D. A., E. A. Hoiby, L. O. Froholm, and P. Brandtzaeg. 1996. Polymerase chain reaction for case ascertainment of meningococcal meningitis: application to the cerebrospinal fluids collected in the course of the Norwegian meningococcal serogroup B protection trial. Scand. J. Infect. Dis. 28:149-153. [DOI] [PubMed] [Google Scholar]
- 12.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, A. J. Fox, and E. B. Kaczmarski. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Voe, I. W., and J. E. Gilchrist. 1973. Release of endotoxin in the cell wall blebs during in vitro growth of Neisseria meningitidis. J. Exp. Med. 138:1156-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gedde-Dahl, T. W., E. A. Hoiby, A. Schillinger, A. Lystad, and K. Bovre. 1983. An epidemiological, clinical and microbiological follow-up study of incident meningococcal disease cases in Norway, winter 1981-1982. Material and epidemiology of the MenOPP project. NIPH Ann. 6:155-168. [PubMed] [Google Scholar]
- 15.Gray, S. J., R. Borrow, and E. B. Kaczmarski. 2001. Meningococcal serology, p. 61-87. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease, methods and protocols. The Humana Press, Totowa, N.J. [DOI] [PubMed]
- 16.Guiver, M., and R. Borrow. 2001. PCR diagnosis, p. 23-39. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease, methods and protocols. The Humana Press, Totowa, N.J.
- 17.Guiver, M., R. Borrow, J. Marsh, S. J. Gray, E. B. Kaczmarski, D. Howells, P. Boseley, and A. J. Fox. 2000. Evaluation of the Applied Biosystems automated Taqman polymerase chain reaction system for the detection of meningococcal DNA. FEMS Immunol. Med. Microbiol. 28:173-179. [DOI] [PubMed] [Google Scholar]
- 18.Hacket, S. J., M. Guiver, J. Marsh, J. A. Sills, A. P. Thomson, E. B. Kaczmarski, and C. A. Hart. 2002. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch. Dis. Child. 86:44-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoiby, E. A., G. Bjune, L. O. Froholm, J. Eng, A. Halstensen, E. Rosenqvist, E. Ronnild, and E. Wedege. 1991. The Norwegian meningococcal serogroup B outer membrane vesicle vaccine protection trials: case tracing, meningococcal antigen detection and serological diagnosis. NIPH Ann. 14:107-121. [PubMed] [Google Scholar]
- 20.Kristiansen, B. E., E. Ask, A. Jenkins, C. Fermer, P. Radstrom, and O. Skold. 1991. Rapid diagnosis of meningococcal meningitis by polymerase chain reaction. Lancet 337:1568-1569. [DOI] [PubMed] [Google Scholar]
- 21.La Scolea, L. J., D. Dryja, D. Sullivan, L. Mosovich, N. Ellerstein, and F. Neter. 1981. Diagnosis of bacteremia in children by quantitative direct plating and a radiometric procedure. J. Clin. Microbiol. 13:478-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzen, D. R., F. Dux, U. Wolk, A. Tsirpouchtsidis, G. Haas, and T. F. Meyer. 1999. Immunoglobulin A1 protease, an exoenzyme of pathogenic Neisseriae, is a potent inducer of proinflammatory cytokines. J. Exp. Med. 190:1049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meijerink, J., C. Mandigers, L. van de Locht, E. Tonnissen, F. Goodsaid, and J. Raemaekers. 2001. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J. Mol. Diagn. 3:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namork, E., and P. Brandtzaeg. 2002. Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet 360:1741. [DOI] [PubMed] [Google Scholar]
- 25.Olcén, P., and H. Fredlund. 2001. Isolation, culture, and identification of meningococci from clinical specimens, p 9-21. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease, methods and protocols. The Humana Press, Totowa, N.J. [DOI] [PubMed]
- 26.Ovstebo, R., K. B. Haug, K. Lande, and P. Kierulf. 2003. PCR-based calibration curves for studies of quantitative gene expression in human monocytes: development and evaluation. Clin. Chem. 49:425-432. [DOI] [PubMed] [Google Scholar]
- 27.Rosenqvist, E., E. A. Hoiby, E. Wedege, K. Bryn, J. Kolberg, A. Klem, E. Ronnild, G. Bjune, and H. Nokleby. 1995. Human antibody responses to meningococcal outer membrane antigens after three doses of the Norwegian group B meningococcal vaccine. Infect. Immun. 63:4642-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seward, R. J., and K. J. Towner. 2000. Evaluation of a PCR-immunoassay technique for detection of Neisseria meningitidis in cerebrospinal fluid and peripheral blood. J. Med. Microbiol. 49:451-456. [DOI] [PubMed] [Google Scholar]
- 29.Sobanski, M. A., R. A. Barnes, and W. T. Coakley. 2001. Detection of meningococcal antigen by latex agglutination, p 41-59. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease, methods and protocols. The Humana Press, Totowa, N.J. [DOI] [PubMed]
- 30.Sullivan, T. D., and L. J. LaScolea. 1987. Neisseria meningitidis bacteremia in children: quantitation of bacteremia and spontaneous clinical recovery without antibiotic therapy. Pediatrics 80:63-67. [PubMed] [Google Scholar]
- 31.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 32.Tzanakaki, G., M. Tsolia, V. Vlachou, M. Theodoridou, A. Pangalis, M. Foustoukou, T. Karpathios, C. C. Blackwell, and J. Kremastinou. 2003. Evaluation of non-culture diagnosis of invasive meningococcal disease by polymerase chain reaction (PCR). FEMS Immunol. Med. Microbiol. 39:31-36. [DOI] [PubMed] [Google Scholar]
- 33.van Deuren, M., P. Brandtzaeg, and J. W. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev. 13:144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zwahlen, A., and F. A. Waldvogel. 1984. Magnitude of bacteremia and complement activation during Neisseria meningitidis infection: study of two co-primary cases with different clinical presentations. Eur. J. Clin. Microbiol. 3:439-441. [DOI] [PubMed] [Google Scholar]