Abstract
From 2002 to 2003, four isolates of Salmonella enterica serotypes Typhimurium, Enteritidis, Blockley, and Panama, isolated in France from patients with gastroenteritis, were found to produce extended-spectrum β-lactamase TEM-52. The study showed the blaTEM-52 gene to be located in a Tn3-like structure and carried by 100- or 32-kb conjugative plasmids.
Salmonella enterica is a major food-borne enteric pathogen in humans worldwide. Wild-type strains of Salmonella are usually susceptible to β-lactams, but an increase in the rate of ampicillin resistance in the last decade with the emergence of S. enterica serotype Typhimurium phage type DT 104 has been observed in developed countries (7). More recently, the emergence of strains of serotype Newport that produce the plasmidic cephamycinase CMY-2 (known as Newport-MDRAmpC) and are resistant to extended-spectrum cephalosporins (ESC) has been reported in the United States (9). Extended-spectrum β-lactamases (ESBLs), which also inactivate the ESC, are very rare in the genus Salmonella. However the number of reported cases in the different serotypes of this organism has been increasing worldwide in recent years (1, 3, 4, 6, 10, 12-14, 18, 20-23). Most ESBLs reported in Salmonella derive from the common plasmid-mediated penicillinases TEM-1, TEM-2, and SHV-1, although there are some other unrelated enzyme groups including CTX-M and PER. We report here the characterization of four TEM-52-ESBL-producing S. enterica strains received at the French National Reference Center for Salmonella in 2002 and 2003. The four isolates described in Table 1 were recovered from stool samples of patients with gastroenteritis. They belonged to four different serotypes: Enteritidis, Typhimurium, Panama, and Blockley. Pulsed-field gel electrophoresis with XbaI used according to a protocol described previously (22) confirmed the isolates to be unrelated (Table 1).
TABLE 1.
Characteristics of TEM-52-producing S. enterica clinical strains under study
| Isolate | Serotype | Antigenic formula | Date of isolation | Location of isolation | Sex/age (yr) of patient | Suspected contaminated food | Recent antecedent hospitalization (<1 mo) | PFGE typea | Size of plasmid(s) carrying blaTEM (kb) |
|---|---|---|---|---|---|---|---|---|---|
| TYP | Typhimurium | 1,4,12:i:1,2 | Feb 2002 | Seine-St-Denis | F/40 | Indian restaurant | No | 4 | 100, 9 |
| ENT | Enteritidis | 9,12:g,m:− | May 2003 | Nord | M/>65 | Unknown | No | 2 | 100, 10 |
| PAN | Panama | 9,12:l,v:1,5 | July 2003 | Pas-de-Calais | M/45 | Unknown | No | 3 | 100 |
| BLO | Blockley | 6,8:k:1,5 | July 2003 | Nord | M/31 | Unknown | Unknown | 1 | 32 |
PFGE, pulsed-field gel electrophoresis.
Antibiotic susceptibility was determined by the disk diffusion method using Mueller-Hinton agar and 32 antibiotic disks (Bio-Rad, Marnes la Coquette, France) according to the recommendations of the French Society of Microbiology (19). MICs of the β-lactams were determined by Etest (AB Biodisk, Solna, Sweden). Escherichia coli ATCC 25922 was used as the control. The four isolates were resistant to ampicillin, ceftriaxone, and ceftazidime by the disk diffusion method. All four isolates were susceptible to cefoxitin, imipenem, aminoglycosides, quinolones, sulfonamides, trimethoprim, chloramphenicol, and tetracycline. The ESBL detection Etest strips and the double disk diffusion test (11) showed an ESBL phenotype for the four isolates. The MICs of the β-lactams are shown in Table 2. Isolates from serotypes Typhimurium (TYP) and Enteritidis (ENT) exhibited a higher level of resistance to ceftazidime and ceftriaxone (MIC > 256 mg/liter) than isolates from serotypes Panama (PAN) and Blockley (BLO) (MIC from 8 to 32 mg/liter).
TABLE 2.
MICs of β-lactams used in this studyf
| β-lactam(s)a | MIC (mg/liter)
|
E. coli C1a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PAN | pPAN-1 | TYP | pTYP-1 | pTYP-2′ | ENT | pENT-1 | pENT-5′ | BLO | pBLO-1 | ||
| Ampicillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | 4 |
| Amoxicillin-CLAb | 4 | 4 | 8 | 4 | 8 | 8 | 4 | 8 | 4 | 4 | 4 |
| Ticarcillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | 2 |
| Ticarcillin-CLAc | 16 | 16 | 64 | 8 | 32 | 64 | 8 | 128 | 16 | 16 | 2 |
| Piperacillin | >256 | >256 | >256 | >256 | >256 | >256 | 256 | >256 | >256 | >256 | 2 |
| Piperacillin-TZBd | 4 | 4 | 4 | 2 | 4 | 4 | 2 | 4 | 4 | 4 | 2 |
| Cefoxitin | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 4 | 2 |
| Ceftazidime | 32 | 16 | >256 | 16 | >256 | >256 | 32 | >256 | 32 | 32 | 0.25 |
| Ceftazidime-CLAe | 0.25 | 0.5 | 0.5 | 0.125 | 0.25 | 1 | 0.125 | 0.5 | 0.5 | 0.5 | NT |
| Ceftriaxone | 8 | 8 | 128 | 8 | 128 | 128 | 8 | 256 | 32 | 32 | 0.06 |
| Cefotaxime-CLAe | 0.06 | 0.06 | 0.125 | 0.03 | 0.03 | 0.125 | 0.03 | 0.03 | 0.25 | 0.25 | NT |
| Cefepime | 2 | 1 | >32 | 1 | 16 | 32 | 2 | 32 | 4 | 4 | 0.06 |
| Aztreonam | 4 | 4 | 32 | 2 | 16 | 16 | 4 | 16 | 4 | 4 | 0.06 |
| Imipenem | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 |
CLA, clavulanic acid; TZB, tazobactam; NT, not tested.
Amoxicillin-CLA (2:1).
CLA, 2 mg/liter.
TZB, 4 mg/liter.
CLA, 4 mg/liter.
MICs of β-lactams (Etest) for TEM-52-producing S. enterica isolates (PAN, TYP, ENT, and BLO), their E. coli C1a transconjugants (pPAN-1, pTYP-1, pTYP-2′, pENT-1, pENT-5′, and pBLO-1), and E. coli C1a are shown.
Crude extracts of β-lactamases were obtained by sonication. Isoelectrofocusing was performed by using a PhastSystem apparatus with PhastGel IEF 3-9 gels (Amersham-Pharmacia Biotech, Freiburg, Germany) as described previously (22). The four isolates produced only one β-lactamase with a pI of 6.0.
Total DNA was extracted by using the InstaGene matrix kit (Bio-Rad). PCR analysis was performed with primers TEM-F (5′-ATAAAATTCTTGAAGACGAAA-3′) and TEM-R (5′-GACAGTTACCAATGCTTAATC-3′) to amplify a 1,080-bp fragment of the blaTEM gene as described previously (22). DNA sequencing of PCR products and deduced amino acid sequence analysis revealed that the β-lactamase was TEM-52, which differed from TEM-1 by three point mutations, Glu104→Lys, Met182→Thr, and Gly238→Ser (Ambler numbering) (2). The DNA sequence of isolate BLO was 100% identical to that of blaTEM-52b (GenBank accession numbers AF12644 and AF027199), while the sequence of the three other isolates was identical to that of blaTEM-52a (GenBank accession number Y13612), except for a silent mutation located at the position Gly78 (GGC→GGT).
A resistance transfer experiment was carried out in liquid medium by using E. coli C1a (nalA) as the recipient strain. Transconjugants were selected on Mueller-Hinton agar supplemented with ceftazidime (2 mg/liter) and nalidixic acid (68 mg/liter). The E. coli transconjugants pPAN-1, pTYP-2′, pENT-5′, and pBLO-1 were obtained for isolates PAN, TYP, ENT, and BLO, respectively, presenting a similar pattern (Table 2) and expressing a β-lactamase with a pI of 6.0. Additionally, transconjugants pTYP-1 and pENT-1 were obtained for isolates TYP and ENT, respectively, presenting a lower resistance to β-lactams (Table 2).
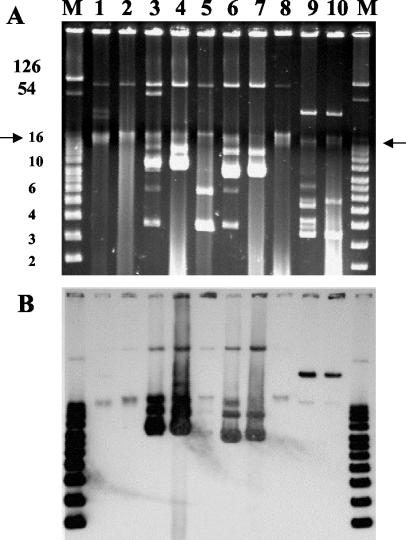
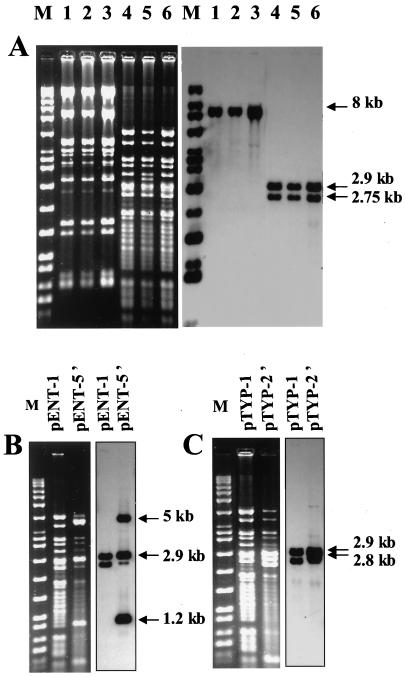
Plasmid DNA was purified by using a QIAGEN (Courtaboeuf, France) Plasmid Midi kit. Molecular sizes of plasmids were determined by using Taxotron software (Institut Pasteur, Paris, France) by comparing them to plasmids of known sizes. Southern hybridization with a PCR-generated probe for blaTEM (1,080 bp) was performed as described previously (7). Extraction of plasmid DNA revealed a 100-kb common plasmid in the isolates PAN, TYP, and ENT and a 32-kb plasmid in isolate BLO, both of which hybridized with probe blaTEM (Fig. 1). Additionally, hybridization signals were observed with 10- and 9-kb plasmids in the more resistant isolates ENT and TYP, respectively (Fig. 1). The 100-kb plasmid was detected in E. coli transconjugants pPAN-1, pTYP-1, pTYP-2′, pENT-1, and pENT-5′, and the 32-kb plasmid was detected in pBLO-1 (Fig. 1). The 10- and 9-kb plasmids were also observed in transconjugants pENT-5′ and pTYP-2′, respectively. The plasmid DNA from transconjugants pPAN-1, pTYP-1, and pENT-1 were compared by restriction endonuclease (EcoRI and PstI; Roche, Mannheim, Germany) and Southern blot (with probe blaTEM) analyses. The plasmids had a very similar fingerprint (Fig. 2A). To study the 10- and 9-kb plasmids carrying additional blaTEM genes, Southern hybridization with a blaTEM probe after restriction with PstI was performed on plasmid DNA from transconjugants pTYP-2′ and pENT-5′ that were compared to pTYP-1 and pENT-1, respectively (Fig. 2B and C). The probe hybridized intensively to three bands of 5, 2.9, and 1.2 kb with pENT-5′ and two bands of 2.9 and 2.75 kb with pTYP-2′. Due to the relative amount of 9- or 10- and 100-kb plasmid DNA in pENT-5′ and in pTYP-2′ (Fig. 1A, lanes 4 and 7), these hybridization patterns have been attributed to 9- and 10-kb plasmids. As there is one internal site in blaTEM, these data suggested the existence of two copies of blaTEM in the 9-kb plasmid and three copies in the 10-kb plasmid.
FIG. 1.
(A) Agarose (0.8%) electrophoresis of plasmid DNA from the four Salmonella isolates TYP, ENT, PAN, and BLO and their E. coli transconjugants. (B) Hybridization of plasmid content with a blaTEM probe. The chromosome position is indicated by arrows. A supercoiled DNA ladder (Invitrogen, Groningen, The Netherlands), plus RP4 and pIP173 plasmids which served as the molecular size marker (M) (band sizes are in kilobase pairs), is shown. Lane 1, isolate PAN; lane 2, E. coli transconjugant pPAN-1; lane 3, isolate ENT; lane 4, E. coli transconjugant pENT-5′; lane 5, E. coli transconjugant pENT-1; lane 6, isolate TYP; lane 7, E. coli transconjugant pTYP-2′; lane 8, E. coli transconjugant pTYP-1; lane 9 isolate BLO; lane 10, E. coli transconjugant pBLO-1.
FIG. 2.
Restriction analysis (left panels) and blaTEM hybridization (right panels) of plasmids isolated from E. coli transconjugants. (A) Lane 1, EcoRI-restricted pPAN-1; lane 2, EcoRI-restricted pTYP-1; lane 3, EcoRI-restricted pENT-1; lane 4, PstI-restricted pPAN-1; lane 5, PstI-restricted pTYP-1; lane 6, PstI-restricted pENT-1. (B) PstI-digested plasmids extracted from pENT-1 and pENT-5′. (C) PstI-digested plasmids extracted from pTYP-1, and pTYP-2′. Marker Raoul I (Appligene, Illkirch, France) was used as a molecular size marker.
To determine whether blaTEM-52 was located in Tn3, Tn3-specific PCR was performed on plasmid DNA with the following primers designed on the basis of the sequence of Tn3 in Salmonella (GenBank accession number AB103092): forward primer 5′-CACGAATGAGGGCCGACAGGA-3′ (located at positions 4018 to 4038 in the tnpR gene) and reverse primer 5′-ACCCACTCGTGCACCCAACTG-3′ (located at positions 4492 to 4512 in blaTEM). All transconjugants gave the expected PCR product of 500 bp (data not shown). These results indicated the presence in our isolates of blaTEM-52 in a Tn3-like structure carried by a self-conjugative plasmid. Transposition of one or more copies of blaTEM-52 to other plasmids within a bacterium may explain the higher resistance of isolates TYP and ENT.
TEM-52 ESBL was first reported in a Klebsiella pneumoniae strain isolated in 1996 in France, from hospitalized children originating from Athens, Greece (17). Since 1996, only one isolate of TEM-52-producing E. coli has been identified in France during a national hospital survey (8). In 1997 and 1999, two studies demonstrated TEM-52 to be the most prevalent TEM-type ESBL among E. coli strains in Korea and among enterobacterial species in Italy (15, 16). The first report of TEM-52 in the genus Salmonella described two strains isolated in 1998 from a hospitalized Yugoslavian infant (20). Both strains exhibited different resistance phenotypes attributed to different copy numbers of blaTEM-52. As resistance was not self-transmissible, the authors suggested the location of the ESBL gene on a transposon without experiment. A second report described five isolates of S. enterica serotypes Saintpaul, Stanley, Agona, and Enteritidis producing TEM-52 and recovered between 1995 and 1997 in Korea that came mostly from hospitalized patients (12). The blaTEM-52 genes were carried on conjugative plasmids greater than 180 kb in length with diverse genetic characteristics. The last report concerned TEM-52-producing isolates of serotype Enteritidis from a hospital outbreak (five cases) in Scotland in 2001 and 2002 (23). The blaTEM-52 gene was carried on a 95-kb conjugative plasmid. The emergence of ESBL-producing Salmonella strains in France is a new phenomenon. No ESBLs were detected among human Salmonella isolates in multicenter surveys conducted by a hospital-based network in 1994 (n = 2,622) and 1997 (n = 2,464) (5). More recently, two surveys conducted by the French National Reference Center for Salmonella in 2000 (n = 1,066) and in 2002 (n = 1,140) identified only one human ESBL-producing Salmonella isolate (isolate TYP, this study) in 2002. One hypothesis for the emergence of TEM-52-producing Salmonella is the nosocomial acquisition by the exchange of ESBL genes among enteric bacteria frequently encountered in hospitals and selected by traces of ESC used in humans. An alternative hypothesis that has been suggested in a report of ESBL (SHV-12)-producing Salmonella, and with the emergence of Newport MDRAmpC, is the transmission through the food chain and consequently to antibiotic selection pressure in livestock (6, 9). In our study, the isolates were generally not acquired from a hospital, and we suspected the second hypothesis but we didn't have evidence for it. This hypothesis was strengthened by the detection of six TEM-52-producing Salmonella strains belonging to four different serotypes and isolated in poultry in Belgium during the period of 2001 to 2002 (A. Cloeckaert, personal communication).
This study is the first report of blaTEM-52 in S. enterica serotypes Panama and Blockley and describes TEM-52-producing isolates of Salmonella in France. The study shows that the blaTEM-52 gene is located in a Tn3-like structure and carried by a conjugative plasmid. Both features explain the high level of resistance of some isolates and the spreading of blaTEM-52 among Salmonella of various serotypes.
Acknowledgments
We thank all the corresponding laboratories of the French Salmonella network and, in particular, E. Dehecq (Laboratoire de Microbiologie, Centre Hospitalier Saint-Philibert, Lomme, France) and J. Ficheux (Laboratoire d'Analyses de Biologie Médicale, Dunkerque, France).
REFERENCES
- 1.AitMhand, R., A. Soukri, N. Moustaoui, H. Amarouch, N. ElMdaghri, D. Sirot, and M. Benbachir. 2002. Plasmid-mediated TEM-3 extended-spectrum β-lactamase production in Salmonella typhimurium in Casablanca. J. Antimicrob. Chemother. 49:169-172. [DOI] [PubMed] [Google Scholar]
- 2.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuil, J., A. Brisabois, I. Casin, L. Armand-Lefèvre, S. Frémy, and E. Collatz. 2000. Antibiotic resistance in salmonellae isolated from humans and animals in France: comparative data from 1994 and 1997. J. Antimicrob. Chemother. 46:965-971. [DOI] [PubMed] [Google Scholar]
- 6.Cardinale, E., P. Colbachini, J. D. Perrier-Gros-Claude, A. Gassama, and A. Aidara-Kane. 2000. Dual emergence in food and humans of a novel multiresistant serotype of Salmonella in Senegal: Salmonella enterica subsp. enterica serotype 35:c:1,2. J. Clin. Microbiol. 39:2373-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded β-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 8.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, et al. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta, A., J. Fontana, C. Crowe, B. Bolstorff, A. Stout, S. Van Duyne, M. P. Hoekstra, J. M. Whichard, T. J. Barrett, and F. J. Angulo. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 10.Hammami, A., G. Arlet, S. Ben Redjeb, F. Grimont, A. Ben Hassen, A. Rekik, and A. Philippon. 1991. Nosocomial outbreak of acute gastroenteritis in a neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 β-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 10:641-646. [DOI] [PubMed] [Google Scholar]
- 11.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 12.Lee, K., D. Yong, J. H. Yum, H. H. Kim, and Y. Chong. 2003. Diversity of TEM-52 extended-spectrum β-lactamase-producing nontyphoidal Salmonella isolates in Korea. J. Antimicrob. Chemother. 52:493-496. [DOI] [PubMed] [Google Scholar]
- 13.Makanera, A., G. Arlet, V. Gautier, and M. Manai. 2003. Molecular epidemiology and characterization of plasmid-encoded β-lactamases produced by Tunisian clinical isolates of Salmonella enterica serotype Mbandaka resistant to broad-spectrum cephalosporins. J. Clin. Microbiol. 41:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morosini, M. I., J. Blazquez, M. C. Negri, R. Canton, E. Loza, and F. Baquero. 1996. Characterization of a nosocomial outbreak involving an epidemic plasmid encoding for TEM-27 in Salmonella enterica subspecies enterica serotype Othmarschen. J. Infect. Dis. 174:1015-1020. [DOI] [PubMed] [Google Scholar]
- 15.Pai, H., S. Lyu, J. H. Lee, J. Kim, Y. Kwon, J.-W. Kim, and K. W. Choe. 1999. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J. Clin. Microbiol. 37:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perilli, M., E. Dell'Amico, B. Segatore, M. R. de Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poyart, C., P. Mugnier, G. Quesne, P. Berche, and P. Trieu-Cuot. 1998. A novel extended-spectrum TEM-type β-lactamase (TEM-52) associated with decreased susceptibility to moxalactam in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 42:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revathi, G., K. P. Shannon, P. D. Stapleton, B. K. Jain, and G. L. French. 1998. An outbreak of extended-spectrum, β-lactamase-producing Salmonella senftenberg in a burns ward. J. Hosp. Infect. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 19.Soussy, C. J., G. Carret, J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M. H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, and J. Sirot. 2000. Comité de l'Antibiogramme de la Société Française de Microbiologie. Communiqué 2000-2001. Pathol. Biol. 48:832-871. [PubMed] [Google Scholar]
- 20.Vahaboglu, H., S. Dodanli, C. Eroglu, R. Ozturk, G. Soyletir, I. Yildirim, and V. Avkan. 1996. Characterization of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1 producing isolates and evidence for nosocomial plasmid exchange by a clone. J. Clin. Microbiol. 34:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vahaboglu, H., M. Fuzi, S. Cetin, S. Gundes, E. Ujhelyi, F. Coskunkan, and O. Tansel. 2001. Characterizations of extended-spectrum β-lactamase (TEM-52)-producing strains of Salmonella enterica serovar Typhimurium with diverse resistance phenotypes. J. Clin. Microbiol. 39:791-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weill, F.-X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (SHV-12 like)-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yates, C. M., D. J. Brown, G. F. S. Edwards, and S. G. B. Amyes. 2004. Detection of TEM-52 in Salmonella enterica serovar Enteritidis isolated in Scotland. J. Antimicrob. Chemother. 53:407-408. [DOI] [PubMed] [Google Scholar]