Abstract
Accumulating evidence suggests that various neurodegenerative diseases, including Alzheimer’s disease (AD), are linked to cytotoxic diffusible aggregates of amyloid proteins, which are metastable intermediate species in protein misfolding. This study presents the first site-specific structural study on an intermediate called amylospheroid (ASPD), an AD-derived neurotoxin composed of oligomeric amyloid-β (Aβ). Electron microscopy and immunological analyses using ASPD-specific “conformational” antibodies established synthetic ASPD for the 42-residue Aβ(1–42) as an excellent structural/morphological analogue of native ASPD extracted from AD patients, the level of which correlates with the severity of AD. 13C solid-state NMR analyses of approximately 20 residues and interstrand distances demonstrated that the synthetic ASPD is made of a homogeneous single conformer containing parallel β-sheets. These results provide profound insight into the native ASPD, indicating that Aβ is likely to self-assemble into the toxic intermediate with β-sheet structures in AD brains. This approach can be applied to various intermediates relevant to amyloid diseases.
A variety of neurological disorders, such as Alzheimer’s disease (AD) and Parkinson’s disease (PD), are associated with the misfolding of disease-specific amyloid proteins. Recent evidence has identified diffusible amyloid intermediates that occur during in the course of amyloid misfolding as more potent toxins in amyloid diseases than amyloid fibrils;1−4 these toxic amyloid intermediate species include oligomers (2–100mers) and larger metastable assemblies of amyloid proteins. Despite their increasing importance, the intrinsically instable and heterogeneous nature of the amyloid intermediates have made it an intractable problem to define their detailed structural features, relationship with amyloid fibrils, and pathogenic functions. Early studies using electron microscopy and atomic force microscopy identified spherical assembles with a diameter ranging from 5 to 20 nm in amyloid proteins such as Alzheimer’s amyloid β protein (Aβ) and Parkinson’s α-synuclein (αSyn).1−5 Thus, intense efforts have focused on elucidating the detailed structural features of amyloid intermediates for Aβ, αSyn, and other disease-related proteins by solid-state NMR (SSNMR) and other biophysical methods.5−15 Nevertheless, site-specific structural features of amyloid intermediates have been difficult to achieve for a majority of species, which include relatively well characterized intermediates of Aβ, such as amyloid β-derived diffusible ligand (ADDL),2 amylospheroid (ASPD),1 Aβ*56,16 globulomer,17 and small oligomers (2–6mers).18−20 To date, no atomic-level structures have been obtained for toxic amyloid intermediates of any disease-specific amyloid proteins other than protofibrils, which contain antiparallel β-sheets.21 More importantly, almost no structural data are currently available for pathologically relevant native amyloid intermediates derived from patients.
Here, we present a new approach to gain detailed NMR-based structural insight of AD-derived native amyloid intermediates through studying ASPD, which is a notable diffusible assembly of Aβ originating from AD patient brains.22 ASPD represents a class of highly toxic spherical amyloid intermediates, which have a diameter of 10–15 nm based on transmission electron microscopy (TEM) analysis.1 Our previous studies found that AD-derived ASPD is pathologically relevant to AD because native ASPD samples isolated from patient brains are toxic to human neurons and their level in AD patient brains correlates well with the pathological severity of AD.22 Despite its increasing importance, structural features of ASPD are, to a large extent, unknown. A recent study indicated that in vitro reconstituted synthetic ASPD for the 42-residue Aβ(1–42) shares essential characteristics with native ASPD based on their neurotoxicity and morphology.22 The similarities between synthetic and native ASPDs in structural and morphological aspects were also suggested by “conformation-specific” antibodies targeting ASPD as well as by TEM studies.22 Here, we analyzed the detailed structural features of synthetic ASPD, which serves as a structural and functional analogue for AD-derived ASPD, by SSNMR, a vital structural tool for amyloid aggregates.6,8,23
We first assessed whether the in vitro reconstituted ASPD used for this study was similar to AD-derived native ASPD based on morphology as well as immuno-reactivity to anti-ASPD antibodies. The native ASPDs, with a 10–15 nm diameter, were collected from soluble brain extracts from patients diagnosed with AD using an immuno-precipitation assay with the conformation-specific monoclonal antibody haASD1, which specifically recognizes the ASPD surface.22 A control experiment of the same sample incubated with mouse IgG did not bind any spherical species (Figure S1 in the Supporting Information (SI)). For the SSNMR analysis, synthetic ASPD samples were prepared by incubating Aβ(1–42) at a concentration of ∼50 μM in F12 medium as previously described (see the Materials and Methods section and SI for details).22 The TEM image of the prepared synthetic ASPD sample shows relatively homogeneous spherical morphologies having an average diameter of 11.0 ± 2.1 nm (n = 65) (Figure 1A).22,24 We then compared the synthetic ASPD with the native ASPD; the data show that the spherical morphology and the size of the synthetic ASPD sample used for this study are similar to those of the native ASPD sample collected from a brain extract of AD patients (Figure 1B), which shows an average diameter of 10.9 ± 1.7 nm (n = 30). It was also confirmed by TEM that the lyophilization or buffer exchange of the ASPD samples required for our NMR analyses did not alter the morphology (Figure S2). In addition, we quantitatively examined the immunological similarities of the ASPD sample prepared for this study to native ASPD by dot blotting (Figure 1C, top) using another anti-ASPD conformation-specific antibody, rpASD1, of polyclonal origin (top row) with control data using a sequence-specific monoclonal antibody against the N-terminal residues Aβ(1–5) (82E1; middle row) for the quantification of Aβ.25 The plot (Figure 1C, bottom) shows the ratio of the reactivity of rpASD1 to the corresponding reactivity of 82E1 in order to indicate the ASPD amount normalized by the Aβ amount. The data clearly demonstrate that the synthetic ASPD (1st column) and lyophilized and rehydrated synthetic ASPD (2nd column) were both recognized by rpASD1 to the same degree as native ASPD (3rd column). The results in Figure 1C suggest that the synthetic ASPD and native ASPD have similarities in their surface structures. In contrast, monomeric Aβ(1–42) (4th column) was barely recognized by rpASD1 under the same conditions. It is known that rpASD1 does not recognize Aβ fibril or ADDL, either.22 Notably, rpASD1 is likely to recognize different epitopes from those recognized by haASD1 used for Figure 1B;22 thus, the result demonstrates quantitative immunological similarity, which is likely orthogonal to that from the immuno-TEM results in Figure 1B. A high level of cytotoxicity was previously reported for both synthetic ASPD and native ASPD on primary rat hippocampal cultures.22 We thus have presented the multiplex immuno-analyses for the first time to examine functional and structural similarities between the freeze-trapped synthetic ASPD and the native oligomer from AD patients in the context of site-specific structural analysis. The results establish the pathological relevance of the synthetic ASPD sample.
Figure 1.
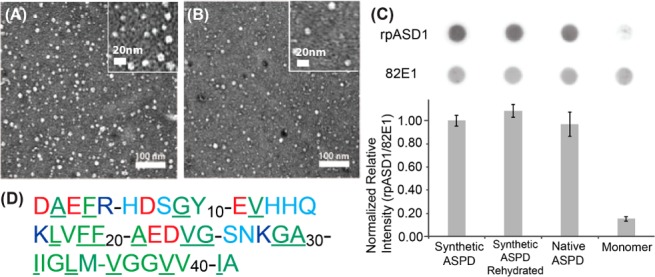
(A) Transmission electron microscopy (TEM) image for a synthetic ASPD sample. The sample was prepared following the protocol used for the sample preparation for the NMR analysis. (B) The corresponding TEM image for a native ASPD sample collected by the immuno-precipitation assay using the haASD antibody1 from a brain extract of an AD patient. (C) A dot blot assay detected by (top) rpASD1 and (middle) 82E1 antibodies for the synthetic ASPD samples without (1st column) and with (2nd column) lyophilization and subsequent rehydration compared to control experiments on a native ASPD sample from an AD patient (3rd column) and monomeric Aβ(1–42) (4th column). (C, bottom) The ratio of the ASPD amount detected by rpASD1 divided by the Aβ amount detected by 82E1 for the synthetic ASPD, native ASPD, and monomer samples. The value was normalized to that of synthetic ASPD. (D) Amino acid sequence of Aβ(1–42), where the residue names are color coded by the type of amino acids as follows: negatively charged (red), positively charged (blue), hydrophilic (cyan), and hydrophobic (green) residues. The underlined residues are those inspected in the present SSNMR analysis.
Despite the effectiveness, these immunological data do not provide any specific structural information beyond the similarity level. Thus, in order to identify site-specific structural features of ASPD, we performed 2D 13C/13C correlation SSNMR experiments on four synthetic ASPD samples having different 13C-labeling schemes (Figure 2; see Table S1 for a list of the samples). The amino acid sequence of Aβ(1–42) is shown with the isotope-labeled sites (underlined) that were inspected in the NMR analysis (Figure 1D). The 2D 13C/13C correlation SSNMR spectra (Figure 2) show promising results for the first attempt at structural analysis of synthetic ASPD. All of the 13C-labeled sites, except for Phe-4, were successfully assigned on the basis of amino acid specific exchange patterns, as shown by the color-coded lines (residues in the inset; see Table S2 for the assignments). The spectra display reasonably narrow line widths (1.9–3.7 ppm), even after considerable Gaussian convolution (1.3–1.5 ppm) for sensitivity enhancement. The natural line widths are comparable to or slightly broader than those observed for lyophilized amyloid fibrils.6 Since it is known that a line width or a 13C chemical shift distribution reflects conformational heterogeneity, the reasonably narrow line widths suggest that the synthetic ASPD is likely to have a well-ordered structure that is comparable to an amyloid fibril, which is known to have a highly ordered structure. Except for some specific residues, such as Leu-17, Val-24, and Ile-31, most of the residues had a single set of major cross peaks for a directly bonded 13C/13C pair. This suggests that Aβ in the ASPD samples has a well-ordered single conformer for the majority of the inspected residues, indicating that Aβ misfolds into a specific form in ASPD. The lack of multiple forms of Aβ in ASPD based on the SSNMR analysis supports the notion that ASPD is a homo-oligomer with a high structural order. These results also demonstrate that ASPD is not polymorphric, but rather morphologically and conformationally homogeneous to a large extent.
Figure 2.
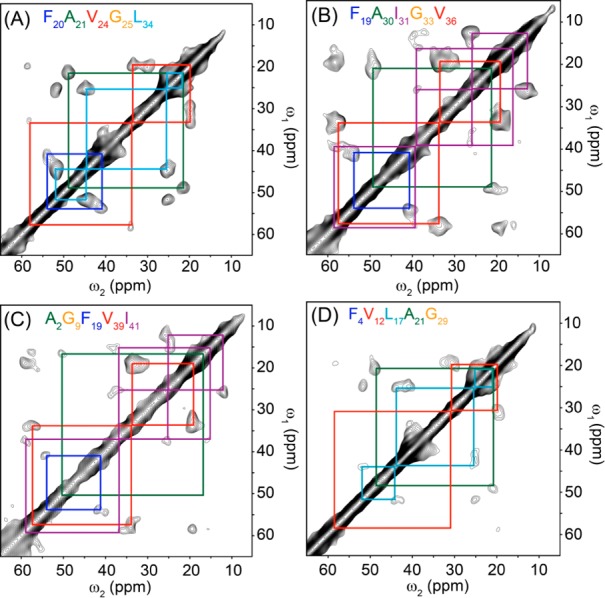
2D 13C/13C SSNMR spectra of ASPD samples for four differently 13C- and 15N-labeled Aβ(1–42) with a short-range DARR mixing (50 ms)26 at a spinning speed of 20 kHz. The Aβ(1–42) samples were labeled with uniformly 13C- and 15N-labeled amino acids at (A) Phe-20, Ala-21, Val-24, Gly-25, Leu-34; (B) Phe-19, Ala-30, Ile-31, Gly-33, Val-36; (C) Ala-2, Gly-9, Phe-19, Val-39, Ile-41; and (D) Phe-4, Val-12, Leu-17, Ala-21, Gly-29. The signals were collected with a t1 period of (A) 5 ms, (B,C) 4 ms, and (D) 3 ms, and a t2 period of 10 ms. The spectrum was processed with Gaussian line broadening of (A,D) 1.3 ppm or (B,C) 1.5 ppm on both dimensions, with linear prediction on t1 to 6 ms. The experimental times were (A,B) 4.9 days, (C) 5.5 days, and (D) 8.2 days.
The results are intriguing, considering that even amyloid fibrils of Aβ, which are the end products in misfolding, typically involve polymorphs.17 Also, a previous SSNMR study on a small toxic Aβ(1–42) oligomer indicated that the protein has a disordered structure.8 Taken together, these results provide the first direct evidence that a highly ordered conformer is formed through spontaneous misfolding of Aβ(1–42) in an amyloid intermediate that has pathological relevance to AD. The N-terminal residues (Ala-2, Phe-4, and Val-12) had weaker signal intensities compared to the corresponding amino acids at other sites, such as Phe-19, Ala-21, and Val-36 (Figure 2A,B). This finding indicates that the N-terminal region of Aβ in ASPD may be disordered with some residual structural order. These results are consistent with a recent finding that the anti-ASPD conformation-specific antibodies, such as rpASD1, recognize the N-terminal residues, which are likely to be exposed to the surface.22 These antibodies are also likely to recognize other regions of Aβ in ASPD, including its hydrophobic regions, which have well-defined conformations, as discussed below.22 The evidence of a single conformer for ASPD with high structural order explains how high affinity (Kd = (0.5–5) × 10–12 M) was achieved for the anti-ASPD antibodies, including rpASD1 and haASD1.
On the basis of the collected 13C chemical shifts (Table S2), we elucidated the conformations of ASPD through a torsion angle analysis with the TALOS software (Figure 3B). The predicted torsion angles suggest that ASPD in vitro predominantly contains a β-sheet motif in the regions. We then performed interstrand 13CO–13CO distance measurements on the ASPD sample prepared with Aβ(1–42) selectively labeled at 13CO in Ala-30 (5.85 ± 0.10 Å) and Val-39 (6.05 ± 0.15 Å) (Figure S3). The obtained interstrand distances of ∼6 Å are consistent with a parallel β-sheet arrangement. This is the first indication that a pathologically relevant amyloid intermediate species has a highly ordered and extended parallel β-sheet structure. The results show an interesting contrast with a recent FT-IR study indicating an antiparallel β-sheet in oligomeric species of Aβ(1–42),7 for which pathological relevance is not established. Our preliminary study shows that the structure of Aβ in ASPD is noticeably different from that in Aβ(1–42) fibril (Figure S4). The interstrand distances of ∼6 Å also deviate from that of ∼5 Å observed for fibrils, suggesting that ASPD may not have an in-register parallel β-sheet arrangement, unlike fibrils.23
Figure 3.
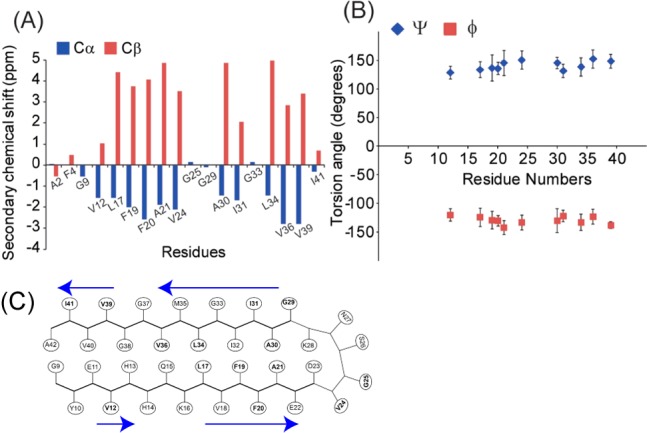
(A) Secondary chemical shift analysis for the 18 residues from the four different ASPD samples. (B) Torsion angle prediction using TALOS software. The predicted torsion angles suggest that (ϕ,ψ) ≈ (−120°, 130°), which is consistent with a β-sheet structure. (C) A possible structural model based on the present data. The residues inspected by SSNMR are shown in bold. The β-sheet regions are represented by blue arrows.
In summary, the present study has revealed that ASPD contains a distinctive parallel β-sheet structure as the primary structural motif with somewhat disordered N-terminal residues. The findings establish ASPD as a promising therapeutic target for AD that is likely to retain a well-defined β-sheet structure in the brain of AD patients. As discussed above, it was previously practically impossible to examine a molecular-level structure of an amyloid intermediate that is linked to the pathology of AD or any other amyloid diseases due to the demanding sample preparation and the lack of a suitable approach for structural analysis. This study has given important clues about the structure of the native amyloid intermediates for the first time. Recently, an excellent study was reported on the brain-derived amyloid fibril of Aβ(1–40), which was prepared with “seed” fibril from a patient affected by AD.27 Our approach provides a novel means to examine pathologically relevant amyloid intermediates, for which “seeding” is not effective, through SSNMR and multiplex analyses with dot-blot, TEM, and immuno-precipitation methods.
Materials and Methods
Aβ(1–42) was synthesized and purified as previously described using solid-phase synthesis with standard Fmoc synthesis and cleavage protocols as well as HPLC purification.1,22 Further details are available in the SI.
The SSNMR experiments were performed following the established procedures used for amyloid intermediates of Aβ(1–40).6 All of the SSNMR experiments were performed with a Varian Infinity-plus or Bruker Avance III SSNMR spectrometer using a home-built 2.5 mm triple-resonance MAS probe at 9.4 T (1H frequency of 400.2 MHz). In the 2D 13C/13C correlation experiments with DARR mixing26 shown in Figure 2, 1.8–3.5 mg samples of isotope-labeled Aβ(1–42) ASPD were used. Other details are in the SI.
The Bioethics Committees of Kyoto University, Institute of Biomedical Research and Innovation, and Niigata University approved all experiments using human subjects. The native ASPDs were collected from concentrated human brain extracts by immunoisolation using the ASPD-specific haASD1 antibody as previously described.22,24 The reactivity of ASPDs with antibodies was estimated by dot blotting using a method previously described.22,24 The details, including the preparation of the brain extract, are provided in the SI.
The nanoscale morphology of the samples was observed by TEM using a JEM-1010 instrument (JEOL, Tokyo, Japan) operated at 100 kV and magnification of 80 000 as previously described.22 Other details, including the grid preparation and image analysis, are found in the SI.
Acknowledgments
This work was supported primarily by the NIH RO1 program (GM 098033) and Alzheimer’s Association IIRG grant (08-91256) for Y.I., and by grants from the Ministry of Health, Labor and Welfare (Research on Nanotechnical Medical) and from the Ministry of Education, Culture, Sports, Science and Technology (Grant-in-Aid for Scientific Research B) to M.H. A portion of the instrumentation efforts required for this study was supported by the NSF (CHE-0957793) and the Dreyfus Foundation for Teacher-Scholar award for Y.I. M.H. thanks Prof. M. Hagiwara at Kyoto University for useful discussion and Prof. A. Kakita at Niigata University for providing the patient brains. Y.I. is grateful to Drs. S. Chimon, C. Jones, and N. Wickramasinghe for their initial efforts on the preparation of Aβ(1–42) at UIC.
Supporting Information Available
Detailed protocols for preparation and characterization of ASPD and additional NMR data. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b03373.
The authors declare no competing financial interest.
Supplementary Material
References
- Hoshi M.; Sato M.; Matsumoto S.; Noguchi A.; Yasutake K.; Yoshida N.; Sato K. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. S.; Chang L.; Viola K. L.; Lacor P. N.; Lambert M. P.; Finch C. E.; Krafft G. A.; Klein W. L. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D. J. Nat. Cell Biol. 2004, 6, 1054. [DOI] [PubMed] [Google Scholar]
- Klein W. L.; Stine W. B.; Teplow D. B. Neurobiol. Aging 2004, 25, 569. [DOI] [PubMed] [Google Scholar]
- Chimon S.; Ishii Y. J. Am. Chem. Soc. 2005, 127, 13472. [DOI] [PubMed] [Google Scholar]
- Chimon S.; Shaibat M. A.; Jones C. R.; Calero D. C.; Aizezi B.; Ishii Y. Nat. Struct. Mol. Biol. 2007, 14, 1157. [DOI] [PubMed] [Google Scholar]
- Stroud J. C.; Liu C.; Teng P. K.; Eisenberg D. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M.; Davis J.; Aucoin D.; Sato T.; Ahuja S.; Aimoto S.; Elliott J. I.; Van Nostrand W. E.; Smith S. O. Nat. Struct. Mol. Biol. 2010, 17, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi-Shad N.; Garfagnini T.; Penco A.; Relini A.; Fogolari F.; Corazza A.; Esposito G.; Bemporad F.; Chiti F. Nat. Struct. Mol. Biol. 2012, 19, 547. [DOI] [PubMed] [Google Scholar]
- De Franceschi G.; Frare E.; Pivato M.; Relini A.; Penco A.; Greggio E.; Bubacco L.; Fontana A.; de Laureto P. P. J. Biol. Chem. 2011, 286, 22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Amo J. M. L.; Fink U.; Dasari M.; Grelle G.; Wanker E. E.; Bieschke J.; Reif B. J. Mol. Biol. 2012, 421, 517. [DOI] [PubMed] [Google Scholar]
- Tay W. M.; Huang D.; Rosenberry T. L.; Paravastu A. K. J. Mol. Biol. 2013, 425, 2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y.; Brender J. R.; Soper M. T.; Krishnamoorthy J.; Zhou Y.; Ruotolo B. T.; Kotov N. A.; Ramamoorthy A.; Marsh E. N. G. Biochemistry 2013, 52, 1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinar D. P.; Balija M. B. G.; Kuegler S.; Opazo F.; Rezaei-Ghaleh N.; Wender N.; Kim H.-Y.; Taschenberger G.; Falkenburger B. H.; Heise H.; Kumar A.; Riedel D.; Fichtner L.; Voigt A.; Braus G. H.; Giller K.; Becker S.; Herzig A.; Baldus M.; Jaeckle H.; Eimer S.; Schulz J. B.; Griesinger C.; Zweckstetter M. EMBO J. 2009, 28, 3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Gallo G.; Korsak M.; Luchinat C.; Mao J.; Ravera E. ChemBioChem 2013, 14, 1891. [DOI] [PubMed] [Google Scholar]
- Lesne S.; Koh M. T.; Kotilinek L.; Kayed R.; Glabe C. G.; Yang A.; Gallagher M.; Ashe K. H. Nature 2006, 440, 352. [DOI] [PubMed] [Google Scholar]
- Yu L. T.; Edalji R.; Harlan J. E.; Holzman T. F.; Pereda L.; Labokovsky B.; Hillen H.; Barghorn S.; Ebert U.; Richardson P. L.; Miesbauer L.; Solomon L.; Bartley D.; Walter K.; Johnson R. W.; Hajduk P. J.; Ojejniczak E. T. Biochemistry 2009, 48, 1870. [DOI] [PubMed] [Google Scholar]
- Roher A. E.; Chaney M. O.; Kuo Y. M.; Webster S. D.; Stine W. B.; Haverkamp L. J.; Woods A. S.; Cotter R. J.; Tuohy J. M.; Krafft G. A.; Bonnell B. S.; Emmerling M. R. J. Biol. Chem. 1996, 271, 20631. [DOI] [PubMed] [Google Scholar]
- Shankar G. M.; Li S. M.; Mehta T. H.; Garcia-Munoz A.; Shepardson N. E.; Smith I.; Brett F. M.; Farrell M. A.; Rowan M. J.; Lemere C. A.; Regan C. M.; Walsh D. M.; Sabatini B. L.; Selkoe D. J. Nat. Med. 2008, 14, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein S. L.; Dupuis N. F.; Lazo N. D.; Wyttenbach T.; Condron M. M.; Bitan G.; Teplow D. B.; Shea J. E.; Ruotolo B. T.; Robinson C. V.; Bowers M. T. Nat. Chem. 2009, 1, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W.; Yau W.-M.; Luo Y.; Mattson M. P.; Tycko R. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A.; Matsumura S.; Dezawa M.; Tada M.; Yanazawa M.; Ito A.; Akioka M.; Kikuchi S.; Sato M.; Ideno S.; Noda M.; Fukunari A.; Muramatsu S.; Itokazu Y.; Sato K.; Takahashi H.; Teplow D. B.; Nabeshima Y.; Kakita A.; Imahori K.; Hoshi M. J. Biol. Chem. 2009, 284, 32895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R. Q. Rev. Biophys. 2006, 39, 1. [DOI] [PubMed] [Google Scholar]
- Matsumura S.; Shinoda K.; Yamada M.; Yokojima S.; Inoue M.; Ohnishi T.; Shimada T.; Kikuchi K.; Masui D.; Hashimoto S.; Sato M.; Ito A.; Akioka M.; Takagi S.; Nakamura Y.; Nemoto K.; Hasegawa Y.; Takamoto H.; Inoue H.; Nakamura S.; Nabeshima Y.; Teplow D. B.; Kinjo M.; Hoshia M. J. Biol. Chem. 2011, 286, 11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi Y.; Sakaguchi G.; Becker A. G.; Gray A. J.; Duff K.; Aisen P. S.; Yamaguchi H.; Maeda M.; Kinoshita N.; Matsuoka Y. Biochem. Biophys. Res. Commun. 2004, 319, 733. [DOI] [PubMed] [Google Scholar]
- Takegoshi K.; Nakamura S.; Terao T. J. Chem. Phys. 2003, 118, 2325. [Google Scholar]
- Paravastu A. K.; Qahwash I.; Leapman R. D.; Meredith S. C.; Tycko R. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


