Abstract
Autism spectrum disorder (ASD) comprise a group of neurodevelopmental disorders characterized by deficits in social and communication capacities and repetitive behaviors. Increasing neuroscientific evidence indicates that the neuropathology of ASD is widespread and involves epigenetic regulation in the brain. Differentially expressed miRNAs in the peripheral blood from autism patients were identified by high-throughput miRNA microarray analyses. Five of these miRNAs were confirmed through quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. A search for candidate target genes of the five confirmed miRNAs was performed through a Kyoto encyclopedia of genes and genomes (KEGG) biological pathways and Gene Ontology enrichment analysis of gene function to identify gene regulatory networks. To the best of our knowledge, this study provides the first global miRNA expression profile of ASD in China. The differentially expressed miR-34b may potentially explain the higher percentage of male ASD patients, and the aberrantly expressed miR-103a-3p may contribute to the abnormal ubiquitin-mediated proteolysis observed in ASD.
Introduction
Autism spectrum disorder (ASD) comprise a group of chronic neurodevelopmental disorders characterized by social and language impairments and restricted and repetitive interests and behaviors [1]. ASD includes autistic disorder, Asperger's syndrome, and pervasive developmental disorder not otherwise specified (PDD-NOS)[2]. Patients with ASD vary greatly in terms of clinical manifestation and may also show associated medical comorbidities. Symptoms begin to appear at the age of three years, and affected individuals often require constant care from family members or professionals [3–5]. The Autism and Developmental Disabilities Monitoring (ADDM) Network calculated the prevalence of 8-year-old ASD as 14.7% based on the data from 11 ADDM Network sites in the USA obtained in 2010 [6]. The prevalence rates have increased significantly in the past decade, which may be due to the comprehensive effect of prenatal risk factors, environmental pollution, and heritable factors. It is one of the most heritable of the common neurodevelopmental diseases. Until now, it has been difficult to understand how diverse genetic susceptibilities translate to a clinical phenotype with so many genomic loci contributing to heterogeneous functions [7]. Thus far, studies have focused more on genetic factors, but only a few studies have investigated the role of miRNA in autism spectrum disorder [8–10].
MicroRNAs (miRNAs) are small endogenous non-coding regulatory RNAs (typically 21–23 nucleotides) that function as posttranscriptional regulators of gene expression [11, 12]. These are known to play a critical role in neurodevelopment, metabolism, neuroplasticity, apoptosis, and other fundamental neurobiological processes. By complementarily base-pairing with the 3’untranslated region (3’UTR) of specific target mRNAs, miRNA can regulate gene expression [13, 14]. Recent studies have demonstrated that miRNAs are present in human body fluids, such as serum, plasma, and cerebrospinal fluid. [15–18]. Several recent studies have reported differential miRNA expression in autism spectrum disorder. However, no specific study of regulatory miRNA expression and the gene regulatory networks of aberrant miRNAs has focused on autism spectrum disorder in China. Therefore, we performed the first investigation of the miRNA expression profile in the peripheral blood of Chinese autism patients and the gene regulatory networks of aberrant miRNAs.
Materials and Methods
Sample collection and RNA purification
5 autism patients (1 female and 4 males; mean age: 4.9±1.917) and 5 controls were recruited in microarray analysis study and another 15 patients (2 females and 13 males; mean age: 4.3±1.623) and 15 controls were recruited in qRT-PCR analysis study. The diagnosis of ASD was established in the patients with the use of the Diagnostic and Statistical Manual, Fourth Edition, Text Revision (DSM-IV-TR; American Psychiatric Association, 2000) criteria. Patients show typical ASD clinical symptoms, including deficits in communication and restricted patterns of interests or activities, while social deficits are observed in the school-age patients. Peripheral blood samples were obtained from participants (5ml blood for each) at Xiangya Hospital, and written informed consents were obtained from the parents of all of the participants because the participants were underage and did not have the capability to provide signed written informed consent. This study was approved by the Institutional Review Board and the Ethics Committee at Xiangya Hospital, Central South University (Changsha, China).
The total RNA was isolated using the Trizol method (Life Technologies, USA) according to the manufacturer’s instructions. The quality of RNA was examined using an Agilent Bioanalyzer (Santa Clara, CA, USA). The RNA quantity and purity were assessed using a K5500 micro-spectrophotometer (Kaiao, China). Values of A260/A28≥1.5 and A260/A230≥1 indicate acceptable RNA purity, and a value of RIN (RNA Integrity Number)≥7 obtained through the Agilent 2200 RNA assay indicates acceptable RNA integrity (Agilent, USA). Genomic DNA contamination was evaluated by gel electrophoresis. The total RNA was stored at -80°C until use.
miRNA expression profiling by microarray
RiboArray miDETECT Human Array (A10101-1-12-19, 1×12K) microarrays were used to screen the miRNA expression profile (RIBOBIO, China). The microarrays contained 2,578 assay probes corresponding to the entire set of annotated human and nonhuman primate miRNA sequences from miRBase20.0. The internal control of the microarray assay was from RiboArray internal controls database referenced from multiple public probe database including Exiqon miRCURY LNA, GeneChip miRNA Array and Agilent miRNA. In this study, five autism patients aged 2.5 to 7 years of age and five age-matched controls were enrolled, and all of these were used as unique samples. For hybridization, 2.5 μg of total RNA labeled with Cy5 was used for each sample. To yield adequate microarray results, the labeling efficiency should be between 1.0 and 3.6. After overnight hybridization and washing, the fluorescence images were scanned using a GenePix 4000B laser scanner (Molecular Device, USA) and digitized using the R software. After normalization using the quantile normalization method [19], the p-values were calculated with the Rank Product Method [20]. miRNAs with p<0.05 were selected for cluster analysis. The raw data is available at Gene Expression Omnibus (accession number: GSE67979).
miRNA qRT-PCR analysis
The total RNA was reverse transcribed to cDNA in a volume of 20 μL according to the manufacturer’s instructions. The five most differentially expressed miRNAs in the 15 autism patients and 15 health controls were screened for further validation by Bulge-Loop miRNA qRT-PCR assays (CFX96, Bio-Rad, USA). Has-miR-16-5p was used as an internal reference in the qRT-PCR experiments [15]. The miRNA qRT-PCR Primer Set (RiboBio Co. Guangzhou, China) was used to detect and quantify the expression of the five microRNAs (let-7a-5p, let-7d-5p, miR-34b-3p, miR-103a-3p, and miR-1228-3p) according to the manufacturer’s instructions [21]. Every qRT-PCR assay was performed in triplicate in a volume of 20 μL containing 1 μL of cDNA. The miRNA expression level was calculated using the 2-ΔΔCt method [11].
Investigation of gene regulatory networks based on miRNA expression
To validate the five most differentially expressed miRNAs validated by qRT-PCR analysis, several specific algorithms have been designed to predict the target genes of miRNAs. We first used four publicly available databases, namely TargetScan, miRanda, CLIP-Seq, and miRDB, for the prediction of potential target genes. Only those candidates that appeared in at least three databases were cinsidered candidate miRNA target genes. A Kyoto encyclopedia of genes and genomes (KEGG) biological pathway (http://www.genome.jp/) analysis and a Gene Ontology enrichment analysis of gene function were used to identify the gene networks that may be modulated by the dysregulated miRNAs. Subsequently, miRNA transcription factors were identified through Chip-Seq next-generation genome sequencing and based on data included in the ENCODE and GEO databases [22, 23]. Integrated miRNA regulatory networks were then constructed based on the above analysis.
Statistical analysis
The microarray data were normalized using a quantile normalization method. The p-values were calculated with the Rank Product Method [19, 20], and differences with p< 0.01 were considered statistically significant. For analysis of the qRT-PCR results, a Wilcoxon rank-sum test was used, and differences with p< 0.05 was considered statistically significant [11]. Fisher’s exact test (p< 0.05) was used to identify significant biological pathways, and the false discovery rates (FDR) were used as significant corrections [24]. A hypergeometric distribution was used for the Gene Ontology enrichment analysis of gene function, and p< 0.01 was considered statistically significant [25].
Result
Microarray assay of dysregulated miRNA expression in autism spectrum disorder
The RNA extracted from the peripheral blood was analyzed using a RiboArray miDETECT Human Array. After normalization with the quantile normalization method, the hybridization data revealed 24 upregulated and 20 downregulated miRNAs (Table 1; more details in S1 Table). Interestingly, the dysregulated miR-15a and miR-15b previously discovered in autistic cerebellar cortices were found in this study to be also dysregulated in the peripheral blood [9]. miR-103, miR-92, and miR-19-2 have been reported to be aberrantly expressed in autistic lymphoblasts (Table 2), whereas the related miR-103a-3p, miR-92a-3p and miR-19b-3p were found to be abnormally expressed in autistic peripheral blood [8, 10, 26]. Thus, the differential expression of these miRNAs in the peripheral blood may partially reflect systemic changes in ASD.
Table 1. The differentially expressed miRNAs obtained from the microarray data a .
| Up-regulated miRNAs | Down-regulated miRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA ID | Accession numberb | Mean ± SD | p-value | miRNA ID | Accession numberb | Mean ± SD | p-value | ||
| controls | patients | controls | patients | ||||||
| hsa-miR-1273c | MIMAT0015017 | 10.815±1.807 | 12.326±1.528 | 0 | hsa-miR-451a | MIMAT0001631 | 13.097±1.846 | 12.108±1.518 | 1.48E-05 |
| hsa-miR-4299 | MIMAT0016851 | 11.472±1.572 | 12.776±0.801 | 2.96E-05 | hsa-miR-16-5p | MIMAT0000069 | 14.303±1.807 | 13.610±2.370 | 0.000751 |
| hsa-miR-5739 | MIMAT0023116 | 10.619±1.356 | 11.541±2.344 | 0.000336 | hsa-miR-940 | MIMAT0004983 | 12.604±0.781 | 11.832±1.196 | 0.000914 |
| hsa-miR-6086 | MIMAT0023711 | 12.118±0.749 | 13.092±0.393 | 0.000479 | hsa-miR-574-3p | MIMAT0003239 | 13.543±1.213 | 12.967±1.083 | 0.001201 |
| hsa-miR-494 | MIMAT0002816 | 13.875±2.658 | 14.836±1.261 | 0.000687 | hsa-let-7d-5p | MIMAT0000065 | 12.501±1.392 | 11.774±1.540 | 0.001215 |
| hsa-miR-4270 | MIMAT0016900 | 14.535±0.369 | 15.402±0.295 | 0.000805 | hsa-let-7a-5p | MIMAT0000062 | 12.607±1.515 | 12.005±1.543 | 0.001448 |
| hsa-miR-642a-3p | MIMAT0020924 | 11.565±0.856 | 12.497±0.394 | 0.00081 | hsa-let-7f-5p | MIMAT0000067 | 11.268±1.840 | 10.931±1.573 | 0.001882 |
| hsa-miR-4516 | MIMAT0019053 | 12.816±2.250 | 13.213±2.188 | 0.001107 | hsa-miR-92a-3p | MIMAT0000092 | 11.667±0.915 | 11.041±1.159 | 0.003172 |
| hsa-miR-4436a | MIMAT0018952 | 10.036±0.814 | 10.981±0.136 | 0.001546 | hsa-miR-3613-3p | MIMAT0017991 | 11.611±0.516 | 11.029±0.540 | 0.004125 |
| hsa-miR-1246 | MIMAT0005898 | 11.168±1.758 | 12.040±0.808 | 0.002055 | hsa-miR-20a-5p | MIMAT0000075 | 12.332±1.069 | 11.799±0.993 | 0.004466 |
| hsa-miR-575 | MIMAT0003240 | 11.310±0.485 | 12.085±0.340 | 0.002821 | hsa-miR-1228-3p | MIMAT0005583 | 12.391±1.506 | 12.095±1.673 | 0.005944 |
| hsa-miR-4721 | MIMAT0019835 | 11.715±0.259 | 12.376±0.544 | 0.003765 | hsa-miR-3935 | MIMAT0018350 | 14.062±1.749 | 13.863±1.483 | 0.006354 |
| hsa-miR-483-5p | MIMAT0004761 | 11.676±1.349 | 12.407±0.485 | 0.004377 | hsa-miR-4700-3p | MIMAT0019797 | 11.320±0.338 | 10.762±0.225 | 0.006453 |
| hsa-miR-1249 | MIMAT0005901 | 11.148±0.327 | 11.549±1.296 | 0.004407 | hsa-miR-15b-5p | MIMAT0000417 | 11.050±1.161 | 10.733±1.234 | 0.006527 |
| hsa-miR-4443 | MIMAT0018961 | 9.865±0.774 | 10.524±0.494 | 0.004501 | hsa-miR-15a-5p | MIMAT0000068 | 11.759±1.233 | 11.300±1.294 | 0.006942 |
| hsa-miR-921 | MIMAT0004971 | 10.094±0.352 | 10.745±0.427 | 0.005608 | hsa-miR-4436b-5p | MIMAT0019940 | 11.854±0.924 | 11.438±0.585 | 0.007426 |
| hsa-miR-34b-3p | MIMAT0004676 | 10.272±1.012 | 10.779±0.696 | 0.005909 | hsa-miR-4665-5p | MIMAT0019739 | 11.620±1.406 | 11.521±1.133 | 0.007441 |
| hsa-miR-6125 | MIMAT0024598 | 13.084±2.164 | 13.256±1.945 | 0.006018 | hsa-miR-19b-3p | MIMAT0000074 | 11.257±1.140 | 10.889±1.059 | 0.008103 |
| hsa-miR-4669 | MIMAT0019749 | 11.311±0.407 | 11.954±0.842 | 0.006897 | hsa-miR-103a-3p | MIMAT0000101 | 12.390±1.097 | 11.965±1.118 | 0.009121 |
| hsa-miR-34c-3p | MIMAT0004677 | 10.318±0.923 | 10.887±0.795 | 0.007624 | hsa-miR-195-5p | MIMAT0000461 | 10.982±1.699 | 10.901±1.378 | 0.009279 |
| hsa-miR-4728-5p | MIMAT0019849 | 11.756±0.617 | 12.280±0.495 | 0.008063 | |||||
| hsa-miR-564 | MIMAT0003228 | 10.076±0.278 | 10.679±0.497 | 0.00831 | |||||
| hsa-miR-574-5p | MIMAT0004795 | 10.807±1.760 | 11.333±0.890 | 0.00873 | |||||
| hsa-miR-4788 | MIMAT0019958 | 12.324±0.978 | 12.940±0.483 | 0.009634 | |||||
a: More details are presented in S1 Table.
b: Accession numbers were obtained from miRBase database.
Table 2. The results of the present study vs. previous autism microRNA studies.
| miRNA ID | Present result | Previous report | Type of sample | Reference |
|---|---|---|---|---|
| hsa-miR-16-5p | down-regulated | down-regulated | lymphoblastoid cell lines | Sarachana et al. [8] |
| hsa-miR-103 | down-regulated | up-regulated | lymphoblastoid cell lines | Sarachana et al. [8] |
| hsa-miR-451 | down-regulated | up-regulated | lymphoblastoid cell lines | Sarachana et al. [8] |
| miR-19b-3p | down-regulated | up-regulated | serum | Mundalil et al. [26] |
| miR-195-5p | down-regulated | up-regulated | serum | Mundalil et al. [26] |
| lymphoblastoid cell lines | Sarachana et al. [8] | |||
| hsa-miR-15a | down-regulated | up-regulated | cerebellar cortex | Abu-Elneel et al. [9] |
| hsa-miR-15b | down-regulated | up-regulated | cerebellar cortex | Abu-Elneel et al. [9] |
| hsa-miR-92a-3p | down-regulated | down-regulated | lymphoblastoid cell lines | Talebizadeh et al. [10] |
A subset of differentially expressed miRNAs was selected for clustering analysis. The correlation of the expression profiles between the biological replicates and treatment conditions was demonstrated through unsupervised hierarchical clustering analysis (S1 File).
qRT-PCR confirmation of the expression of selected miRNAs
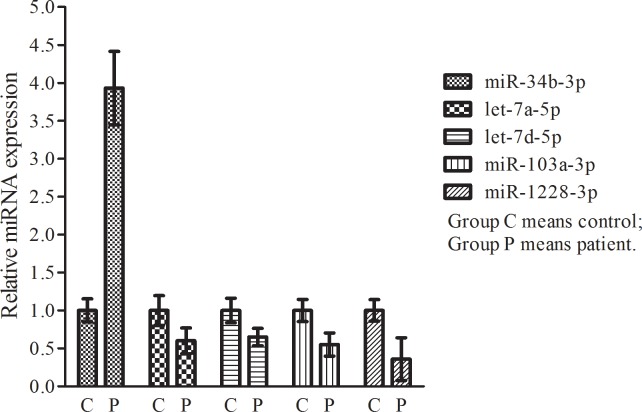
Based on the dysregulated miRNA profiles obtained from the microarray analysis combined with recent research on the function of miRNAs, the five most aberrantly expressed miRNAs were selected for further qRT-PCR validation. As a result, the miR-34b expression level was significantly increased, whereas the levels of miR-let-7a, miR-let-7d, miR-103a, and miR-1228 were decreased (Fig 1, p< 0.05).
Fig 1. qRT-PCR confirmation of the expression of selected miRNAs.
The levels of the five differentially expressed five miRNAs were statistically significant. The miR-34b expression level was significantly increased, whereas the levels of miR-let-7a, miR-let-7d, miR-103a, and miR-1228 were decreased.
Investigation of gene regulatory networks based on miRNA expression
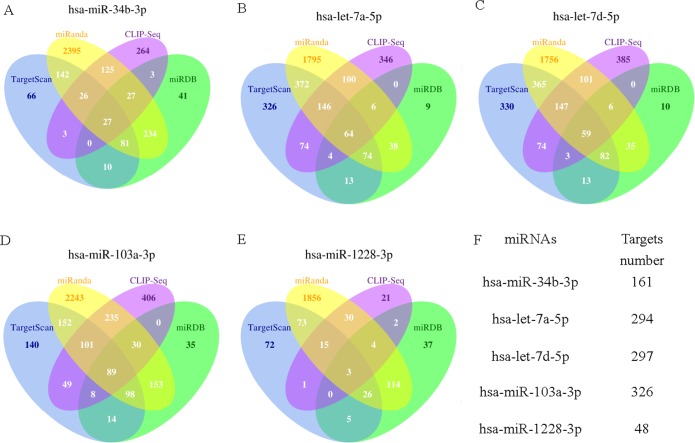
It has been predicted that the regulation of miRNAs is not a simple process. More than one target in different functional categories can be modulated by one miRNA, and more than one miRNA may target the same specific gene [10, 18]. Thus, the statistical data for the miRNA candidate target genes predicted by the four network databases (TargetScan, miRanda, CLIP-Seq, and miRDB) are presented in Fig 2. The Kyoto encyclopedia of genes and genomes (KEGG) biological pathway database was employed to extract the most relevant target genes on different biological pathways to predict the underlying connections among the target genes and their associated functions. The three most significant pathways for the five miRNAs were taken into account (Table 3). A Gene Ontology enrichment analysis of gene function was performed to fully explore the specific functions of the target genes. Interestingly, the results generated by the KEGG and GO analyses indicated correlations between the candidate targets of the miRNAs and autism and revealed other neurological functions previously and newly reported to be related to ASD, such as circadian rhythm-mammal [27], ubiquitin-mediated proteolysis (UPS) [28], dopaminergic synapse [29, 30], MAPK signaling pathway, development of primary male sexual characteristics and male sex differentiation in male gonad development.
Fig 2. Prediction of target genes.
Four databases were used for the prediction of target genes: TargetScan, miRanda, CLIP-Seq, and miRDB. Venn Diagrams A, B, C, D, and E show the intersection of each miRNA target gene predicted by various databases, and intersection of the target genes obtained from at least three databases. F indicates the total number of candidate target genes for each miRNA.
Table 3. Kyoto encyclopedia of genes and genomes (KEGG) biological pathway analysis for the five miRNAs.
| miRNA | Term | ID a | P-Value | Genes b |
|---|---|---|---|---|
| hsa-let-7a-5p | p53 signaling pathway | hsa04115 | 0.006623744 | RRM2,MDM4,CDKN1A,CASP3,THBS1 |
| MAPK signaling pathway | hsa04010 | 0.00890767 | NGF,CASP3,CHUK,MAP4K4,DUSP1,PLA2G3,DUSP4, | |
| NRAS,MAP4K3,GFBR1,PAK1 | ||||
| TGF-beta signaling pathway | hsa04350 | 0.017204351 | GDF6,E2F5,TGFBR1,THBS1,ACVR1C | |
| hsa-let-7d-5p | Protein digestion and absorption | hsa04974 | 0.001653889 | COL1A2,COL3A1,COL4A1,COL24A1,OL1A1,COL14A1, |
| SLC16A10 | ||||
| ECM-receptor interaction | hsa04512 | 0.007451271 | COL1A2,COL3A1,COL4A1,COL24A1,THBS1,COL1A1 | |
| MAPK signaling pathway | hsa04010 | 0.00776506 | NGF,CASP3,CHUK,MAP4K4,DUSP1,PLA2G3,DUSP4, | |
| MAP4K3,NRAS,FAS,TGFBR1,PAK1 | ||||
| hsa-miR-34b-3p | Lysosome | hsa04142 | 0.003264022 | LAMP1,AP3D1,GNS,M6PR,SCARB2 |
| RIG-I-like receptor signaling pathway | hsa04622 | 0.020271307 | AZI2,DDX3X,CYLD | |
| Tight junction | hsa04530 | 0.024135052 | MRAS,MLLT4,CTNNA1,CASK | |
| hsa-miR-103a-3p | Ubiquitin mediated proteolysis | hsa04120 | 0.001387411 | BTRC,FBXW7,UBE4A,HERC2,UBE2J1,CUL4A,WWP1, |
| CDC23,UBE2Q1 | ||||
| Circadian rhythm-mammal | hsa04710 | 0.002514562 | NPAS2,PRKAB2,BTRC,CLOCK | |
| Dopaminergic synapse | hsa04728 | 0.00395313 | KIF5A,PPP2R3C,KIF5C,ITPR1,SCN1A,CLOCK,PPP2R5C,PLCB1 | |
| hsa-miR-1228-3p | Adherens junction | hsa04520 | 0.012562355 | PTPN1,TJP1 |
| Insulin signaling pathway | hsa04910 | 0.043243871 | PTPN1,TSC1 | |
| Dorso-ventral axis formation | hsa04320 | 0.05440918 | ETV6 |
a: The ID numbers were obtained from the KEGG Pathway database.
b: The genes indicate the target genes in the pathway.
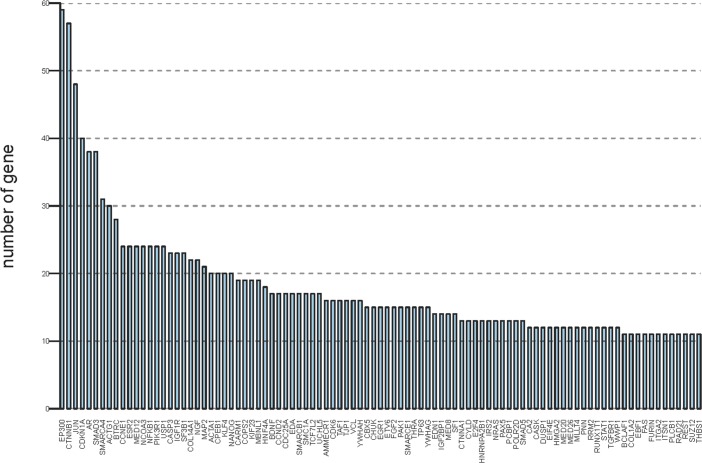
We also included miRNA transcription factors in our study. Therefore, based on the above-described results, the S2 File shows the complex and integrated miRNA regulatory networks, which present the relationship between the miRNAs and mRNAs and the relationship between the miRNAs and miRNA transcription factors. Additionally, we calculated the number of associated genes for both candidate target genes and transcription factor genes in the miRNA regulatory interaction networks and ranked the number of associated genes, which was greater than 10 and is shown in Fig 3.
Fig 3. Importance of target genes and transcription factor genes.
The horizontal axis indicates the names of target genes and transcription factor genes, and the vertical axis indicates the numbers of associated genes. The ranking was performed according to the number of associated genes, more than 10 of which could be shown.
Discussion
Autism spectrum disorder (ASD) is one of the most heritable neurodevelopmental diseases with a markedly increasing incidence in China. The characteristics of ASD include repetitive behaviors accompanying deficits in social interaction and communication [31]. Because the diagnosis of ASD mainly relies on psychometric evaluations, its diagnosis is often difficult and subjective. Therefore, molecular evidence for early and objective ASD diagnosis particularly non-invasive miRNA/gene biomarkers from the peripheral blood and genetic associations or interactions, is indispensable.
As described above, miRNAs are small non-coding regulatory RNAs that play an important role in fundamental neurobiological processes, particularly neurodevelopmental diseases. In Rett syndrome, multiple aberrantly expressed miRNAs, including miR-30a/d, miR-381, and miR-495, negatively regulate target gene translation [32]. In Down syndrome samples, five miRNAs (miR-155, miR-802, miR-125b-2, let-7c, and miR-99a) have been found to be overexpressed in fetal hippocampus and exclusively expressed in neurons [33]. In Fragile X syndrome, miR-367 negatively regulates FXR1P expression in human cell lines, and a loss of FXR1P downregulates brain-specific miRNAsthat are critical in neurodevelopment [34, 35].
A series of recent studies have shown that miRNAs in serum and plasma are considered biomarkers for several diseases [36]. Additionally, several studies have identified the expression profiles of miRNAs in ASD, and some of these studies presented data from lymphoblastoid cell cultures [5, 8, 10]. One of these studies included the analysis of samples from the cerebella of ASD patients [9]. We explored the miRNA expression profiles and the gene regulatory networks of aberrantly expressed miRNAs in autism spectrum disorder in China. In this study, five miRNAs (miR-103a, miR-34b, miR-let-7a, miR-let-7d, and miR-1228) were confirmed to be aberrantly expressed in the peripheral blood of ASD patients.
miR-34 modulates aging and neurodegeneration in Drosophila (dme-mir-34 and has-mir-34b are orthologous) [37], and miR-34b is elevated in spinocerebellar ataxia type 3/Machado-Joseph disease (SCA3/MJD) as demonstrated in our previous study [11]. These data illustrate the roles of miR-34b in both neurodevelopmental and neurodegenerative diseases. Mutiple results obtained in the current study indicate a relationship between miR-34b and ASD. The candidate target genes of miR-34b are mainly associated with central nervous system neuron development, long-term memory and tight junction pathways whose breakdown may result in diseases such as systemic inflammatory response syndrome (SIRS), inflammatory bowel disease, and autism [38]. Additionally, the strong association between the candidate target genes of miR-34b and ASD implies that miR-34b may play roles in the physiopathology of ASD. A linkage between autism and the EIF4E region on chromosome 4q has been found in genome-wide linkage studies [39], and functional variants modulating EIF4E expression have been previously indicated as risk factors for ASD [40]. Dysregulation of eukaryotic translation initiation factor 4E (eIF4E)-dependent translational control may contribute to ASD-like phenotypes [41]. MAP2 encodes a protein that is enriched in dendrites and is implicated in determining and stabilizing dendritic shape during neuron development. Depleted MAP2 neuronal expression and reduced dendrite numbers have been found in autistic adults [42]. In addition, the MAP2 gene has been identified a good candidate for the generation of a preserved speech variant (PSV) in a patient with autism and Rett-like features [43]. Interestingly, the candidate target genes of miR-34b also affect the development of primary male sexual characteristics, male sex differentiation and male gonad development, which may explain the higher percentage of males in the ASD group.
miR-103 and miR-107 have been reported to be highly expressed in brain tissue [44]. Sarachana et al. previously hypothesized that the dysregulation of miR-103/7 may result in abnormal lipid and fatty acid metabolism in ASD [8]. Several findings of our study provide plausible evidence that miR-103a-3p plays a role in the physiopathological mechanism of ASD. The candidate target genes of miR-103a-3p reveal close relationships with the dysfunctional physiological pathways related to ASD, such as central nervous system development, neuron projection morphogenesis and skeletal muscle tissue and organ development, and with various pathways, including ubiquitin-mediated proteolysis (UPS), circadian rhythm-mammal [8, 26] and dopaminergic synapse [28, 29]. Directly, HECT-type E3 ubiquitin ligase (UBE3A) dysfunction appears to induce a large spectrum of pediatric cognitive dysfunctions, such as autism and Rett and Angelman syndromes, depending on the mechanism and severity of UBE3A loss [27]. Indirectly, UPS is involved in neural functions and co-morbid diseases with ASD, such as synaptic dysfunctions and muscle diseases. Ubiquitination is a highly specific method for manipulating protein expression at the neural synapse [45, 46] and overloading or other perturbations of ubiquitin-dependent proteolysis is likely involved in inclusion myopathies [27]. Based on the results of various previous studies [8], we hypothesized that the aberrantly expressed miR-103a-3p may contribute to abnormal ubiquitin-mediated proteolysis in ASD. Specifically, the candidate target gene BTRC is involved not only in the mammal circadian rhythm and mammal and ubiquitin-mediated proteolysis but also in the Wnt signaling pathway [47], all of which have been acknowledged to play roles in ASD. Additionally, one candidate target gene of miR-103a-3p, BDNF, encodes brain-derived neurotrophic factor (BDNF). Increasing evidence suggests a direct or indirect involvement of BDNF in autism [48], and this factor plays a key role in neuronal survival, morphology, differentiation and synaptic strength [49]. It is increased in some ASD patients as well as in some animal models of ASD [50, 51].
Other candidate miRNAs have also associated with neuronal physiology and pathology. A previous study suggested that embryonic development may be relevant to ASD [8]. In addition, miR-let-7a expression have been found to be high in early human embryonic tissues and abruptly decreases thereafter, which suggests that miR-let-7a plays critical roles in early embryonic development [52]. Therefore, miR-let-7a may be indirectly associated with ASD through the embryonic development process. miR-let-7d suppresses neural stem cell proliferation by reducing TLX expression in neural stem cells, which may be a novel strategy for identifying potential interventions in relevant neurological diseases [53]. Notably, the candidate target genes of both miR-let-7a and miR-let-7d are associated with the MAPK signaling pathway, which directly or indirectly play roles in the physiopathology of ASD. Ghahramani Seno et al. previously suggested that mitogen-activated protein kinases (MAPK) may be involved in the development of autism [5]. In addition, several genes in the MAPK signaling pathway have been reported to be relevant to ASD. NGF encodes nerve growth factor (NGF) and influences inflammatory responses and neuron projection development. The serum NGF concentrations have been reported to be significantly higher in autistic children [54]. Another analysis suggests that NGF is a potential risk gene for nonverbal communication (NVC) impairments [55]. Recent studies have implied an association between mitochondrial dysfunction and ASD [56]. The protein levels of caspase-3, which is encoded by CASP3, are also increased in ASD patients [57]. NRAS plays roles in the regulation of long-term neuronal synaptic plasticity and transmission and is considered a candidate gene for autism. This finding may provide a novel insight into the etiology of autism [58].
Interestingly, the candidate target genes of miR-1228-3p were found to not be involved in brain-specific or brain-related functions but involved in the process of hepatocyte growth factor (HGF) production. A recent study reported that decreased HGF levels are found in autistic children and suggested that the serum HGF concentration may be a useful biomarker of autistic children [59]. Additionally, our findings show an association between the miR-1228-3p target genes and the insulin signaling pathway, which are hypothesized to contribute to the development of autism in genetically susceptible individuals by promoting PI3K/Tor pathway activation in neurons [60].
Taken together, four of the five aberrantly expressed miRNAs are related to the brain, suggesting that the gene expression differences observed in the peripheral blood may indicate similar alterations in the brain that are likely attribute to a system-scale differential expression of miRNAs.
Conclusion
In summary, we investigated the global miRNA expression profiles of ASD in China, confirming that the use of alternative and more easily accessible peripheral blood for the study of miRNA expression in ASD is valid. The differentially expressed miR-34b may potentially explain why ASD is markedly more common in males than in females. The aberrantly expressed miR-103a-3p may contribute to abnormal ubiquitin-mediated proteolysis in ASD. Thus, these five miRNAs are beneficial for better understanding ASD-specific miRNA expression profiles in peripheral blood and provide new insights into future researches for ASD.
Supporting Information
(XLSX)
An unsupervised hierarchical cluster analysis of 150 significantly differentially expressed miRNAs among all of the autistic individuals (A6, A357, A14, A15, A16) and controls (4, 567, 28, 29, 30) shows the distinct miRNA expression pattern of the two groups (p < 0.05).
(PDF)
The image shows not only the relationship between miRNAs and mRNAs or transcriptional factors (TFs) but also the interactive relationships between the proteins of the target genes. Orange rhombi: miRNAs, blue circles: mRNAs, aubergine circles: TFs, pink lines: miRNA-mRNA interactions, blue lines: mRNA-TF interactions, green lines: miRNA-TF interactions.
(PDF)
Acknowledgments
The authors are grateful to all of the subjects for their participation in our study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Basic Research Program (973 Program) (Nos. 2012CB944601, 2012CB517902 and 2011CB510002 to Hong Jiang), the National Natural Science Foundation of China (Nos. 81410308019, 81471156, 81271260, 30971585 to Hong Jiang; No 31401135 to Rong Qiu), Hunan Funds for Distinguished Young Scientists (No. 14JJ1008 to Hong Jiang), Xinjiang Natural Science Foundation (No. 201318101-4 to Hong Jiang), Natural Science Foundation of Hunan Province (No 2012SK3194 to Wei Zhuang), the Undergraduate Innovation Project of Central South University (No. YB13028, 201410533324 to Hong Jiang), Graduate Innovation Project of Central South University (No. 2014zzts078 to Zhao Chen), Research projects of the First People's Hospital of Chenzhou (No. N2014-017, to Fengzhen Huang) and High-level medical personnel of Hunan province “225” Project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kolevzon A, Gross R, Reichenberg A (2007) Prenatal and perinatal risk factors for autism: a review and integration of findings. Arch Pediatr Adolesc Med 161(4): 326–333. [DOI] [PubMed] [Google Scholar]
- 2. Dawson S, Glasson EJ, Dixon G, Bower C (2009) Birth defects in children with autism spectrum disorders: a population-based, nested case-control study. Am J Epidemiol 169(11): 1296–1303. 10.1093/aje/kwp059 [DOI] [PubMed] [Google Scholar]
- 3. Goyal DK, Miyan JA (2014) Neuro-immune abnormalities in autism and their relationship with the environment: a variable insult model for autism. Front Endocrinol (Lausanne) 5: 29 10.3389/fendo.2014.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeJong H, Bunton P, Hare DJ (2014) A systematic review of interventions used to treat catatonic symptoms in people with autistic spectrum disorders. J Autism Dev Disord 44(9): 2127–2136. 10.1007/s10803-014-2085-y [DOI] [PubMed] [Google Scholar]
- 5. Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G, et al. (2011) Gene and miRNA expression profiles in autism spectrum disorders. Brain Res 1380: 85–97. 10.1016/j.brainres.2010.09.046 [DOI] [PubMed] [Google Scholar]
- 6. Wingate M, Kirby RS, Pettygrove S, Cunniff C, Schulz E, Ghosh T, et al. (2014) Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ 63(2): 1–21. [PubMed] [Google Scholar]
- 7. Holt R, Monaco AP (2011) Links between genetics and pathophysiology in the autism spectrum disorders. EMBO Mol Med 3(8): 438–450. 10.1002/emmm.201100157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarachana T, Zhou R, Chen G, Manji HK, Hu VW (2010) Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med 2(4): 23 10.1186/gm144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. (2008) Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 9(3): 153–161. 10.1007/s10048-008-0133-5 [DOI] [PubMed] [Google Scholar]
- 10. Talebizadeh Z, Butler MG, Theodoro MF (2008) Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res 1(4): 240–250. 10.1002/aur.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi Y, Huang F, Tang B, Li J, Wang J, Shen L, et al. (2014) MicroRNA profiling in the serums of SCA3/MJD patients. Int J Neurosci 124(2): 97–101. 10.3109/00207454.2013.827679 [DOI] [PubMed] [Google Scholar]
- 12. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2): 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aw S, Cohen SM (2012) Time is of the essence: microRNAs and age-associated neurodegeneration. Cell Res 22(8): 1218–1220. 10.1038/cr.2012.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11(9): 597–610. 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 15. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127(1): 118–126. 10.1002/ijc.25007 [DOI] [PubMed] [Google Scholar]
- 16. Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. (2008) Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis 14(1): 27–41. [DOI] [PubMed] [Google Scholar]
- 17. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. (2008) Serum microRNAs are promising novel biomarkers. PLoS One 3(9): e3148 10.1371/journal.pone.0003148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sevcikova S, Kubiczkova L, Sedlarikova L, Slaby O, Hajek R (2013) Serum miR-29a as a marker of multiple myeloma. Leuk Lymphoma 54(1): 189–191. 10.3109/10428194.2012.704030 [DOI] [PubMed] [Google Scholar]
- 19. Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19(2): 185–193. [DOI] [PubMed] [Google Scholar]
- 20. Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J (2006) RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22(22): 2825–2827. [DOI] [PubMed] [Google Scholar]
- 21. Li YQ, Zhang MF, Wen HY, Hu CL, Liu R, Wei HY, et al. (2013) Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics (Sao Paulo) 68(1): 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, et al. (2011) A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9(4): e1001046 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arvey A, Agius P, Noble WS, Leslie C (2012) Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res 22(9): 1723–1734. 10.1101/gr.127712.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1): 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rivals I, Personnaz L, Taing L, Potier MC (2007) Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23(4): 401–407. [DOI] [PubMed] [Google Scholar]
- 26. Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, et al. (2014) Serum microRNA profiles in children with autism. Mol Autism 5: 40 10.1186/2040-2392-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corbett BA, Mendoza S, Abdullah M, Wegelin JA, Levine S (2006) Cortisol circadian rhythms and response to stress in children with autism. Psychoneuroendocrinology 31(1): 59–68. [DOI] [PubMed] [Google Scholar]
- 28. Lehman NL (2009) The ubiquitin proteasome system in neuropathology. Acta Neuropathol 118(3): 329–347. 10.1007/s00401-009-0560-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, et al. (2014) Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature 511(7508): 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Penzes P, Buonanno A, Passafaro M, Sala C, Sweet RA (2013) Developmental vulnerability of synapses and circuits associated with neuropsychiatric disorders. J Neurochem 126(2): 165–182. 10.1111/jnc.12261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh AS, Chandra R, Guhathakurta S, Sinha S, Chatterjee A, Ahmed S, et al. (2013) Genetic association and gene-gene interaction analyses suggest likely involvement of ITGB3 and TPH2 with autism spectrum disorder (ASD) in the Indian population. Prog Neuropsychopharmacol Biol Psychiatry 45: 131–143. 10.1016/j.pnpbp.2013.04.015 [DOI] [PubMed] [Google Scholar]
- 32. Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, et al. (2010) Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A 107(42): 18161–18166. 10.1073/pnas.1005595107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, et al. (2008) Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun 370(3): 473–477. 10.1016/j.bbrc.2008.03.120 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Cheever A, Blackwell E, Ceman S (2010) Fragile X protein family member FXR1P is regulated by microRNAs. Rna 16(8): 1530–1539. 10.1261/rna.2022210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu XL, Zong R, Li Z, Biswas MH, Fang Z, Nelson DL, et al. (2011) FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J Neurosci 31(39): 13705–13709. 10.1523/JNEUROSCI.2827-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scholer N, Langer C, Dohner H, Buske C, Kuchenbauer F (2010) Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature. Exp Hematol 38(12): 1126–1130. 10.1016/j.exphem.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 37. Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, et al. (2012) The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482(7386): 519–523. 10.1038/nature10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z, Li N, Neu J (2005) Tight junctions, leaky intestines, and pediatric diseases. Acta Paediatr 94(4): 386–393. [DOI] [PubMed] [Google Scholar]
- 39. Neves-Pereira M, Muller B, Massie D, Williams JH, O'Brien PC, Hughes A, et al. (2009) Deregulation of EIF4E: a novel mechanism for autism. J Med Genet 46(11): 759–765. 10.1136/jmg.2009.066852 [DOI] [PubMed] [Google Scholar]
- 40. Waltes R, Gfesser J, Haslinger D, Schneider-Momm K, Biscaldi M, Voran A, et al. (2014) Common EIF4E variants modulate risk for autism spectrum disorders in the high-functioning range. J Neural Transm 121(9): 1107–1116. 10.1007/s00702-014-1230-2 [DOI] [PubMed] [Google Scholar]
- 41. Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. (2013) Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493(7432): 371–377. 10.1038/nature11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mukaetova-Ladinska EB, Arnold H, Jaros E, Perry R, Perry E (2004) Depletion of MAP2 expression and laminar cytoarchitectonic changes in dorsolateral prefrontal cortex in adult autistic individuals. Neuropathol Appl Neurobiol 30(6): 615–623. [DOI] [PubMed] [Google Scholar]
- 43. Pescucci C, Meloni I, Bruttini M, Ariani F, Longo I, Mari F, et al. (2003) Chromosome 2 deletion encompassing the MAP2 gene in a patient with autism and Rett-like features. Clin Genet 64(6): 497–501. [DOI] [PubMed] [Google Scholar]
- 44. Wilfred BR, Wang WX, Nelson PT (2007) Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 91(3): 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwartz AL, Ciechanover A (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 49: 73–96. 10.1146/annurev.pharmtox.051208.165340 [DOI] [PubMed] [Google Scholar]
- 46. Yi JJ, Ehlers MD (2005) Ubiquitin and protein turnover in synapse function. Neuron 47(5): 629–632. [DOI] [PubMed] [Google Scholar]
- 47. Martin PM, Yang X, Robin N, Lam E, Rabinowitz JS, Erdman CA, et al. (2013) A rare WNT1 missense variant overrepresented in ASD leads to increased Wnt signal pathway activation. Transl Psychiatry 3: e301 10.1038/tp.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Halepoto DM, Bashir S, L ALA (2014) Possible role of brain-derived neurotrophic factor (BDNF) in autism spectrum disorder: current status. J Coll Physicians Surg Pak 24(4): 274–278. [PubMed] [Google Scholar]
- 49. Binder DK, Scharfman HE (2004) Brain-derived neurotrophic factor. Growth Factors 22(3): 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koh JY, Lim JS, Byun HR, Yoo MH (2014) Abnormalities in the zinc-metalloprotease-BDNF axis may contribute to megalencephaly and cortical hyperconnectivity in young autism spectrum disorder patients. Mol Brain 7: 64 10.1186/s13041-014-0064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Connolly AM, Chez M, Streif EM, Keeling RM, Golumbek PT, Kwon JM, et al. (2006) Brain-derived neurotrophic factor and autoantibodies to neural antigens in sera of children with autistic spectrum disorders, Landau-Kleffner syndrome, and epilepsy. Biol Psychiatry 59(4): 354–363. [DOI] [PubMed] [Google Scholar]
- 52. Lozoya T, Dominguez F, Romero-Ruiz A, Steffani L, Martinez S, Monterde M, et al. (2014) The Lin28/Let-7 system in early human embryonic tissue and ectopic pregnancy. PLoS One 9(1): e87698 10.1371/journal.pone.0087698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao C, Sun G, Ye P, Li S, Shi Y (2013) MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci Rep 3: 1329 10.1038/srep01329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dincel N, Unalp A, Kutlu A, Ozturk A, Uran N, Ulusoy S (2013) Serum nerve growth factor levels in autistic children in Turkish population: a preliminary study. Indian J Med Res 138(6): 900–903. [PMC free article] [PubMed] [Google Scholar]
- 55. Lu AT, Yoon J, Geschwind DH, Cantor RM (2013) QTL replication and targeted association highlight the nerve growth factor gene for nonverbal communication deficits in autism spectrum disorders. Mol Psychiatry 18(2): 226–235. 10.1038/mp.2011.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rossignol DA, Frye RE (2012) A review of research trends in physiological abnormalities in autism spectrum disorders: immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol Psychiatry 17(4): 389–401. 10.1038/mp.2011.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Siniscalco D, Sapone A, Giordano C, Cirillo A, de Novellis V, de Magistris L, et al. (2012) The expression of caspases is enhanced in peripheral blood mononuclear cells of autism spectrum disorder patients. J Autism Dev Disord 42(7): 1403–1410. 10.1007/s10803-011-1373-z [DOI] [PubMed] [Google Scholar]
- 58. Xia K, Guo H, Hu Z, Xun G, Zuo L, Peng Y, et al. (2014) Common genetic variants on 1p13.2 associate with risk of autism. Mol Psychiatry 19(11): 1212–1219. 10.1038/mp.2013.146 [DOI] [PubMed] [Google Scholar]
- 59. Russo AJ, Krigsman A, Jepson B, Wakefield A (2009) Decreased Serum Hepatocyte Growth Factor (HGF) in Autistic Children with Severe Gastrointestinal Disease. Biomark Insights 4: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stern M (2011) Insulin signaling and autism. Front Endocrinol (Lausanne) 2: 54 10.3389/fendo.2011.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
An unsupervised hierarchical cluster analysis of 150 significantly differentially expressed miRNAs among all of the autistic individuals (A6, A357, A14, A15, A16) and controls (4, 567, 28, 29, 30) shows the distinct miRNA expression pattern of the two groups (p < 0.05).
(PDF)
The image shows not only the relationship between miRNAs and mRNAs or transcriptional factors (TFs) but also the interactive relationships between the proteins of the target genes. Orange rhombi: miRNAs, blue circles: mRNAs, aubergine circles: TFs, pink lines: miRNA-mRNA interactions, blue lines: mRNA-TF interactions, green lines: miRNA-TF interactions.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.