Abstract
Adult antisocial behavior (AAB) is moderately heritable, relatively common and has adverse consequences for individuals and society. We examined the molecular genetic basis of AAB in 1379 participants from a case–control study in which the cases met criteria for alcohol dependence. We also examined whether genes of interest were expressed in human brain. AAB was measured using a count of the number of Antisocial Personality Disorder criteria endorsed under criterion A from the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV). Participants were genotyped on the Illumina Human 1M BeadChip. In total, all single-nucleotide polymorphisms (SNPs) accounted for 25% of the variance in AAB, although this estimate was not significant (P=0.09). Enrichment tests indicated that more significantly associated genes were over-represented in seven gene sets, and most were immune related. Our most highly associated SNP (rs4728702, P=5.77 × 10−7) was located in the protein-coding adenosine triphosphate-binding cassette, sub-family B (MDR/TAP), member 1 (ABCB1). In a gene-based test, ABCB1 was genome-wide significant (q=0.03). Expression analyses indicated that ABCB1 was robustly expressed in the brain. ABCB1 has been implicated in substance use, and in post hoc tests we found that variation in ABCB1 was associated with DSM-IV alcohol and cocaine dependence criterion counts. These results suggest that ABCB1 may confer risk across externalizing behaviors, and are consistent with previous suggestions that immune pathways are associated with externalizing behaviors. The results should be tempered by the fact that we did not replicate the associations for ABCB1 or the gene sets in a less-affected independent sample.
Introduction
Adult antisocial behavior (AAB), which is broadly defined as a ‘pervasive pattern of disregard for and violation of the rights of others',1 is relatively common and is associated with adverse consequences for both individuals and societies. According to prevalence estimates from the 2001–2002 National Epidemiologic Survey on Alcohol and Related Conditions, 3.63% (95% confidence interval=3.34–3.92) of adult Americans qualify for an antisocial personality disorder (ASPD) diagnosis, and men are affected at a greater rate than women.2 Findings from the all-male Vietnam Era Twin Registry indicate that ASPD is highly heritable, with genetic factors accounting for 69% of the variation in diagnoses.3 Twin and adoption studies further indicate that dimensional measures of AAB are moderately heritable, with genetic factors accounting for ~40% of the variation in these behaviors.4, 5, 6 These figures are similar to the results from a meta-analysis of 51 twin and adoption studies from child, adolescent and adult samples, which found that additive genetic influences accounted for 32% of the variance in antisocial behavior.7
A number of studies of AAB have found statistically significant genetic associations or gene-by-environment interactions with measures of early environmental adversity for variants in candidate genes or gene regions (for example, 5-HTTLPR, COMT and MAOA).8, 9, 10 A recent meta-analysis found evidence for associations between antisocial behavior (measured across a range of ages) and variation in the serotonin transporter gene SLC6A4 (that is, the 5-HTTLPR polymorphism) and the monoamine oxidase A (MAOA) gene.11 Although there has been longstanding controversy surrounding failures to replicate and publication bias for candidate gene approaches,12 it was only recently that researchers began to systematically examine how common genetic variation across the genome is associated with AAB. No single-nucleotide polymorphisms (SNPs) met genome-wide significance in the genome-wide association study (GWAS) by Tielbeek et al.13 of non-diagnostic categorical measures of AAB in two Australian samples. The most highly associated gene to emerge from their analysis was DYRK1A (P=8.7 × 10−5) on chromosome 21, which is a candidate gene for developmental disabilities.14 In this same study, attempts to replicate the associations between seven candidate genes or polymorphisms from the literature (DAT1, DRD2, DRD4, 5-HTTLPR, COMT, MAOA and C1QTNF7) and AAB as a categorical variable were unsuccessful. Likewise, studies in a community-based sample reported no significant associations between a closely related behavioral disinhibition phenotype and rare nonsynonymous exonic or common SNPs from across the genome.15, 16
Additional screens of high-risk samples that are likely to be enriched for AAB are needed to detect genetic associations. Our goal in the present study was to fill this gap and examine the molecular genetic basis of AAB in such a sample. We examined SNP heritability of a dimensional measure of AAB, followed by a GWAS, gene-based tests and gene set enrichment tests. Participants were part of a case–control sample in which cases met the criteria for alcohol dependence. Alcohol-use disorders are phenotypically and genetically correlated with AAB and ASPD,17, 18 suggesting that this sample is likely to be enriched for variants contributing to AAB. We then examined whether genes of interest were expressed in human brain to evaluate the biological plausibility that identified genes would be associated with a behavioral outcome.
Materials and methods
Sample and participants
Participants came from the Collaborative Study on the Genetics of Alcoholism (COGA).19 The primary goal of COGA is to identify genes involved in alcohol dependence and related disorders. Alcohol-dependent probands were identified through alcohol treatment programs at seven US sites and were invited to participate if they had a sufficiently large family (usually sibships >3 with parents available) with two or more members in the COGA catchment area. Community probands and their families were recruited through driver's license records, mailings to randomly selected employees and students at a university, and attendees at medical and dental clinics. The institutional review boards at all sites approved this study and written consent was obtained from all participants. The present study included the participants in the European-American subset from the COGA case–control GWAS sample (in which cases met Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) alcohol dependence criteria20) for whom adult antisocial behavior interview data were also available (n=1379; 739 (54%) male and 640 (46%) female). European ancestry was determined using a principal-component-based analysis in PLINK.20 The average age at assessment was 43.8 years (s.d.=11.7; range=18–79).
AAB
AAB was measured using a count of the number of ASPD criteria endorsed under criterion A from the DSM-IV.1 In view of our modest sample size, we chose to use a more powerful dimensional phenotype rather than a less powerful diagnostic phenotype. The seven criteria included: engaging in illegal activities; deceitfulness; impulsivity and failing to plan ahead; irritability and aggressiveness; disregard for the safety of self and others; consistent irresponsibility at work or with finances; and lack of remorse. Criterion counts were obtained from items in the reliable (with a kappa=0.70 for ASPD diagnoses) and valid Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA).21, 22 We note that 203 (15%) of participants were administered an early version of the SSAGA that included a skip pattern whereby participants were asked the ASPD questions only if they had endorsed two or more alcohol dependence/abuse, marijuana dependence/abuse or other drug dependence/abuse symptoms. In total, 42 participants (representing 3% of the overall sample) were not asked the ASPD questions because of the skip pattern.
Genotyping and imputation
Participants were genotyped on the Illumina Human 1M BeadChip (San Diego, CA, USA); detailed genotyping and quality control information for this sample can be found in Edenberg et al.20 The European-American and African-American participants in the case–control sample were imputed together (n=1884). Prior to imputation, monomorphic SNPs (that is, SNPs with a minor allele frequency <0.0001), SNPs with missingness >2%, SNPs with Hardy–Weinberg equilibrium P<1 × 10−6 and SNPs that did not map to the Hg19 reference genome were filtered out, leaving 936 240 SNPs. Phasing of entire chromosomes or chromosome arms was performed using SHAPEIT.23, 24 Genotype imputation of additional SNPs was carried out with IMPUTE2 v.2.031 using the March 2012 release (v3) of the 1000 Genomes Project data (www.1000genomes.org25). Imputation analysis was performed for genomic windows of 5 Mb with an overlap interval of 500 kb between adjacent segments. Following the recommendations of the authors of IMPUTE2, we did not limit our imputation procedure to European reference samples. Monomorphic sites were excluded.
Analytic plan
We used genome-wide complex trait analysis (GCTA26) to estimate the extent to which autosomal common genetic variation accounted for variance in AAB. GCTA estimates the genetic relationships among individuals in a sample from genome-wide SNP data, and then uses a mixed-linear modeling framework to estimate heritability. The Illumina 1M BeadChip provides sufficient coverage of variation across the genome for the purposes of estimating common SNP heritability using GCTA; accordingly, we used non-imputed genotypic data for this analysis.
For the GWAS, the imputed gene dosage data were analyzed in PLINK.27 Covariates included: cohort (a four-category variable based on year of birth that was included to control for potential cohort effects: 1896–1929; 1930–1949; 1950–1969; and after 1970), age at assessment, sex and the first four principal components derived from a principal component-based analysis performed in PLINK on the measured genotypic data to cluster the samples along with HapMap reference samples (CEU, YRI, CHB and JPT). Results were filtered to include only SNPs with minor allele frequency >0.01 that also met an information criterion of >0.80. In total, 7 287 851 SNPs met these criteria.
We used KGG 2.5 (refs. 28, 29) to conduct gene-based tests of our GWAS results. This gene-based test examines whether the set of SNPs in a gene is associated with the phenotype at a level greater than chance. We used publicly available 1000 Genomes Phase 1 version 3 (European subsample) linkage disequilibrium (LD) files to build the ‘analysis genome' by position, for autosomes only. We included extended gene lengths of 5 kb at both the 5′ and 3′ ends. SNPs in high LD (r2>0.9) were connected; those in low LD (r2 <0.02) were considered independent. We used the hybrid set-based test for genome-wide association studies29 and report both the uncorrected P-values as well as the q-value based on a Benjamini and Hochberg30 false discovery rate. q-values <0.05 are considered genome-wide significant. Following this, we used i-GSEA4GWAS31 to assess enrichment across canonical pathways (defined by the authors of the i-GSEA4GWAS program) and gene ontologies. In total, 23 153 genes were submitted along with P-values (which were log-transformed by i-GSEA4GWAS). We set the minimum number of genes per category to 20, and the maximum to 200.
Genes of interest (that is, meeting a P-value threshold of less than 1 × 10−4) were compared with microarray data from nine brain regions (prefrontal cortex, cerebral cortex, thalamus, visual cortex, hippocampus, amygdala, caudate nucleus, putamen and cerebellum) from post-mortem tissue from two males and two females, which included an alcoholic and a control of each sex, to determine whether they were expressed in the brain.32 These nine regions were chosen to provide broad coverage across the brain. The expression values for our genes of interest did not substantially vary across these regions, and we therefore report the maximum of the average expression value. A gene was presumed to be expressed in the brain if the maximum expression level across the nine regions was higher than 16.0, because this is above the background signal for the arrays.
Robustness check and pleiotropy analyses
The sample used in the present study was originally recruited as part of a case–control GWAS of alcohol dependence. To check that our results were not simply driven by the phenotypic association between AAB and alcohol dependence, we examined whether the genome-wide significant gene to emerge from our analysis of AAB remained at least nominally significant after statistically controlling for DSM-IV alcohol dependence case–control status using a gene-based test. We also conducted a series of post hoc gene-based pleiotropy analyses to examine whether the genome-wide significant gene to emerge from our analysis showed evidence for association across a broader range of genetically correlated externalizing phenotypes.18 To pursue this, we examined association between our gene of interest and DSM-IV criterion counts for four major drug classes: alcohol dependence, cannabis dependence, cocaine dependence and opioid dependence. Because these were separate analyses of correlated phenotypes, a Bonferroni correction would be too stringent. We therefore used a nominal P-value cutoff of P<0.05 as evidence for association.
Replication study
Independent replication was tested in the COGA family-based GWAS (fGWAS) sample. The fGWAS sample includes 118 European-American COGA families densely affected with alcohol dependence (at least 3+ affected members) for whom genome-wide association data are available.33 The 270 individuals from the COGA case–control sample who were also in the fGWAS sample were removed from the replication data set. In total, AAB data from 1796 individuals in the fGWAS sample were available for analysis. Of these, 823 (46%) participants were male and 973 (54%) were female. The average age at assessment was 36.2 years (s.d.=15.2; range=18–88). AAB was measured using a count of the number of endorsed ASPD criteria under criterion A from the DSM-IV, as for our initial analysis. Association analyses in this sample were run using GWAF,34 which accounts for familial nesting and genetic distance using a kinship matrix. Covariates included cohort, age at assessment, sex and the first four principal components to control for genetic ancestry. Unimputed genotypic data were used.
Results
In the case–control sample, participants endorsed an average of 2.56 AAB criteria (s.d.=2.26; range=0–7). A total of 621 (45%) participants (468 males and 153 females) met criterion A for ASPD, defined by endorsement of three or more of the seven criteria. Males had higher AAB criterion counts than females (male mean=3.45, female mean=1.53; P<0.0001). Age was negatively associated with AAB criterion counts (r=−0.29, P<0.0001), indicating that older participants endorsed fewer lifetime AAB criteria. As expected, individuals classified as alcohol-dependent cases in the sample had higher AAB criterion counts (mean 3.8, s.d.=2.0) than those without alcohol dependence (mean 0.67, s.d.=0.96, P<0.0001).
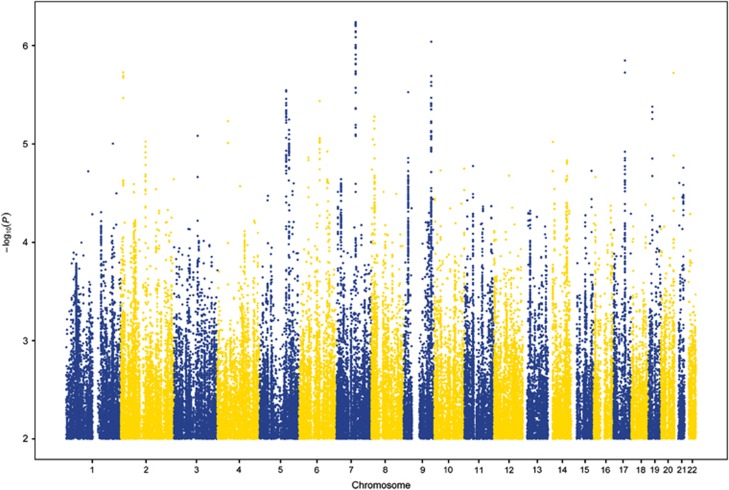
According to GCTA, the heritability of AAB was moderate, but the estimate did not reach statistical significance (h2=0.25, s.e.=0.20, P=0.09). The GWAS results are summarized in a Manhattan plot in Figure 1 and the quantile–quantile plot is shown in Supplementary Figure S1. The genomic inflation factor was acceptable, λ=1.004, suggesting that technical issues and population stratification did not inflate the results. No single SNP met the strict genome-wide significance threshold (P⩽5 × 10−8). The most highly associated SNP was rs4728702 (P=5.8 × 10−7), located in the protein-coding gene adenosine triphosphate-binding cassette, sub-family B (MDR/TAP), member 1 (ABCB1) on chromosome 7. The top SNP results (P⩽5 × 10−6) from this analysis are summarized in Table 1.
Figure 1.
Association results from the genome-wide association study of adult antisocial behavior. On the x axis are single-nucleotide polymorphism (SNP) positions for chromosomes 1–22. On the y axis are negative logarithms (base 10) of the P-values for each SNP, whereby higher values indicate smaller P-values.
Table 1. Top SNPs (P<5 × 10-6) from a GWAS of adult antisocial behavior.
| SNP | CHR | POS | EFF | NEFF | AF | BETA | P |
|---|---|---|---|---|---|---|---|
| rs4728702 | 7 | 87180678 | A | T | 0.43 | −0.36 | 5.77E−07 |
| rs1016793 | 7 | 87199182 | G | A | 0.59 | 0.37 | 5.91E−07 |
| rs1128503 | 7 | 87179601 | A | G | 0.43 | −0.36 | 5.97E−07 |
| rs4728700 | 7 | 87171659 | T | C | 0.45 | −0.36 | 6.08E−07 |
| rs868755 | 7 | 87189930 | T | G | 0.41 | −0.37 | 6.15E−07 |
| rs10276036 | 7 | 87180198 | C | T | 0.43 | −0.36 | 6.22E−07 |
| rs3789244 | 7 | 87181849 | G | T | 0.43 | −0.36 | 6.22E−07 |
| rs2235026 | 7 | 87182882 | T | C | 0.43 | −0.36 | 6.32E−07 |
| rs2235021 | 7 | 87199264 | C | A | 0.57 | 0.36 | 7.15E−07 |
| rs2235020 | 7 | 87199265 | T | A | 0.57 | 0.36 | 7.30E−07 |
| rs2235046 | 7 | 87174066 | T | C | 0.45 | −0.35 | 8.20E−07 |
| rs10985923 | 9 | 126038684 | G | T | 0.93 | 0.71 | 9.10E−07 |
| rs6959435 | 7 | 87158185 | G | T | 0.43 | −0.36 | 9.82E−07 |
| rs2373586 | 7 | 87157583 | A | C | 0.43 | −0.36 | 9.85E−07 |
| rs62817660 | 7 | 87200529 | G | A | 0.57 | 0.35 | 1.03E−06 |
| rs12539098 | 7 | 87200639 | T | C | 0.57 | 0.35 | 1.04E−06 |
| rs11975994 | 7 | 87192731 | G | A | 0.43 | −0.35 | 1.05E−06 |
| rs1016794 | 7 | 87198459 | G | A | 0.57 | 0.35 | 1.05E−06 |
| rs2520464 | 7 | 87201086 | C | T | 0.57 | 0.35 | 1.06E−06 |
| rs2032582 | 7 | 87160618 | A | C | 0.45 | −0.35 | 1.23E−06 |
| rs1202167 | 7 | 87197059 | C | T | 0.57 | 0.35 | 1.37E−06 |
| rs6504898 | 17 | 52427312 | G | T | 0.56 | −0.36 | 1.42E−06 |
| rs1202168 | 7 | 87195962 | G | A | 0.57 | 0.35 | 1.53E−06 |
| rs1202169 | 7 | 87195850 | T | C | 0.57 | 0.35 | 1.55E−06 |
| rs2235013 | 7 | 87178626 | C | T | 0.51 | −0.34 | 1.83E−06 |
| rs2235033 | 7 | 87179143 | A | G | 0.51 | −0.34 | 1.83E−06 |
| rs12704364 | 7 | 87181175 | C | T | 0.51 | −0.34 | 1.83E−06 |
| rs6961665 | 7 | 87181418 | C | A | 0.51 | −0.34 | 1.83E−06 |
| rs58738000 | 2 | 11704426 | C | G | 0.92 | −0.66 | 1.87E−06 |
| rs6504902 | 17 | 52433609 | T | C | 0.56 | −0.35 | 1.88E−06 |
| rs4810138 | 20 | 57144231 | G | C | 0.33 | 0.38 | 1.90E−06 |
| rs2235027 | 7 | 87182779 | G | T | 0.51 | −0.34 | 1.91E−06 |
| rs10234411 | 7 | 87164892 | T | A | 0.45 | −0.34 | 1.94E−06 |
| rs4148738 | 7 | 87163049 | C | T | 0.45 | −0.34 | 1.96E−06 |
| rs68152859 | 2 | 11699747 | G | A | 0.92 | −0.67 | 2.02E−06 |
| rs10985911 | 9 | 126007069 | T | C | 0.93 | 0.68 | 2.04E−06 |
| rs5020877 | 2 | 11694370 | A | G | 0.92 | −0.67 | 2.12E−06 |
| rs142285425 | 9 | 125951669 | A | G | 0.93 | 0.67 | 2.34E−06 |
| rs10985900 | 9 | 125954349 | T | C | 0.93 | 0.67 | 2.34E−06 |
| rs7870519 | 9 | 125917716 | A | T | 0.93 | 0.67 | 2.34E−06 |
| rs62579000 | 9 | 125973531 | C | T | 0.93 | 0.67 | 2.34E−06 |
| rs10985892 | 9 | 125908794 | T | C | 0.93 | 0.67 | 2.35E−06 |
| rs10985908 | 9 | 125986335 | C | T | 0.93 | 0.67 | 2.35E−06 |
| rs62578996 | 9 | 125919594 | G | A | 0.93 | 0.66 | 2.56E−06 |
| rs10808072 | 7 | 87176463 | A | G | 0.51 | −0.33 | 2.69E−06 |
| rs55980995 | 5 | 121875565 | A | G | 0.88 | −0.52 | 2.84E−06 |
| rs6893509 | 5 | 121860590 | T | G | 0.89 | −0.55 | 2.92E−06 |
| rs28368138 | 9 | 21207334 | C | G | 0.95 | −0.82 | 2.97E−06 |
| rs1202165 | 7 | 87197594 | G | A | 0.59 | 0.35 | 3.00E−06 |
| rs12002466 | 9 | 125865899 | T | C | 0.93 | 0.65 | 3.40E−06 |
| rs180996880 | 2 | 11693084 | C | A | 0.92 | −0.65 | 3.41E−06 |
| rs74602468 | 5 | 121863973 | C | T | 0.90 | −0.56 | 3.48E−06 |
| rs67242082 | 6 | 94334202 | A | G | 0.86 | −0.48 | 3.66E−06 |
| rs76585835 | 5 | 121867413 | G | A | 0.90 | −0.56 | 3.83E−06 |
| rs41277128 | 9 | 125610975 | G | A | 0.93 | 0.66 | 3.85E−06 |
| rs7033878 | 9 | 125618080 | T | G | 0.93 | 0.66 | 3.88E−06 |
| rs62580884 | 9 | 125619738 | C | A | 0.93 | 0.66 | 3.88E−06 |
| rs10985798 | 9 | 125624520 | G | C | 0.93 | 0.66 | 3.88E−06 |
| rs62580922 | 9 | 125631594 | T | C | 0.93 | 0.66 | 3.88E−06 |
| rs10985807 | 9 | 125652376 | C | T | 0.93 | 0.66 | 3.88E−06 |
| rs78303085 | 9 | 125869322 | T | A | 0.93 | 0.66 | 3.96E−06 |
| rs62578960 | 9 | 125869287 | G | A | 0.93 | 0.66 | 3.97E−06 |
| rs80233205 | 5 | 121871869 | G | A | 0.90 | −0.56 | 4.03E−06 |
| rs55919124 | 5 | 121880314 | T | A | 0.90 | −0.56 | 4.05E−06 |
| rs62580923 | 9 | 125635317 | A | T | 0.93 | 0.65 | 4.13E−06 |
| rs6413435 | 19 | 18497137 | G | A | 0.83 | −0.44 | 4.18E−06 |
| rs6948766 | 7 | 87191246 | G | A | 0.52 | −0.33 | 4.32E−06 |
| rs11763872 | 7 | 87217215 | T | C | 0.49 | 0.33 | 4.40E−06 |
| rs7034822 | 9 | 125621808 | G | A | 0.93 | 0.65 | 4.46E−06 |
| rs12378760 | 9 | 125661283 | T | C | 0.93 | 0.65 | 4.46E−06 |
| rs17149293 | 5 | 121864140 | G | T | 0.90 | −0.55 | 4.53E−06 |
| rs76809806 | 5 | 121873792 | G | A | 0.90 | −0.55 | 4.66E−06 |
| rs12979706 | 19 | 18495424 | G | A | 0.83 | −0.44 | 4.75E−06 |
| rs12514901 | 5 | 121878119 | T | G | 0.89 | −0.54 | 4.84E−06 |
| rs78479583 | 5 | 121871427 | G | A | 0.90 | −0.55 | 4.96E−06 |
Abbreviations: AF, allele frequency of the effect allele; BETA, regression coefficient; CHR, chromosome; EFF, effect allele; GWAS, genome-wide association study; NEFF, non-effect allele; POS, position (in base pairs); SNP, single-nucleotide polymorphism.
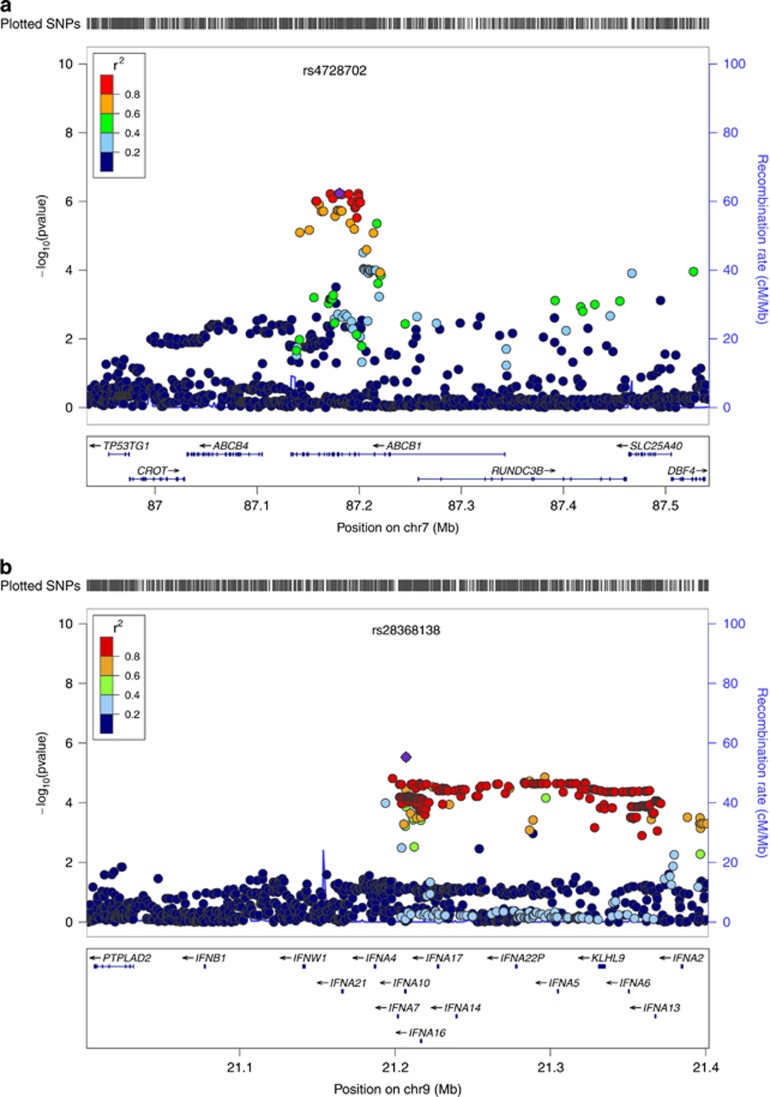
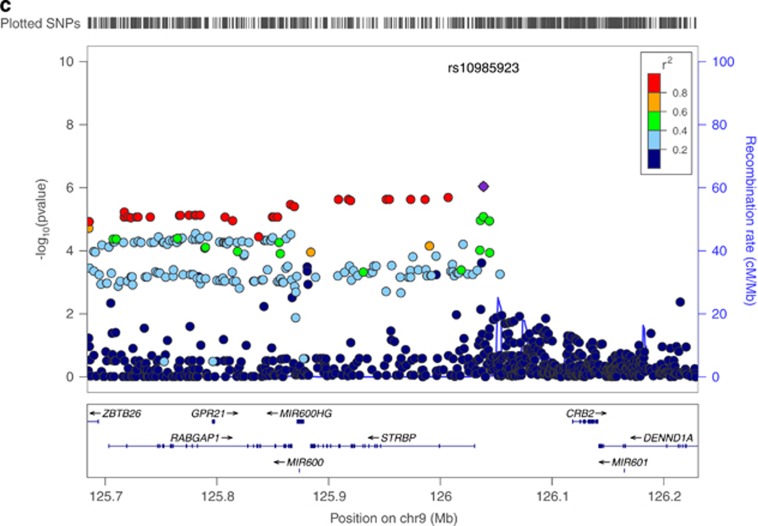
In our gene-based test, ABCB1 met the threshold for genome-wide significance (q-value=0.03). This gene was robustly expressed in brain tissue (maximum expression value across nine brain regions=210). The regional association plot for this gene is shown in Figure 2a. The 17 genes from this analysis meeting a P-value threshold of less than 1 × 10−4 are listed in Table 2, along with their expression values. Two clusters of genes on chromosome 9 appear in this list. As shown in Figures 2b and c, four of these genes are genes for type I interferon (IFNA7, IFNA10, IFNA16 and IFNA17) on chromosome 9p, and five others (STRBP, MIR600, RABGAP1, MIR600HG and ZBTB26) appear in a cluster on chromosome 9q.
Figure 2.
(a) Association results from the genome-wide association study (GWAS) of adult antisocial behavior (AAB) for a 200-kb region surrounding rs4728702 in ABCB1 on chromosome 7. (b) Association results from the GWAS of AAB for a 200-kb region surrounding rs28368138 on chromosome 9p. Four genes for type I interferon (IFNA7, IFNA10, IFNA16 and IFNA17) depicted here reached nominal significance (P-value threshold of less than 1 × 10−4) in gene-based tests. (c) Association results from the GWAS of AAB for a 200-kb region surrounding rs10985923 on chromosome 9q. Five genes (STRBP, MIR600, RABGAP1, MIR600HG and ZBTB26) depicted here reached nominal significance (P-value threshold of less than 1 × 10−4) in gene-based tests. All plots were constructed using LocusZoom.35 Shown on the x axis of all figures are position numbers, and on the y axis are the negative logarithms (base 10) of the P-values for each single-nucleotide polymorphism (SNP), whereby higher values indicate smaller P-values, and local estimates of recombination rates. SNPs are color coded to visualize the haplotype structure in this region, with orange and red coloring indicating SNPs higher in linkage disequilibrium (LD) and blue and green indicating SNPs lower in LD.
Table 2. Top genes associated with adult antisocial behavior.
| Gene name | Chromosome | Gene P-value | Gene q-value | Brain exp |
|---|---|---|---|---|
| ABCB1 | 7 | 1.42E−06 | 0.033 | 210 |
| CD46 | 1 | 1.28E−05 | 0.297 | 451 |
| ABCB5 | 7 | 2.06E−05 | 0.476 | — |
| IFNA7 | 9 | 2.34E−05 | 0.542 | — |
| MIR600 | 9 | 2.53E−05 | 0.584 | NA |
| MIR600HG | 9 | 2.53E−05 | 0.584 | NA |
| STRBP | 9 | 3.55E−05 | 0.821 | 359 |
| GREB1 | 2 | 3.85E−05 | 0.891 | 163 |
| RC3H2 | 9 | 3.88E−05 | 0.897 | 498 |
| ZBTB6 | 9 | 4.28E−05 | 0.989 | 190 |
| IFNA17 | 9 | 4.55E−05 | 1.000 | — |
| RABGAP1 | 9 | 7.97E−05 | 1.000 | 1228 |
| NKAIN3 | 8 | 8.04E−05 | 1.000 | 281 |
| OLIG2 | 21 | 8.11E−05 | 1.000 | 155 |
| ZBTB26 | 9 | 8.24E−05 | 1.000 | 153 |
| IFNA16 | 9 | 8.52E−05 | 1.000 | — |
| IFNA10 | 9 | 9.68E−05 | 1.000 | — |
Brain exp. is the maximum of average expression for nine brain regions; ‘—' is below background; NA indicates measurement not available because the gene was not present on array.
Notes: Gene P-value and gene q-value refer to the uncorrected P-values and the q-values based on a Benjamini and Hochberg false discovery rate for the gene-based tests. A gene was presumed to be expressed in the brain if the maximum expression level across the nine regions was higher than 16.0 because this is above the background signal for the arrays. Expression ranges from 5–20,140; median=108; 90th percentile=484. For comparison, the neurotransmitter receptor gene HTR1B (serotonin 1D beta receptor) that is normally expressed in the brain has a maximum expression of 182.
Enrichment analyses indicated that the genes meeting more stringent significance criteria were more likely to fall into seven known canonical pathways and gene ontologies that are summarized in Table 3. Most of these gene sets are immune related. Many of the same genes were represented across multiple gene sets, and thus these sets are not independent.
Table 3. Canonical pathways and gene ontologies.
| Gene set name | Gene set q-value |
|---|---|
| Cytokine activity | <0.0001 |
| Jak-STAT signaling pathway | <0.0001 |
| Toll-like receptor signaling pathway | 0.0005 |
| Antigen processing and presentation | 0.0008 |
| Hematopoietin interferon class D200 domain cytokine receptor binding | 0.002 |
| Natural killer cell-mediated cytotoxicity | 0.005 |
| Tryptophan metabolism | 0.05 |
Robustness check and pleiotropy analyses
In our robustness check, the association between ABCB1 variation and AAB was attenuated after controlling for alcohol dependence, but did not disappear (P=1.3 × 10−2). Gene-based tests do not have corresponding effect sizes, but the beta coefficient for our most significantly associated SNP (rs4728702) was reduced from −0.36 to −0.17. The sample averages for the four drug classes analyzed as part of our pleiotropy analyses are shown in Supplementary Table S1. The results from a series of gene-based tests indicated that ABCB1 variation was associated with alcohol dependence criteria (P=7.1 × 10-5) and cocaine dependence criteria (P=0.01), but not cannabis dependence criteria (P=0.19) or opioid dependence criteria (P=0.50).
Replication attempt
The sample average for AAB in the COGA fGWAS sample was 1.76 criteria (s.d.=1.76; range=0–7), which was significantly lower than in the case–control data set (unpaired t-test: t(3137)=11.21, P<0.01). A total of 498 (28%) participants (330 males and 168 females) met criterion A for ASPD. There was no evidence for replication of ABCB1 with AAB in the fGWAS sample (P=0.88), nor did any of the other genes or gene sets from Tables 2 and 3 replicate at P<0.05.
Discussion
Historically, molecular genetic research on AAB has been limited to the examination of a small number of candidate genes with purported biological relevance; only recently have researchers begun to conduct atheoretical genome-wide scans for this phenotype.13, 15, 16 In our genome-wide investigation, we found that autosomal SNPs accounted for ~25% of the variation in a dimensional measure of AAB. Although this estimate was not statistically significant (P=0.09), which is likely attributable to our modest sample size, it maps nicely to meta-analytic findings that additive genetic influences account for 32% of the variation in antisocial behavior.7 Our finding also maps to recent GCTA analyses in a community-based sample, where it was found that common genetic variation accounted for 26% (P=0.002) of the variation in a behavioral disinhibition phenotype.16
No SNP reached genome-wide significance in our GWAS of AAB. Our most associated SNP, rs4728702, was located in ABCB1 on chromosome 7. In our gene-based tests, ABCB1 was significant at the genome-wide level; however, we did not find an association for this gene in our replication sample. In expression analyses, we also found that ABCB1 is robustly expressed in human brain. This provides some biologically plausible evidence that ABCB1 variation could be associated with behavioral outcomes. ABCB1 codes for a member of the adenosine triphosphate-binding cassette transporters, ABCB1 or P-glycoprotein, which transport molecules across cellular membranes and also across the blood–brain barrier. ABCB1 is considered a pharmacogenetic candidate gene in view of ABCB1 transporters' ability to change drug pharmacokinetics.
Variation in ABCB1 has been previously associated with a number of psychiatric phenotypes, including opioid36 and cannabis37 dependence, as well as with treatment outcomes for depression38 and addiction.39 The related rodent gene, Abcb1a, is differentially expressed in three brain regions (accumbens shell, central amygdala and ventral tegmental area) of alcohol-preferring animals compared with non-preferring animals.40, 41 Furthermore, ethanol exposure changes ABCB1 expression. An in vitro study of human intestinal cells found that ethanol exposure increased messenger RNA ABCB1 expression level, and that these increases were maintained even after a week of ethanol withdrawal.42 Similarly, ABCB1 expression was increased in lymphoblastoid cell lines following ethanol exposure,32 and in rodents, Abcb1a expression was increased in the nucleus accumbens of alcohol-preferring rats following alcohol exposure.43
Taken as a whole, this pattern suggests that ABCB1 has pleiotropic effects across a number of externalizing spectrum behaviors/disorders, and that its expression is affected by ethanol exposure. The former is consistent with findings from the twin and molecular genetics literature, demonstrating that common externalizing disorders and behaviors (for example, alcohol dependence, other drug abuse or dependence, AAB and conduct disorder) share genetic influences,18, 44 and that this shared genetic factor is highly heritable (h2=80%).45 Supplementary analyses in our own sample were consistent with this hypothesis, and we found evidence that ABCB1 variation was associated with alcohol and cocaine dependence criterion counts. However, we did not find associations between ABCB1 and marijuana or opioid dependence criterion counts.
We also found evidence for enrichment (q-values⩽0.05) across multiple canonical pathways and gene ontologies including cytokine activity, Jak-STAT signaling pathway, toll-like receptor signaling pathway, antigen processing and presentation, cytokine receptor binding and natural killer cell-mediated cytotoxicity. Although the immediate biological relevance of these categories to AAB is not clear, these enrichment findings include many immune-related pathways and may be best interpreted in light of the associations among AAB and alcohol, cannabis, cocaine and opioid dependence criterion counts in the sample. Immune and inflammatory pathways have been hypothesized to be associated with psychiatric disorders across the internalizing and externalizing spectra.46 For example, it is known that alcohol alters cytokine activity,47 induces changes in neuroimmune signaling in the brain48 and that alcohol dependence is associated with low-grade systemic inflammation.49 Likewise, the monocytes of individuals who are cocaine dependent show decreased expression of tumor necrosis factor-α and interleukin-6 proinflammatory cytokines in response to a bacterial ligand relative to controls.50 Four of the top genes (based on P-values) to emerge in our analysis are genes for type I interferon (IFNA7, IFNA10, IFNA16 and IFNA17), which reside in a cluster on chromosome 9p. Previous studies demonstrate that interferon A treatment of hepatitis C patients can induce multiple psychiatric symptoms including depression51 and impulsivity.52 Although we did not find significant enrichment for these pathways in our replication sample, these results add preliminary evidence to a growing literature that variation in genes in immune-relevant pathways may predispose individuals to AAB and closely related behaviors.
The present study expands upon the initial AAB GWAS by Tielbeek et al.13 as well as more recently published GWAS of a behavioral disinhibition phenotype15, 16 in two important ways. First, we used a case–control sample where the cases met criteria for alcohol dependence. By virtue of the association between alcohol dependence and AAB, and the relatively high rates of individuals meeting clinical cutoffs for criterion A for ASPD in the present sample (63% of males and 24% of females) compared with American population-based prevalence estimates, it is likely that the sample was enriched for genetic variants predisposing individuals toward externalizing spectrum behaviors such as AAB. Previous work indicates that the genetic influences on AAB completely overlap with the genetic influences on alcohol dependence, other drug abuse/dependence and conduct disorder—that is, AAB does not have unique genetic influences above and beyond those shared with these other externalizing disorders.18 In view of this, gene identification efforts for AAB are likely to be more successful in more severely affected samples or in samples where participants high in AAB also tend to have comorbid alcohol or substance-use disorders, such as the COGA sample. In contrast, for example, only 6% of the participants in the Tielbeek et al.13 community-based sample met their non-diagnostic AAB case criteria. This sample may also have had low rates of comorbid alcohol and other drug diagnoses, limiting the ability to find genome-wide significant effects. Second, we used a dimensional measure of AAB, which is more powerful than a binary diagnostic variable, and better represents the underlying dimensional structure of AAB.53 These differences may explain, in part, why we were able to detect a significant genetic association in the present sample.
Our study should be interpreted in the context of several limitations. First, our sample size was relatively small. Second, because the COGA case–control alcohol dependence sample is highly affected by AAB, the findings emerging from our study may not generalize to lower-risk populations or other types of high-risk populations. Our null replication attempt may be attributable, in part, to the replication sample being relatively less affected than the discovery sample. There are other instances where genetic associations for externalizing behaviors have replicated within highly affected samples, but not less-affected samples. For example, GABRA2 is associated with alcohol dependence in samples where alcohol-dependent cases came from clinically recruited samples and families densely affected by alcoholism,54, 55, 56 but not community-based samples.57 A sample recruited for this purpose is likely to be enriched for genetic variation that predisposes individuals to a range of externalizing behavior problems, including AAB;18 however, whether our findings generalize to other populations at high risk for AAB (for example, incarcerated inmates58) is unknown. Third, because we limited the current analyses to European-Americans, our results may not generalize to other racial and ethnic groups.
Fourth, similar to all psychiatric outcomes, antisocial behavior has a developmental component, and evidence from the twin literature suggests that there are genetic influences on adolescent and adult antisocial behavior that are distinct from genetic influences on child antisocial behavior.4 The degree to which the genetic associations documented here for AAB are also associated with child or adolescent antisocial behavior is not clear. The results from this study provide an empirical starting point for subsequent developmental analyses to examine these questions. Fifth, there are likely to be aspects of the environment that moderate genetic influences on AAB that we did not explicitly examine here but that may be valuable to pursue in subsequent studies. Finally, our genome-wide association approach examined only common genetic variation. There is suggestive evidence that rare nonsynonymous exonic SNPs account for 14% (P=0.05) of the variance in a behavioral disinhibition phenotype.16 As rare variant-genotyping arrays and whole-genome sequencing become more widely available and cost effective, our understanding of the genetics of AAB will improve.
In summary, our goal in this study was to take an atheoretical approach to investigate the molecular genetic basis of AAB in a high-risk sample. The heritability of AAB was 25%, although this estimate did not differ significantly from zero. No SNP reached strict genome-wide significance, but gene-based tests identified an association between ABCB1 and AAB. Expression analyses further indicated that ABCB1 is robustly expressed in the brain, providing some evidence that variation in this gene could be related to a behavioral outcome. Previously documented associations between variants in ABCB1 and other drugs of abuse suggest that ABCB1 may confer general risk across a range of externalizing behaviors, rather than risk that is unique to AAB. This was consistent with post hoc analyses in our sample, where we found that variation in ABCB1 was associated with DSM-IV alcohol and cocaine dependence criteria. These pieces of evidence suggest that ABCB1 may be a gene of interest for further study. We also found enrichment of several immune-related canonical pathways and gene ontologies, which is consistent with previous suggestions that immune and inflammatory pathways are associated with externalizing spectrum behaviors. As a whole, our study goes beyond the candidate gene approach typically taken in studies of AAB, and implicates a gene and gene sets for which there is convergent evidence from other lines of research. These findings, although novel and promising, would benefit from direct replication.
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B Porjesz, V Hesselbrock, H Edenberg and L Bierut, includes ten different centers: University of Connecticut (V Hesselbrock); Indiana University (HJ Edenberg, J Nurnberger Jr and T Foroud); University of Iowa (S Kuperman and J Kramer); SUNY Downstate (B Porjesz); Washington University in St. Louis (L Bierut, A Goate, J Rice and K Bucholz); University of California at San Diego (M Schuckit); Rutgers University (J Tischfield); Texas Biomedical Research Institute (L Almasy), Howard University (R Taylor) and Virginia Commonwealth University (D Dick). Other COGA collaborators include: L Bauer (University of Connecticut); D Koller, S O'Connor, L Wetherill, X Xuei (Indiana University); Grace Chan (University of Connecticut); S Kang, N Manz, (SUNY Downstate); J-C Wang (Washington University in St. Louis); A Brooks (Rutgers University); and F Aliev (Virginia Commonwealth University). A Parsian and M Reilly are the National Institute on Alcohol Abuse and Alcoholism (NIAAA) Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the NIAAA and the National Institute on Drug Abuse. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1RR024992 from the National Center for Research Resources, a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Funding support for GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438) and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease' (HHSN268200782096C). This work was also supported by F32AA022269 (Salvatore), K01AA021399 (Edwards) and K02AA018755 (Dick). We thank Kim Doheny and Elizabeth Pugh from CIDR and Justin Paschall from the NCBI dbGaP staff for valuable assistance with genotyping and quality control in developing the data set available at dbGaP.
Disclaimer
This publication is solely the responsibility of the authors and does not necessarily represent the official view of the funders.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) Washington, DC; 1994. [Google Scholar]
- Grant BF, Hasin DS, Stinson FS, Dawson DA, Chou SP, Ruan WJ, et al. Prevalence, correlates, and disability of personality disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. J Clin Psychiatry. 2004;65:948–958. doi: 10.4088/jcp.v65n0711. [DOI] [PubMed] [Google Scholar]
- Fu Q, Heath AC, Bucholz KK, Nelson E, Goldberg J, Lyons M, et al. Shared genetic risk of major depresson, alcohol dependence, and marijuana dependence: Contribution of antisocial personality disorder in men. Arch Gen Psychiatry. 2002;59:1125–1132. doi: 10.1001/archpsyc.59.12.1125. [DOI] [PubMed] [Google Scholar]
- Jacobson KC, Prescott CA, Kendler KS. Sex differences in the genetic and environmental influences on the development of antisocial behavior. Dev Psychopathol. 2002;14:395–416. doi: 10.1017/s0954579402002110. [DOI] [PubMed] [Google Scholar]
- Torgersen S, Czajkowski N, Jacobson K, Reichborn-Kjennerud T, Roysamb E, Neale MC, et al. Dimensional representations of DSM-IV cluster B personality disorders in a population-based sample of Norwegian twins: a multivariate study. Psychol Med. 2008;38:1617–1625. doi: 10.1017/S0033291708002924. [DOI] [PubMed] [Google Scholar]
- Lyons MJ, True WR, Eisen SA, Goldberg J, Meyer JM, Faraone SV, et al. Differential heritability of adult and juvenile antisocial traits. Arch Gen Psychiatry. 1995;52:906–915. doi: 10.1001/archpsyc.1995.03950230020005. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Waldman ID. Genetic and environmental influences on antisocial behavior: a meta-analysis of twin and adoption studies. Psychol Bull. 2002;128:490–529. [PubMed] [Google Scholar]
- Douglas K, Chan G, Gelernter J, Arias AJ, Anton RF, Poling J, et al. 5-HTTLPR as a potential moderator of the effects of adverse childhood experiences on risk of antisocial personality disorder. Psychiat Genet. 2011;21:240–248. doi: 10.1097/YPG.0b013e3283457c15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley DL, Eaves LJ, Wormley B, Silberg JL, Maes HH, Kuhn J, et al. Childhood adversity, monoamine oxidase A genotype, and risk for conduct disorder. Arch Gen Psychiatry. 2004;61:738–744. doi: 10.1001/archpsyc.61.7.738. [DOI] [PubMed] [Google Scholar]
- Fowler T, Langley K, Rice F, van den Bree MBM, Ross K, Wilkinson LS, et al. Psychopathy trait scores in adolescents with childhood ADHD: the contribution of genotypes affecting MAOA, 5HTT and COMT activity. Psychiat Genet. 2009;19:312–319. doi: 10.1097/YPG.0b013e3283328df4. [DOI] [PubMed] [Google Scholar]
- Ficks CA, Waldman ID. Candidate genes for aggression and antisocial behavior: a meta-analysis of association studies of the 5HTTLPR and MAOA-uVNTR. Behav Genet. 2014;44:427–444. doi: 10.1007/s10519-014-9661-y. [DOI] [PubMed] [Google Scholar]
- Duncan L, Keller MC. A critical review of the first ten years of measured gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielbeek JJ, Medland SE, Benyamin B, Byrne EM, Heath AC, Madden PA, et al. Unraveling the genetic etiology of adult antisocial behavior: a genome-wide association study. PLoS One. 2012;7:e45086. doi: 10.1371/journal.pone.0045086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle B, Elizalde C, Galceran J, Becker W, Tejedor FJ. The MNB/DYRK1A protein kinase: neurobiological functions and Down syndrome implications. J Neural Transm Suppl. 2003;67:129–137. doi: 10.1007/978-3-7091-6721-2_11. [DOI] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, et al. A genome-wide association study of behavioral disinhibition. Behav Genet. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Feng S, Miller MB, Hicks BM, Pankratz N, Abecasis GR, et al. Rare nonsynonymous exonic variants in addiction and behavioral disinhibition. Biol Psychiatry. 2014;75:783–789. doi: 10.1016/j.biopsych.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton WM, Conway KP, Stinson FS, Colliver JD, Grant BF. Prevalence, correlates, and comorbidity of DSM-IV antisocial personality syndromes and alcohol and specific drug use disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. J Clin Psychiatry. 2005;66:677–685. doi: 10.4088/jcp.v66n0602. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit MA, et al. The collaborative study on the genetics of alcoholism. Alcohol Health Res W. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA—a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JL, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Kwan JS, Sham PC. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91:478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010;38:W90–W95. doi: 10.1093/nar/gkq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Brooks AI, Deng L, Liang L, Wang JC, Kapoor M, et al. Ethanol treatment of lymphoblastoid cell lines from alcoholics and non-alcoholics causes many subtle changes in gene expression. Alcohol. 2014;48:603–610. doi: 10.1016/j.alcohol.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatr. 2013;18:1218–1224. doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer B, Erb R, Pavlic M, Ulmer H, Giacomuzzi S, Riemer Y, et al. Association of polymorphisms in pharmacogenetic candidate genes (OPRD1, GAL, ABCB1, OPRM1) with opioid dependence in European population: a case-control study. PLoS One. 2013;8:e75359. doi: 10.1371/journal.pone.0075359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyamina A, Bonhomme-Faivre L, Picard V, Sabbagh A, Richard D, Blecha L, et al. Association between ABCB1 C3435T polymorphism and increased risk of cannabis dependence. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:1270–1274. doi: 10.1016/j.pnpbp.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Lin KM, Chiu YF, Tsai IJ, Chen CH, Shen WW, Liu SC, et al. ABCB1 gene polymorphisms are associated with the severity of major depressive disorder and its response to escitalopram treatment. Pharmacogenet Genomics. 2011;21:163–170. doi: 10.1097/FPC.0b013e32833db216. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Levran O, Proudnikov D, Nielsen DA, Kreek MJ. Search for genetic markers and functional variants involved in the development of opiate and cocaine addiction and treatment. Ann N Y Acad Sci. 2010;1187:184–207. doi: 10.1111/j.1749-6632.2009.05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, et al. Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol. 2013;47:517–529. doi: 10.1016/j.alcohol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, et al. Gene expression in the ventral tegmental area of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Pharmacol Biochem Behav. 2012;102:275–285. doi: 10.1016/j.pbb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theile D, Schmidt TT, Haefeli WE, Weiss J. In-vitro evaluation of chronic alcohol effects on expression of drug-metabolizing and drug-transporting proteins. J Pharm Pharmacol. 2013;65:1518–1525. doi: 10.1111/jphp.12124. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, et al. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG. Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet. 2013;43:97–107. doi: 10.1007/s10519-013-9584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96:684–695. [PubMed] [Google Scholar]
- Kelley KW, Dantzer R. Alcoholism and inflammation: neuroimmunology of behavioral and mood disorders. Brain Behav Immun. 2011;25:S13–S20. doi: 10.1016/j.bbi.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Bechara R, Brown LA, Guidot DM, Mandrekar P, Oak S, et al. Cytokines and alcohol. Alcohol Clin Exp Res. 2006;30:720–730. doi: 10.1111/j.1530-0277.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- Zou JY, Crews FT. Release of neuronal HMGB1 by ethanol through decreased HDAC activity activates brain neuroimmune signaling. PLoS One. 2014;9:e87915. doi: 10.1371/journal.pone.0087915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Cani PD, Neyrinck AM, Starkel P, Jamar F, Mikolajczak M, et al. Role of intestinal permeability and inflammation in the biological and behavioral control of alcohol-dependent subjects. Brain Behav Immun. 2012;26:911–918. doi: 10.1016/j.bbi.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Irwin MR, Olmos L, Wang M, Valladares EM, Motivala SJ, Fong T, et al. Cocaine dependence and acute cocaine induce decreases of monocyte proinflammatory cytokine expression across the diurnal period: autonomic mechanisms. J Pharmacol Exp Ther. 2007;320:507–515. doi: 10.1124/jpet.106.112797. [DOI] [PubMed] [Google Scholar]
- Cicek IE, Cicek E, Kayhan F, Uguz F, Erayman I, Kurban S, et al. The roles of BDNF, S100B, and oxidative stress in interferon-induced depression and the effect of antidepressant treatment in patients with chronic viral hepatitis: a prospective study. J Psychosom Res. 2014;76:227–232. doi: 10.1016/j.jpsychores.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Fabregas BC, Abreu MN, Dos Santos AK, Moura AS, Carmo RA, Teixeira AL. Impulsiveness in chronic hepatitis C patients. Gen Hosp Psychiatry. 2014;36:261–265. doi: 10.1016/j.genhosppsych.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Marcus DK, Ruscio J, Lilienfeld SO, Hughes KT. Converging evidence for the latent structure of antisocial personality disorder: consistency of taxometric and latent class analyses. Crim Justice Behav. 2008;35:284–293. [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, et al. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, et al. Variations in GABRA2, encoding the α2 subunit of the GABA-A receptor are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Irons DE, Iacono WG, Oetting WS, Kirkpatrick RM, Vrieze SI, Miller MB, et al. Gamma-aminobutyric Acid system genes-no evidence for a role in alcohol use and abuse in a community-based sample. Alcohol Clin Exp Res. 2014;38:938–947. doi: 10.1111/acer.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazel S, Danesh J. Serious mental disorder in 23 000 prisoners: a systematic review of 62 surveys. Lancet. 2002;359:545–550. doi: 10.1016/S0140-6736(02)07740-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.