Abstract
Tuberculosis continues to be a major killer disease, despite an all-out effort launched against it in the postgenomic era. We describe here the population structure of Mycobacterium tuberculosis strains, as revealed by a chromosome-wide scan of fluorescent amplified fragment length polymorphisms (FAFLPs), for more than 1,100 independent isolates from 11 different countries. The bacterial strains were genotyped based on a total of 136 ± 1 different FAFLP markers at the genome sequence interface, with details on IS6110 profiles, drug resistance status, clinicopathological observations, and host status integrated into the analysis process. The strains were found to cluster with possible geographic affinities, including the parameters of host species type, IS6110 profile, and drug susceptibility status. Of the five most commonly amplified fragment sets (or amplitypes), type A predominated in strains of mixed origin, deposited in The Netherlands; type B was exclusively observed for Indian isolates; type C was found mainly in strains from Peru and Australia; and types D and E predominated in European strains from France and Italy. The amplitypes were independent of certain large sequence polymorphisms representing two important deletions, TbD1 and Rd9. It appears that M. tuberculosis has a high genomic diversity with a possible geographic evolution. This may have occurred due to specific genomic deletions and synonymous substitutions selected rigorously against host defenses and environmental stresses on an evolutionary timescale. The genotypic data reported here are additionally significant for genotype-phenotype correlations and for determining whether pathogen diversity is a reflection f the host population diversity.
Mycobacterium tuberculosis, the underlying cause of tuberculosis, infects almost every third person in the world and has been regarded as the most successful pathogen in the history of the diseases of mankind. The accumulation of changes in the genomic content, occurring through gene acquisition and loss, is the major underlying (historical) event in the emergence of fit and successful strain variants in the M. tuberculosis complex (13, 14). A few predominant genotypes circulating throughout the world are responsible for the major outbreaks of the recent past, and these belong to the so-called Beijing, Haarlem, and African clusters (5, 13, 21, 23). These major strain groups have been classified based on repetitive element IS6110 genotyping and spoligotyping patterns and have been described as the predominant pathotypes in the world. Although the M. tuberculosis genome contains several repetitive elements, only a few are polymorphic and these have not been rigorously studied (18). The abundance of polymorphisms related to the mobility of repetitive elements, coupled with the restricted number of single-nucleotide polymorphisms, indicates that transposition and homologous recombination are the major events contributing to the diversity of M. tuberculosis strains (21). In addition, polymorphisms seen with different molecular markers reveal a high degree of mutual association. This supports the hypothesis that M. tuberculosis has a strong clonal population structure (21).
The study of the molecular epidemiology of tubercle bacilli changed after the availability of genome sequence data in the public domain (8). Recently, DNA microarrays have been used for comparative genomics with different M. tuberculosis clones for which clinical and epidemiological information was available (4, 10). The deletion patterns described recently suggest that there is substantial genomic variability among different M. tuberculosis genotypes in the world (13). It is likely that new genotypes do indeed exist but that they go largely unnoticed because of the nonavailability of high-resolution genomic tools. Juxtaposing host diversity with the bacterial population structure could throw some light on evolutionarily significant genomic events, such as the eukaryotic-prokaryotic gene fusion (9). Based on the presence or absence of an M. tuberculosis-specific deletion (TbD1), a new evolutionary scenario for the evolution of the M. tuberculosis complex and the origin of human tuberculosis has been proposed (7).
The currently available typing systems designed for molecular epidemiology (3) are not capable of classifying strains on the basis of the whole genome, including various evolutionary changes and random base substitutions. There is a need for a genome sequence-based (11) classification of predominantly rampant strains that will also assist global efforts aimed at controlling this deadly disease. These issues, coupled with our continued interest in the evolution of M. tuberculosis genotypic diversity (1, 2, 17), have led to the present study, which aimed at revisiting the global population diversity of M. tuberculosis. We analyzed the nature of the global diversity of tubercle bacilli, with an emphasis on evolutionary genomics, by whole-genome fingerprinting and genotyping of M. tuberculosis isolates from different world populations followed by analyses of large sequence polymorphisms representing two important deletions, TbD1 and Rd9 (7). We used fluorescent amplified fragment length polymorphism (FAFLP) typing (1, 2, 11, 17), a fluorescent form of AFLP analysis (24), as a stand-alone, portable approach with a valid phylogenetic basis (12, 15, 16). Polymorphisms in band patterns were mapped to specific loci, allowing the individual strains and isolates to be genotyped or differentiated based on the alleles they carry. The five major clusters representing the corresponding FAFLP patterns observed in this study should help us to understand the worldwide spread and partitioning of M. tuberculosis genotypes and the possible evolutionary background of this organism. Also, it should be possible to identify informative markers for the identification and epidemiology of different M. tuberculosis populations on a global scale. Such markers are undeniably the most highly resolving tools for studying host and environmental impacts on pathogen evolution and could be developed as genotype-phenotype databases (17).
MATERIALS AND METHODS
Bacterial strains and DNAs.
A total of 1,124 independent strains of M. tuberculosis from India, The Netherlands, Australia, Peru, France, Italy, and other regions were selected for FAFLP analysis (Table 1). For this study, the isolates were randomly picked based on geographical relevance, without any consideration of microbiological or biochemical parameters. A few strains from the M. tuberculosis complex group other than M. tuberculosis were also studied. About 99% of the strains were from human hosts. Blind-coded DNA samples from each of the strains were obtained for genomic analysis. The standard reference strains M. tuberculosis H37Rv (virulent strain), M. tuberculosis Erdman, M. bovis AN5, M. bovis ATCC 27290, M. bovis ATCC 27291, M. bovis BCG, seal bacillus, M. microti, and M. africanum were also used as controls at various levels. Blind-coded samples of M. tuberculosis isolated in The Netherlands (n = 150) were kindly provided by Kristin Kremer and D. van Soolingen of The National Institute for Public Health and the Environment, Bilthoven, The Netherlands. One hundred ten isolates of this origin were mainly obtained from the immigrant population in The Netherlands, and the majority of these isolates were singly or multiply resistant. The remaining 40 isolates were mainly obtained from the native Dutch population and were drug sensitive. Genomic DNAs for FAFLP analysis were prepared from all isolates by a previously described standard method (22).
TABLE 1.
Distribution of M. tuberculosis strains according to FAFLP amplitypes
| Geographic region | No. of strains analyzed | Strain characteristicsa | Amplitype(s) (no. of strains with amplitype) |
|---|---|---|---|
| India | 800 | Mostly MDR, low-copy-number IS6110 group, TbD1 and Rd9 regions intact | B (755), A (45) |
| Peru | 100 | Drug sensitive, high-copy-number IS6110 group | C (100) |
| The Netherlands, Arabia, Turkey, Morocco | 110 | Drug-resistant strains isolated in The Netherlands | A (110) |
| The Netherlands | 40 | Dutch strains, sensitive to all drugs, some resistant to isoniazid and rifampin | A (30) |
| D (5) | |||
| E (5) | |||
| France | 20 | Mostly with a unique RFLP cluster, TbD1 and Rd9 regions intact | D (6) |
| E (14) | |||
| Italy | 25 | M. bovis, Rd9 region intact | C (5) |
| E (20) | |||
| Australia | 20 | M. tuberculosis complex (human), Rd9 region intact | C (19) |
| D (1) | |||
| Canada | 5 | M. tuberculosis complex (bovine) | C (5) |
| Vietnam | 2 | Beijing genotype, IS6110 negative | C (2) |
| Tanzania | 2 | African genotype | C (2) |
MDR, multidrug resistant.
Adapters, primers, and preselective and selective amplification.
The sequences for the EcoRI adapter were 5′ CTCGTAGACTGCGTACC 3′ and 3′ CATCTGACGCATGGTTAA 5′ while those for the MseI adapter were 5′ GACGATGAGTCCTGAG 3′ and 3′ TACTCAGGACTCAT 5′ (24). The nonselective forward primer for the MseI adapter site was unlabeled. The reverse primers for the EcoRI adapter site contained a selective base at the 3′ end (A, G, C, or T) and were labeled with a fluorophore (FAM, JOE, NED, or TAMRA). These primers were obtained commercially (AFLP microbial fingerprinting kit; Applied Biosystems). The restriction-ligation reactions, preselective and selective amplifications, and gel separation of the samples were performed as described earlier (1, 2).
Computer-assisted genotypic analysis of FAFLP data.
Based on computer modeling with the chosen restriction enzymes, the M. tuberculosis H37Rv sequence data (8) were divided into various categories based on sizes (in base pairs). Genotyper software (Applied Biosystems) was used on these categories to allow comparisons of the FAFLP fragment data for field isolates. Before this, the predicted fragments were used for homology searches at the Institute for Genome Research server (http://www.tigr.org) and locus information was obtained from the TubercuList database sever at the Institut Pasteur (http://genolist.pasteur.fr/TubercuList). Based on the presence or absence of monomorphic and polymorphic bands or peaks, different FAFLP profiles were identified as amplitypes. These amplitypes were color coded, tiled, and superimposed in various ways to estimate the marker size in base pairs, the fluorescence intensity (peak height), data points on the gel, and the frequency of monomorphic (unique) bands. The bands were sized and genotyped for all of the isolates within the user-defined categories of marker size in base pairs. The presence or absence of markers within the categories was scored by a user-defined Genotyper macro that generated final output in the form of a binary table for all of the samples. Phylogenetic trees were generated (1, 2) from the binary data to delineate divergence and relatedness among the amplitypes. All of the amplitypes were deposited in the AmpliBASE MT database (http://210.212.212.4/) (17).
Analysis of LSPs by PCR-based identification of TBd1 and Rd9 deletions.
Representative samples from different FAFLP clusters were selected for large sequence polymorphism (LSP) analysis by PCR-based genotyping for the presence and absence of the TbD1 and Rd9 regions (7). Two hundred isolates originating from India (n = 96), France (n = 20), Australia (n = 13), Italy (n = 20), Peru (n = 21), and The Netherlands (n = 30) were subjected to deletion typing. PCR amplifications using genomic DNA samples were carried out with flanking as well as internal primers, as described by Brosch et al. (7).
RESULTS
Distribution of FAFLP markers in the genome.
In silico predictive computer methods using the genome sequence (8) of M. tuberculosis H37Rv revealed a total of 136 (±1) fragments of sizes from 50 to 500 bp upon digestion with the MseI and EcoRI enzymes. The four primer combinations used (EcoRI+A and MseI+0, EcoRI+G and MseI+0, EcoRI+C and MseI+0, and EcoRI+T and MseI+0) generated a total of 136 or 137 differently sized fragments ranging in size from 50 to 500 bp. Sixty-one (44.85%) of these fragments were polymorphic. The A-selective primer combination (EcoRI+A and MseI+0) produced 34 of the 136 fragments (25%), 9 of which were discriminatory (26.47% of the A-selective fragments and 14.7% of the total number of polymorphic fragments produced). The C-selective primer combination (EcoRI+C and MseI+0) produced 34 of the 136 fragments (25%), 16 of which were discriminatory (47.05% of the C-selective fragments and 26.22% of the total number of polymorphic fragments produced). The G-selective primer combination (EcoRI+G and MseI+0) produced 49 of the 136 fragments (36.02%), 29 of which were discriminatory (59.18% of the G-selective fragments and 47.54% of the total number of polymorphic fragments produced). The T-selective primer combination (EcoRI+T and MseI+0) produced 19 or 20 of the 136 fragments (14.7%), 7 of which were discriminatory (35% of the T-selective fragments and 11.47% of the total number of polymorphic fragments produced). Genomic FAFLP profiles of M. bovis strains were highly similar to the M. tuberculosis amplitypes (∼95% band sharing) and were therefore analyzed according to the in silico restricted map of strain H37Rv.
Geographic differences in predominant M. tuberculosis genotypes.
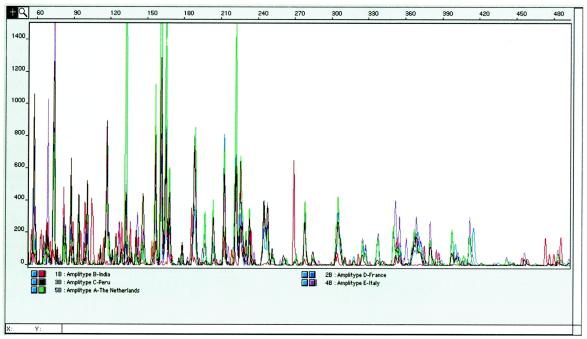
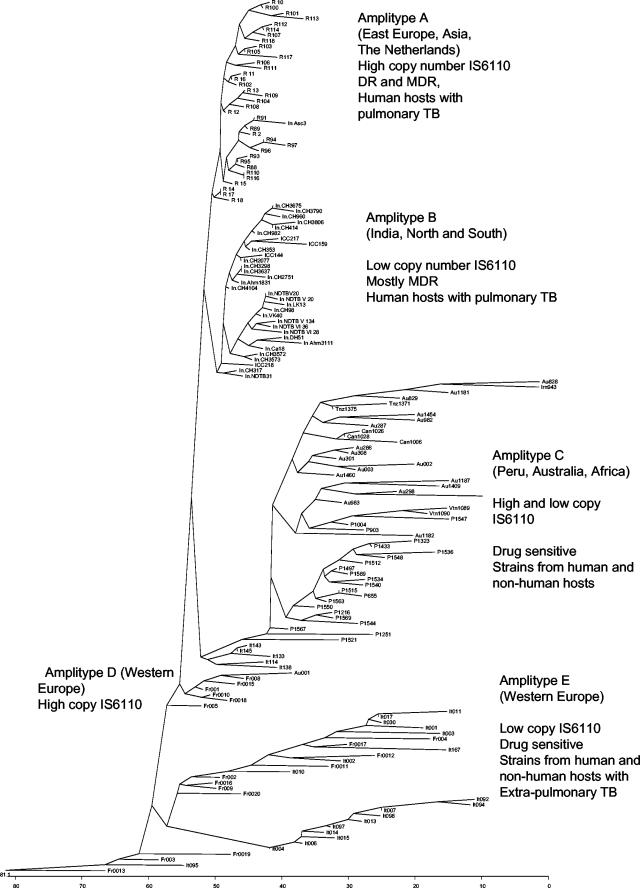
The FAFLP data were phenetically analyzed for regional and geographic affinities. The distribution of various markers in the analyzed strains was determined by computer-assisted genotyping. As many as 36 important markers derived from the M. tuberculosis H37Rv chromosome were found to be nonuniformly distributed geographically and were found to be the basis of phylogenetic clustering. In addition to this, several new markers with no available locus information were amplified. This was more pronounced in the case of Indian strains. Genotypic analysis was therefore also extended to score for these new markers. Representative clustering data are shown in Fig. 1 and 2, and the full set of polymorphic markers is summarized in Table 2. In particular, type A motifs (amplitype A) predominated in strains from The Netherlands but were never seen in other European strains from France and Italy. A few (at least 10%) of the Indian strains also revealed similarity to the amplitype A strains from The Netherlands. Similarly, type B fragment sets were observed for Indian strains and some European strains. Almost all 800 strains tested from Indian patients invariably carried nearly similar genotypes and clustered very closely, with an average genetic distance of ∼20%. Strains from some parts of the Indian States of Gujarat and Rajasthan showed divergent amplitypes compared with the other isolates from India. However, this did not significantly alter the clustering pattern of type B strains. The type C amplitype was found to represent a more diverse group of strains, mainly those from Peru, Australia, Italy, and France. The average genetic distance within this cluster was as high as ∼50%. The Peruvian isolates formed a distinct subcluster within the type C cluster. A few of the Peruvian isolates clustered along with Vietnamese strains with IS6110 null genotypes. Canadian and Tanzanian isolates branched out as separate lineages within the type C cluster, but they were found to be genetically linked to the Australian strains. Italian strains clustering within type C all carried a single copy of the IS6110 element and were of a bovine origin.
FIG. 1.
Genescan analysis output for five different strains that are predominant in different geographical areas of the world. FAFLP profiles for each of the representative strains were termed amplitypes. These amplitypes were color coded and were superimposed or extrapolated to compare the data points representing important loci. The horizontal scale indicates the sizes of the traces in base pairs. Peak heights as a function of the amplification products are indicated by the vertical scale.
FIG. 2.
Phylogenetic clustering of M. tuberculosis isolates. A neighbor-joining tree was generated from differences in Genotyper output read as binary data. The lower scale represents the genetic distances between the isolates and/or amplitypes. MDR, multidrug resistant; TB, tuberculosis.
TABLE 2.
Details of polymorphic FAFLP markers used for genome-wide sampling of biogeographic differences among predominant strains from across the world
| AFLP marker | Primer pair | Genomic coordinates | Locus | Annotation |
|---|---|---|---|---|
| 90 | MseI+0/EcoRI+G | 2278594-2278651 | Rv2031c | 14-kDa antigen (Hsp16.3) |
| 95 | MseI+0/EcoRI+G | 3042461-3042523 | Rv2729c | Conserved hypothetical protein |
| 133 | MseI+0/EcoRI+G | 3732652-3732753 | Rv3343c | Member of the PPE family |
| 162 | MseI+0/EcoRI+G | 922607-922735 | Rv0830 | Conserved hypothetical protein |
| 183 | MseI+0/EcoRI+G | 3507201-3507351 | Rv0883c | Conserved hypothetical protein |
| 2 | MseI+0/EcoRI+G | 2874632-2874797 | Rv1451 | Cytochrome C oxidase assembly factor |
| 204 | MseI+0/EcoRI+G | 3734250-3734419 | Rv3343c | Member of the PPE family |
| 231 | MseI+0/EcoRI+G | 152456-152654 | Rv0126 | Trehalose synthase |
| 261 | MseI+0/EcoRI+G | 2819664-2819893 | Rv2504c | ScoA, energy metabolism(fermentation) |
| 272 | MseI+0/EcoRI+G | 1920645-1920883 | Rv1696 | RecN |
| 285 | MseI+0/EcoRI+G | 2274225-2274477 | Rv2027c | Regulatory protein, sensor histidine kinase |
| 307 | MseI+0/EcoRI+G | 1900195-1900468 | Rv1675c | Putative transcriptional regulator |
| 326 | MseI+0/EcoRI+G | 1664358-1664652 | Rv1475c | Aconitase |
| 351 | MseI+0/EcoRI+G | 1011015-1011333 | Rv0907 | Penicillin binding protein |
| 356 | MseI+0/EcoRI+G | 2563840-2564163 | Rv2291 | Thiosulfate sulfurtransferase |
| 366 | MseI+0/EcoRI+G | 1220493-1220826 | Rv3159c | Host cell receptor binding protein |
| 373 | MseI+0/EcoRI+G | 4343205-4343544 | Rv3868 | Conserved hypothetical protein |
| 388 | MseI+0/EcoRI+G | 3730329-3730683 | Rv2888c | Probable amidase |
| 403 | MseI+0/EcoRI+G | 3271384-3271754 | Rv2935 | PpsE phenolpthiocerol synthesis (PksF) |
| 457 | MseI+0/EcoRI+G | 3465375-3465799 | Rv3096 | Hypothetical protein |
| 158 | MseI+0/EcoRI+C | 2910836-2910963 | Rv2584c | Adenine phosphoribosyl transferase |
| 205 | MseI+0/EcoRI+C | 4371591-4371763 | Rv3887c | Probable membrane protein |
| 213 | MseI+0/EcoRI+C | 848749-848929 | Rv0755c | Host cell receptor binding protein/PPE |
| 248 | MseI+0/EcoRI+C | 507-720 | Rv1018c | UDP N-acet-glucose pyrophosphorylase |
| 279 | MseI+0/EcoRI+C | 3197899-3198145 | Rv2888c | Probable amidase |
| 339 | MseI+0/EcoRI+C | 4034016-4034323 | Rv3592 | Conserved hypothetical protein |
| 399 | MseI+0/EcoRI+C | 37096-37462 | Rv0035 | Putative amino acid activating protein |
| 413 | MseI+0/EcoRI+C | 4105241-4105622 | Rv3665c | Dipeptide transport system permease |
| 485 | MseI+0/EcoRI+C | 3791264-3791717 | Rv3377c | Conserved hypothetical protein cyclase |
| 198 | MseI+0/EcoRI+A | 1306776-1306943 | Rv1175c | 2-4,Dienoyl coA reductase |
| 246 | MseI+0/EcoRI+A | 3527665-3527877 | Rv3159c | Host cell receptor binding protein/PPE |
| 254 | MseI+0/EcoRI+A | 4387695-4387914 | Rv3902c | Hypothetical protein |
| 264 | MseI+0/EcoRI+A | 961543-961773 | Rv0862c | Conserved hypothetical protein |
| 337 | MseI+0/EcoRI+A | 4224186-4224491 | Rv3778c | NifS-related protein |
| 411 | MseI+0/EcoRI+A | 1696775-1697153 | Rv1507c | Conserved hypothetical protein |
| 384 | MseI+0/EcoRI+T | 1036526-1036880 | Rv0929 | Phosphate transport protein PstC |
Amplitypes D and E were mainly generated from French and Italian isolates. Some of the strains used were from patients with extrapulmonary infections, although most were from pulmonary tuberculosis patients. No correlation of the amplitype with patterns of disease versus treatment outcome (or drug resistance) for a given region was found.
Correlation between FAFLP analysis and IS6110 typing patterns.
Many of the strains of Indian origin were low-copy-number IS6110 types, and about 10% of the strains carried only a single copy of IS6110. About 80% of the isolates were from North India, mainly from Delhi, Chandigarh, and Agra. The remaining 20% of the isolates were from Western India, mainly isolated from Jaipur and Ahmedabad. The division of the Indian cluster (type B) into distinct subclusters of low- and high-copy-number IS6110 strains was more remarkable. This was indeed based on specific genomic signatures revealed by high-copy-number IS6110 isolates as well as low-copy-number IS6110 isolates. The correlation of IS6110 profiles with the overall clustering of the strains we studied was very significant in the case of the Indian cluster. The microbiological, biochemical, and molecular genetic characteristics of many of the isolates from this cluster have already been published (19, 20).
FAFLP analysis also subdivided the large type C cluster into several subclusters. Many of the strains of this type had one to four copies of the IS6110 element. The branching out of the Canadian, Tanzanian, and Vietnamese strains was more remarkable, as these strains carried only one copy or no copies of the IS6110 element. Interestingly, however, strains of Italian origin carried only a single IS6110 copy, yet 15 of the 20 strains analyzed clustered with French strains which carried 8 to 14 copies of the element. Similarly, most of the strains isolated in The Netherlands displayed multicopy IS6110 profiles and these clustered together. This indicates that the clustering occurred mostly based on the pattern of chromosomal markers that have shown a strong geographic bias, so much so that the IS6110 copy number could not influence regional clustering to a large extent. This is a very significant observation in the context of the molecular epidemiology of M. tuberculosis vis-à-vis IS6110-based typing methods.
LSPs in different FAFLP clusters.
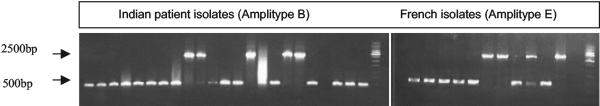
LSP analysis using primers specific for flanking and internal regions of the TbD1 and Rd9 elements was performed with representative samples from India (amplitype B), Italy (amplitype E), Australia (amplitype C), Peru (amplitype C), and France (amplitype E). TbD1, as well as the Rd9 region, was found to be present in 13 of the 96 strains from India (13.54%). Similarly, these were also intact in 4 (20%) of the 20 French strains analyzed. Despite this similarity in the intactness of evolutionarily significant genomic landmarks (Fig. 3), the French and Indian strains clustered separately with geographically closer strains. Four of the 13 (30.76%) human-derived strains of the M. tuberculosis complex from Australia revealed the absence of the TbD1 and Rd9 regions from the genome. The TbD1 region was also absent from 3 of the 20 (15%) Italian M. bovis strains. These strains, however, did not cluster with the Australian strains. All of the Peruvian and Dutch isolates tested revealed normal TbD1 and RD9 profiles, but these isolates clustered separately by FAFLP profiling. Therefore, the TbD1 and Rd9 analysis clearly indicates that the repertoire of genomic rearrangements scanned by FAFLPs is far more specific than are LSPs. It appears that LSPs occur rather randomly in some (15 to 30%) strains across the continent, irrespective of the host status and geographic location.
FIG. 3.
TbD1 LSPs in representative Indian and French isolates. PCR products of 500 bp indicate a deletion of the TbD1 region from the genome, whereas products of 2,500 bp indicate the intactness of the region.
DISCUSSION
It has long been speculated that the genomes of M. tuberculosis strains from across the world are remarkable for their lack of genetic heterogeneity and that most of the strains are similar at the nucleotide sequence level. The absence of silent base substitutions in M. tuberculosis has been interpreted as indicating that M. tuberculosis is a relatively young species, perhaps 15,000 to 20,000 years old (21). Other explanations, such as an increased fidelity of replication or the existence of clonal sweeps that purge variability, have been speculated upon (13), but there is no supporting evidence for them. Many of the evolutionarily important events analyzed to date mainly focused on the bottlenecks acquired by the M. tuberculosis chromosome on an evolutionary timescale and whether M. bovis was the progenitor of M. tuberculosis or vice versa (7, 21). We suggest that there is a need for exploration of the ancient geographic circumstances under which M. tuberculosis may have become associated with its human host and subsequently expanded its host tropism amidst pathogen evolution on an evolutionary timescale. Also, an assessment of whether geographic partitioning of the M. tuberculosis gene pool has something to do with the overwhelming human population diversity needs to be fully performed. We tried to test these ideas by using the high-resolution fingerprinting method of FAFLP to survey base substitutions and genetic rearrangements across the genome. This method of genotyping was carefully chosen and modified to achieve a high resolution that was sufficient enough to allow the rapid identification of genomic loci without much ambiguity. This is possible because one can essentially computationally predict, using in silico approaches, the entire pattern of FAFLPs by using the genome sequence text data as a template.
In this study, variations in terms of band sharing, even up to a difference of 1 bp, indicated the discriminatory power of the technique. DNA sequencing of 36 of the 136 ± 1 FAFLP fragments revealed that the genomic diversity in our collection of strains was mostly due to base substitutions or deletions in important genes, such as those for aconitase, amidase, permease, several members of the PE and PPE family, putative regulators of transcription, efflux pumps, hypothetical proteins, conserved hypothetical proteins, etc. (Table 2). Our observations that genotypic differences were indeed due to the availability or abolishment of EcoRI and MseI sites support the idea that the rate of silent substitutions may be higher than was previously expected and that M. tuberculosis may not be evolutionarily as very recent as was previously proposed (21).
We did not analyze multiple-drug-resistant strains with candidate gene mutations separately in the context of resistance type-phenotype correlations, although such strains predominated in the Indian (19, 20) and Dutch (data not shown) clusters. Instead, they were subjected to FAFLP typing in a blinded manner and later analyzed for amplitype-specific, regional representations in the clusters. It would have been very exciting if such strains had carried potential resistance-linked markers. However, we do not know how significant such resistance-associated markers would have been in terms of influencing the current biogeographic clustering. Many of the strains that we analyzed (16) were already characterized by their IS6110 restriction fragment length polymorphisms and were supplied to us as blind-coded DNA samples. We found the IS6110 clusters of epidemiologically related strains to be further subdivided when FAFLP data were used for phylogenetic analyses. The subdivision of Indian clusters containing only one or zero IS6110 copies was more remarkable. One possible reason for geographically related strains to cluster together, independent of their IS6110 profiles, could be the fact that the FAFLP amplitype is essentially a sampled output of various base substitutions and specific deletions across the genome as a whole, taking into consideration various selection pressures that might have operated on the bacterial chromosome in totality, independent of the mobility of the IS6110 element. The amplitype, in its true sense, therefore, is a representative of base substitutions (or deletion events) in a given isolate and is not a reflection of the criteria used for the selection of isolates for the study.
Our analysis of LSPs in representative isolates from different FAFLP clusters was indeed suggestive of a deletion mechanism occurring as a random, not a universal, phenomenon. The evolutionary scenario proposed by Brosch et al. (7), however, is driven by the assumption that large genomic deletions are highly specific and precisely timed phenomena. This does not appear to be the case, however, at least for the >150 isolates that we analyzed. We studied a large representative collection of isolates from all different geographical regions, particularly the Indian, South American, and Australian regions. It is therefore apparent that LSPs had a negligible impact on clustering, which was rather dictated by regional geographic forces, including the environmental impact and host diversity (1), and these dominated over various insertion, deletion, and substitution events across the genome as a whole.
Our “phylogenomic” analyses of strains from diverse populations support the idea that M. tuberculosis may have undergone adaptive evolution as a result of selection at many loci. We suggest that the distinctiveness of M. tuberculosis genotypes in South Asia, Australia, and the West could be due to strain variants of M. tuberculosis that are particularly well adapted to patients in these regions. These variants possibly spread through different populations during different time periods and then adapted. Subsequent recombination has not been sufficiently frequent to break such geographical groupings. In another scenario, genetic rearrangements seem to have occurred on a global scale for most of the motifs studied, such as the 20 variable regions resulting from insertion-deletion events (6) on an evolutionary timescale (7) and some of the candidate gene polymorphisms (21), but the genome sequence has remained broadly conserved at the nucleotide level for some of the genes. In both cases, our data may be significant enough to speculate about the age of the M. tuberculosis genotype in South Asia and the West and to predict the time and circumstances under which M. tuberculosis possibly became associated with humans.
Our findings on genetic differences among the genotypes may be the topic of further investigations to find out the exact role of the polymorphisms detected by FAFLP. Much of our understanding of M. tuberculosis genome plasticity, pathogenicity, and dissemination dynamics is based on strains from industrialized countries. Independent of this, the genomic differences among various isolates from across the world at many loci, as highlighted in this study, should encourage large-scale genomic profiling of strains. Isolates that are indigenous to some of the understudied geographic regions that are relatively unaffected by homogenizing forces such as immigration, international travel, and business tourism should be analyzed on priority. Such a geographic genomics approach may unravel newer possibilities toward our understanding of mycobacterial pathogenesis and the host component. However, since environmental and host factors clearly contribute to the clinical and epidemiologic behavior of strains, these must be carefully integrated into the investigative process.
Acknowledgments
We are thankful to D. van Soolingen, Cristina Gutierrez, and Debby Cousins for providing genomic DNA samples of the strains from The Netherlands, France, and Australia, respectively. The help given by Sugandhan and Suresh Kumar is thankfully acknowledged.
Financial assistance from the Department of Biotechnology, Government of India, the Council for Scientific and Industrial Research (CSIR), The World Health Organization (TDR), and the International Society for Infectious Diseases (ISID) is gratefully acknowledged. This work was supported by core grants to the CDFD from the Department of Biotechnology, Government of India.
REFERENCES
- 1.Ahmed, N., L. Caviedes, M. Alam, K. R. Rao, V. Sangal, P. Sheen, R. H. Gilman, and S. E. Hasnain. 2003. Distinctiveness of Mycobacterium tuberculosis genotypes from human immunodeficiency virus type 1-seropositive and -seronegative patients in Lima, Peru. J. Clin. Microbiol. 4:1712-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, N., M. Alam, A. A. Majeed, S. A. Rahman, A. Cataldi, D. Cousins, and S. E. Hasnain. 2002. Genome sequence based, comparative analysis of the fluorescent amplified fragment length polymorphisms (FAFLP) of tubercle bacilli from seals provides molecular evidence for a new species within the Mycobacterium tuberculosis complex. Infect. Genet. Evol. 2:193-199. [DOI] [PubMed] [Google Scholar]
- 3.Behr, M. A., and P. M. Small. 1997. Molecular fingerprinting of Mycobacterium tuberculosis: how can it help the clinician? Clin. Infect. Dis. 25:806-810. [DOI] [PubMed] [Google Scholar]
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Bifani, P. J., B. B. Plikakytis, and V. Kapur. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-455. [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, A. Pym, K. Eiglmeier, T. Garnier, and S. T. Cole. 2000. Comparative genomics of the mycobacteria. Int. J. Med. Microbiol. 290:143-152. [DOI] [PubMed] [Google Scholar]
- 7.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 39:3537-3544. [DOI] [PubMed] [Google Scholar]
- 9.Gamieldien, J., A. Ptitsyn, and W. Hide. 2002. Eukaryotic genes in Mycobacterium tuberculosis could have a role in pathogenesis and immunomodulation. Trends Genet. 18:5-8. [DOI] [PubMed] [Google Scholar]
- 10.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 11.Goulding, J. N., J. Stanley, N. Saunders, and C. Arnold. 2000. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kursters. 1996. Evaluation of DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 13.Kato-Maeda, M., P. J. Bifani, B. N. Krieswirth, and P. M. Small. 2001. The nature and consequence of genetic variability in Mycobacterium tuberculosis. J. Clin. Investig. 107:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstedt, B., E. Heir, T. Vardund, and G. Kapperud. 2000. Fluorescent amplified polymorphism genotyping of Salmonella enterica subsp. enterica serovars and comparison with pulsed-field gel electrophoresis typing. J. Clin. Microbiol. 38:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majeed, A. A., N. Ahmed, K. R. Rao, S. Ghousunnissa, F. Kauser, B. Bose, H. A. Nagarajaram, V. M. Katoch, D. V. Cousins, L. A. Sechi, R. H. Gilman, and S. E. Hasnain. 2004. AmpliBASE MT: a Mycobacterium tuberculosis diversity knowledge base. Bioinformatics 20:989-992. [DOI] [PubMed] [Google Scholar]
- 18.Poulet, S., and S. T. Cole. 1995. Repeated DNA sequences in mycobacteria. Arch. Microbiol. 163:79-86. [DOI] [PubMed] [Google Scholar]
- 19.Siddiqi, N., M. Shamim, N. K. Jain, A. Rattan, A. Amin, V. M. Katoch, S. K. Sharma, and S. E. Hasnain. 1998. Molecular analysis of multi-drug resistance in Indian isolates of Mycobacterium tuberculosis. Mem. Inst. Oswaldo Cruz 3:589-594. [DOI] [PubMed] [Google Scholar]
- 20.Siddiqi, N., M. Shamim, S. Hussain, R. K. Choudhary, N. Ahmed, Prachee, S. Banerjee, G. R. Savithri, M. Alam, N. Pathak, A. Amin, M. Hanief, V. M. Katoch, S. K. Sharma, and S. E. Hasnain. 2002. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob. Agents Chemother. 46:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Soolingen, D., P. de Haas, P. Hermans, and J. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:199-201. [DOI] [PubMed] [Google Scholar]
- 23.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]