ABSTRACT
The physiological function of biotin requires biotin protein ligase activity in order to attach the coenzyme to its cognate proteins, which are enzymes involved in central metabolism. The model intracellular pathogen Francisella novicida is unusual in that it encodes two putative biotin protein ligases rather than the usual single enzyme. F. novicida BirA has a ligase domain as well as an N-terminal DNA-binding regulatory domain, similar to the prototypical BirA protein in E. coli. However, the second ligase, which we name BplA, lacks the N-terminal DNA binding motif. It has been unclear why a bacterium would encode these two disparate biotin protein ligases, since F. novicida contains only a single biotinylated protein. In vivo complementation and enzyme assays demonstrated that BirA and BplA are both functional biotin protein ligases, but BplA is a much more efficient enzyme. BirA, but not BplA, regulated transcription of the biotin synthetic operon. Expression of bplA (but not birA) increased significantly during F. novicida infection of macrophages. BplA (but not BirA) was required for bacterial replication within macrophages as well as in mice. These data demonstrate that F. novicida has evolved two distinct enzymes with specific roles; BplA possesses the major ligase activity, whereas BirA acts to regulate and thereby likely prevent wasteful synthesis of biotin. During infection BplA seems primarily employed to maximize the efficiency of biotin utilization without limiting the expression of biotin biosynthetic genes, representing a novel adaptation strategy that may also be used by other intracellular pathogens.
IMPORTANCE
Our findings show that Francisella novicida has evolved two functional biotin protein ligases, BplA and BirA. BplA is a much more efficient enzyme than BirA, and its expression is significantly induced upon infection of macrophages. Only BplA is required for F. novicida pathogenicity, whereas BirA prevents wasteful biotin synthesis. These data demonstrate that the atypical occurrence of two biotin protein ligases in F. novicida is linked to distinct roles in virulence and biotin metabolism.
INTRODUCTION
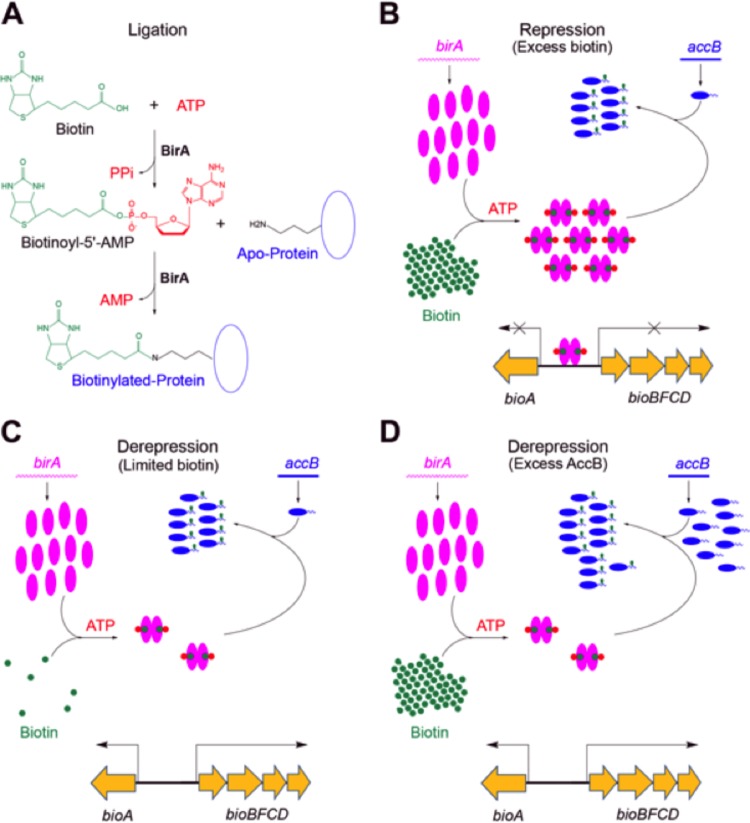
Biotin protein ligases are highly conserved and metabolically essential enzymes that catalyze attachment of the biotin coenzyme (vitamin H) to key protein subunits (or domains) of enzymes of central metabolism (1, 2) (Fig. 1A). Bacterial biotin protein ligases fall into two groups. Group I ligases act solely as biotin attachment enzymes, whereas group II proteins (generally called BirA proteins) are bifunctional proteins that regulate biotin synthesis in addition to catalyzing biotin attachment. The two enzyme groups are readily distinguished by the presence of an N-terminal winged helix-turn-helix domain that facilitates the binding of BirA proteins to the operator sequences of biotin synthetic operons and subsequent repression of transcription and biotin synthesis (1, 3). The group II ligases lack this DNA binding domain and show structural diversity. For example, Bacillus subtilis BirA can be converted to a fully functional group I ligase by deletion of the winged helix-turn-helix domain (3), whereas similar E. coli BirA N-terminal deletions result in ligases of severely compromised activity (4, 5). DNA binding by BirA proteins requires biotinoyl-AMP (biotinoyl-adenylate), the product of the first ligase half reaction (Fig. 1A). The fact that these proteins make their own regulatory ligand allows biotin synthesis to be regulated by both the intracellular biotin concentration and the levels of proteins that are the substrates for biotin attachment (Fig. 1B to D) (2, 6). The two modes of transcriptional derepression act by a common mechanism in that both decrease the levels of the BirA–biotinoyl-AMP complex required to bind the bio operator (7). Biotinylated enzymes are rare, as mammals have only four such proteins and Escherichia coli has only a single biotinylated protein. Typically, an organism (e.g., a mammal, plant, or bacterium) encodes a single biotin protein ligase that modifies each of the biotin-requiring enzymes.
FIG 1 .
Biotin protein ligase reaction and regulation of E. coli biotin operon transcription. (A) Biotin protein ligase activity of BirA and BplA. (B to D) General model of bio operon regulation by BirA. Pink ovals, BirA; tailed blue ovals, AccB; green dots, biotin; green dots with red pentagons, bio-5′-AMP (biotinoyl-5′-adenylate). Panel B shows the transcriptionally repressed state, whereas panels C and D separately show the two modes of derepression of bio operon transcription engendered by either biotin limitation or excess unbiotinylated AccB acceptor protein. Both derepression modes act by decreasing the level of biotinoyl-AMP, the ligand required for operator binding by BirA.
Francisella novicida is a Gram-negative bacterium and model intracellular pathogen that is a rare cause of human disease (8), often used as a surrogate for the category A select agent Francisella tularensis. Our recent studies showed that F. novicida virulence requires biotin synthesis (9). The biotin operons of F. novicida and E. coli have the same gene arrangement, although in both bacteria, the enzymes (BioJ and BioH, respectively) that catalyze the last step in synthesis of the biotin pimelate moiety are encoded outside the operons (see Fig. S1 in the supplemental material) (9).
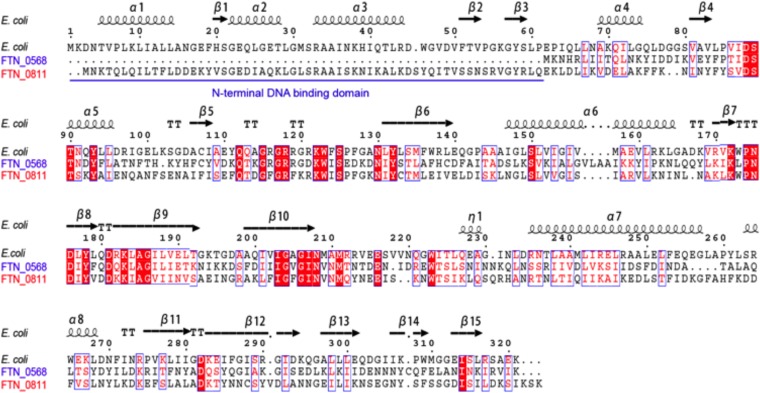
F. novicida differs markedly from E. coli and other bacteria in that it encodes two putative biotin protein ligases. These are FTN_0811, a group II candidate ligase of 320 residues encoded next to the last gene of the biotin operon (albeit divergently transcribed) and FTN_0568, a gene located far from the biotin operon that encodes a group I biotin protein ligase of 260 residues (Fig. 2). The longer protein, which we call BirA, has an N-terminal domain that is readily modeled as a winged helix-turn-helix regulatory domain, whereas the shorter protein, which we name BplA (biotin protein ligase A), lacks this domain (Fig. 2). These observations raised the questions of whether both genes encode active ligase enzymes and, if so, what the physiological rationale for the presence of the unique pair of biotin protein ligases in F. novicida is. We report that both genes encode functional biotin protein ligases, although BplA is a much more robust enzyme than BirA. BplA is required for pathogenesis, whereas BirA appears to function to prevent wasteful biotin synthesis.
FIG 2 .
Sequence alignments of FTN_0568 (BplA, in blue) and FTN_0811 (BirA, in red) with E. coli BirA, the DNA binding domain of which is underlined in blue. The E. coli BirA structural elements are depicted above the alignments.
RESULTS
Both bplA and birA encode active biotin protein ligases.
F. novicida BplA and BirA were first assayed for ligase activity by the ability to permit growth of E. coli strain BM4062 at low biotin concentrations. The high biotin requirement of strain BM4062 is due to a point mutation (birA85) that results in a temperature-sensitive growth phenotype, decreased affinity for biotin, and deficient regulation of biotin operon transcription (10, 11). The strain also contains a bioF::lacZ fusion, which results in biotin auxotrophy and provides a visual assay of bio operon transcription (10).
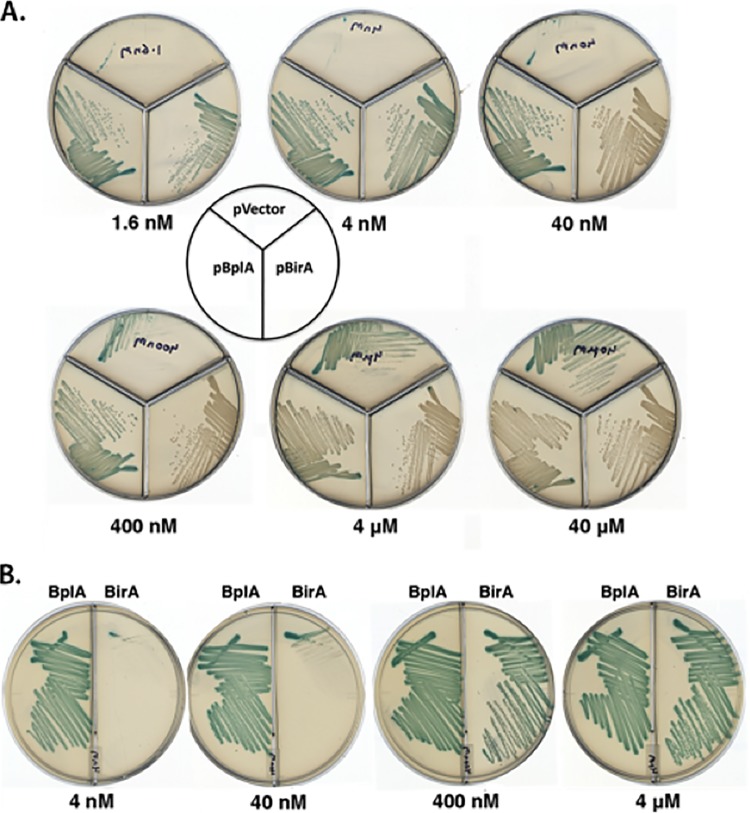
The F. novicida genes were expressed from an arabinose-inducible (paraBAD) promoter in the presence or absence of arabinose. Both proteins allowed growth of strain BM4062 at nonpermissive temperatures and at low biotin concentrations. However, at low biotin concentrations, growth of the strain expressing F. novicida BirA required arabinose induction, whereas the strain expressing F. novicida BplA grew well under conditions (glucose in place of arabinose) that fully repress basal transcription from the paraBAD promoter (Fig. 3) (12). These data argued either that BplA was a much more active ligase than BirA or that BplA was more readily translated in E. coli. To address these possibilities, we constructed and expressed hexahistidine-tagged versions of the two proteins, and upon denaturing gel electrophoresis of crude extracts, we found that BirA and BplA were expressed at very similar levels (data not shown). Thus, the lack of complementation of the E. coli birA strain observed at low biotin concentrations seemed unlikely to be due to poor expression of F. novicida BirA and hence strongly suggested that BplA was a much more robust ligase than BirA.
FIG 3 .
Growth of the derivatives of E. coli strain BM4062 carrying plasmid pBAD322 (empty vector) or pBAD322 derivatives encoding either the F. novicida BplA or BirA ligase. (A) Ligase expression was induced with arabinose; the strain BM4062 derivative with a wild-type arabinose operon was used to avoid arabinose toxicity. The biotin concentrations are shown below each plate. As shown at the higher biotin concentrations in the sectors containing the vector strain, the BirA encoded by the host strain mutant has weak ligase activity (21). (B) The strains were grown in the absence of arabinose induction and in the presence of glucose to repress the basal level of expression from the paraBAD promoter. The original strain BM4062 was used. Strain BM4062 containing the plasmid encoding BplA was streaked in the left-hand sectors, whereas the right-hand sectors contained the BirA-encoding plasmid. The plates were minimal medium M9 supplemented with 0.1% Casamino Acids, 40 µg/ml X-Gal, and 100 µg/ml ampicillin with 0.2% arabinose supplementation (A) and 0.4% glucose supplementation (B). The plates were incubated overnight at either 42°C (A) or 37°C (B).
As expected from its lack of a DNA binding domain, BplA expression had no effect on regulation of the E. coli biotin operon (assayed by X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] hydrolysis) (Fig. 3). However, an unexpected finding was that expression of F. novicida BirA weakly repressed transcription of the E. coli bio operon (Fig. 3). This was most clearly seen upon arabinose induction of F. novicida BirA expression in the presence of 40 nM biotin (Fig. 3A) but was also seen in the absence of induction (the tiny white colonies formed in the presence of 40 nM biotin) (Fig. 3B).
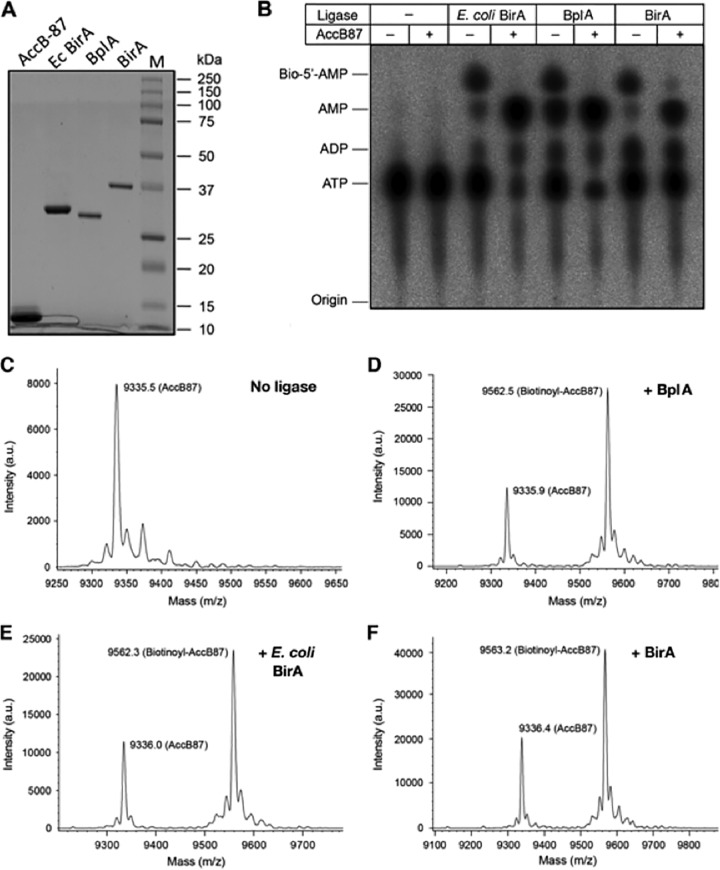
To further compare the two ligases, we purified the hexahistidine-tagged proteins to homogeneity (Fig. 4; also, see Fig. S2A and B in the supplemental material), and peptide mapping showed that each contained a significant number of the tryptic peptides predicted from the DNA sequence (see Fig. S2C and D), thereby confirming the identification of the proteins. Cross-linking with ethylene glycol bis-succinimidylsuccinate indicated that BplA was monomeric in solution (see Fig. S2E), whereas BirA formed a mixture of monomers and dimers (see Fig. S2F; dimers were also seen during purification). An attempt to detect formation of BplA-BirA mixed multimers by cross-linking gave no support for this notion (see Fig. S2G).
FIG 4 .
Enzymatic activities of the F. novicida BplA and BirA proteins. (A) SDS-PAGE analysis of the proteins used in these experiments. M, molecular mass standards. The SDS-PAGE results are consistent with the calculated molecular masses of the hexahistidine-tagged versions of E. coli BirA, F. novicida BirA, and F. novicida BplA, which are 36.1, 38.7, and 32.5 kDa, respectively. (B) TLC assays of biotin protein ligase activities. The synthesis of 32P-labeled biotinoyl-AMP (biotinoyl-adenylate) from [α-32P]ATP was assayed as described in Materials and Methods, analyzed by thin-layer chromatography on glass cellulose plates, and visualized by autoradiography. The reaction products biotinoyl-5′-AMP (bio-5′-AMP), ADP, AMP, and the ATP are indicated in the left margin. The proteins assayed are given at the top of the figure. The abilities of the F. novicida proteins to convert α-32P-labeled ATP to biotinoyl-AMP and subsequently to AMP were tested by addition of the AccB-87 acceptor protein. (C to F) Low-resolution matrix-assisted laser desorption/ionization analyses of the overall conversion of apo-AccB-87 to the biotinylated species catalyzed by the various ligases in overnight incubations as shown on the panels. The calculated masses of apo-AccB-87 and biotinylated AccB-87 are 9,333.8 and 9,560.1, respectively.
The purified hexahistidine-tagged proteins (Fig. 4A) were used to compare the F. novicida BplA and BirA ligases to the well-studied E. coli BirA. We used a thin-layer-chromatographic method that assays conversion of α-32P-labeled ATP and biotin to biotinoyl-AMP (Fig. 4B). This provides a direct assay of the first ligase partial reaction (Fig. 1A) and upon addition of acceptor protein provides an indirect assay of the second ligase partial reaction, transfer of biotin from biotinoyl-AMP to the acceptor protein (Fig. 1A). Addition of acceptor protein results in loss of the intensity of the biotinoyl-AMP spot with the concomitant appearance of AMP and consumption of ATP (Fig. 4B). The increased consumption of ATP results from the fact that in the absence of acceptor, biotinoyl-AMP remains tightly bound within the ligase active site such that only one molecule is formed per molecule of ligase (i.e., biotinoyl-AMP synthesis is not catalytic). Biotin transfer to an acceptor protein allows catalysis, which results in increased conversion of ATP to AMP.
In these assays, the most striking difference between BplA and BirA was that upon addition of the acceptor protein, BplA (and also E. coli BirA) consumed most of the α-32P-labeled ATP, whereas no appreciable increase in ATP consumption was seen in the F. novicida BirA assay (Fig. 4B). This indicates that F. novicida BirA catalyzes biotin attachment more slowly than the other two ligases. Moreover, residual biotinoyl-AMP was seen only in the case of F. novicida BirA (Fig. 4B). The F. novicida BirA reactions also accumulated an appreciable amount of ADP, an off-pathway product. ADP production has been previously observed only in reactions with mutant E. coli BirA proteins having compromised ligase activity (4). However, mass spectrometry showed that given a long incubation time, F. novicida BirA, like BplA and E. coli BirA, could catalyze the full ligase reaction (Fig. 4C to F).
F. novicida contains only a single biotinylated protein.
One possible rationale (albeit unprecedented) for the presence of two biotin protein ligases in F. novicida would be that the enzymes could specifically biotinylate different acceptor proteins. The Francisella species genome annotations list only a single protein as containing a canonical site for biotin attachment, the AccB subunit of acetyl coenzyme A (acetyl-CoA) carboxylase. These annotations seem reliable because the gene is located immediately upstream of a gene that encodes another acetyl-CoA carboxylase subunit, AccC (biotin carboxylase), and this gene arrangement is found in many bacteria, including the well-characterized accB-accC operon of E. coli (13, 14). Moreover, the Francisella AccB proteins include a readily modeled “thumb” structure, which is essential for E. coli AccB function (15) and constitutes a motif allowing small biotinylated proteins to be identified as subunits of acetyl-CoA carboxylase rather than of another biotin-dependent enzyme (16). Note that in E. coli (and the other bacteria tested), accB is an essential gene. Moreover, its biotinylation is essential for acetyl-CoA carboxylase-catalyzed synthesis of malonyl-CoA, the indispensable building block of fatty acid synthesis which cannot be provided by supplementation of growth media.
Notwithstanding the annotations, it remained possible that Francisella species encode a biotinylated protein that lacks a recognizable biotin attachment sequence. To test this possibility, we performed Western blots of extracts of three F. novicida strains using a streptavidin probe and detected only a single biotinylated protein in these extracts (see Fig. S3 in the supplemental material). Consistent with its annotation, this protein had the characteristic and atypical SDS gel mobility first seen for E. coli AccB (13). Although according to amino acid sequencing the E. coli AccB protein is 16.7 kDa, it migrates as though it is considerably larger (ca. 20 to 22 kDa). This anomalous migration is attributed to the extended alanine/proline-rich sequences spanning residues 40 to 70 (13). Hence, F. novicida encodes two enzymes to modify a single acceptor protein. The protein extracts assayed were from F. novicida mutant strains in which the gene encoding either BplA or BirA was disrupted by insertion of a kanamycin resistance cassette or from a mutant lacking the biotin synthetic enzyme BioJ as a control (4). Biotinylated AccB was present in all three extracts, although the band in the strain lacking BplA was considerably fainter than that in the strain lacking BirA (see Fig. S3). Note that E. coli also grows well with only a fraction of its normal level of biotinylated AccB (13, 15). These data together with the enzymatic assays and E. coli complementation data demonstrate that BplA is the major F. novicida biotinylation enzyme, although BirA suffices for growth in the laboratory at some cost in growth rate (see below; also, see Fig. S5 in the supplemental material).
F. novicida BirA binds the E. coli bio operator.
The surprising result that expression of F. novicida BirA weakly repressed transcription of the E. coli bioBFCD operon (Fig. 3) argued that the protein must bind the E. coli bioO operator (Fig. 1). Indeed, sequences related to the E. coli operator sequence are found within the Francisella bioA-bioB intragenic regions (see Fig. S4A in the supplemental material), suggesting that these two divergent bacteria might share some BirA-operator interactions. This was tested by electrophoretic mobility shift assays, which showed that F. novicida BirA bound the minimal E. coli bio operator (see Fig. S4C), although markedly less tightly than its cognate operator (see Fig. S4D). This result is in accord with the results of Fig. 3 and with reverse transcriptase PCR analyses (data not shown), which showed only a 3- to 5-fold repression of E. coli bioBFCD transcription upon high level expression of F. novicida BirA. In contrast, E. coli BirA failed to bind the F. novicida operator (see Fig. S4F), although the BirA preparation bound its cognate operator (see Fig. S4E). The inability to bind the F. novicida operator is expected, because several of the E. coli operator bases shown to interact with E. coli BirA in previous DNA footprinting experiments are absent (17). As expected, BplA showed no binding of the F. novicida sequence (see Fig. S3B).
F. novicida BirA represses bioF expression.
Since BirA, but not BplA, bound the cognate bioO operator in vitro, we tested the in vivo role of each protein in transcriptional regulation of biotin synthesis in F. novicida. We generated deletion mutants lacking either bplA or birA and quantified expression of the representative biotin synthesis gene bioF by quantitative real time-PCR (qRT-PCR) (Fig. 5). The levels of bioF expression were similar in the wild-type and ΔbplA strains but significantly increased in the ΔbirA strain (Fig. 5). Complementation of the ΔbirA mutation with a wild-type copy of the gene restored bioF expression to the wild-type level. These data indicate that BirA acts as a transcriptional repressor of bioF expression in F. novicida, whereas BplA lacks repressor activity.
FIG 5 .
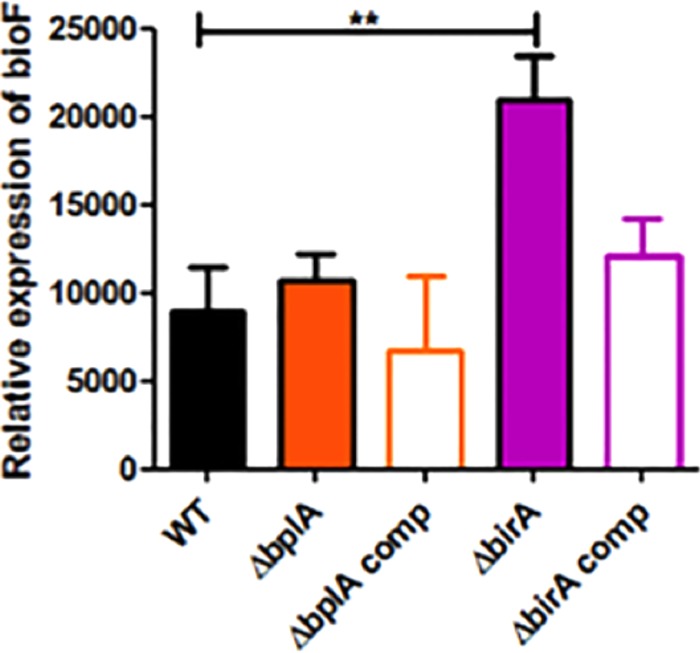
F. novicida BirA represses bioF expression. Expression of bioF in wild-type F. novicida U112 (WT) and the ΔbplA, ΔbirA, bplA, and birA trans complemented (comp) strains was measured by quantitative real-time PCR (qRT-PCR) in relation to the housekeeping gene uvrD. **, P < 0.005.
BplA contributes to F. novicida replication in minimal medium and in macrophages.
Given that biotin protein ligases are essential enzymes of central metabolism, we tested the physiological contributions of BplA and BirA to F. novicida growth. In a defined minimal medium (Chamberlain’s medium [CHB]), the ΔbplA strain exhibited a modest growth defect relative to the wild-type strain (see Fig. S5A in the supplemental material). In contrast, the ΔbirA strain grew as well as the wild-type strain, as did the ΔbplA and ΔbirA complemented strains (see Fig. S5A). Moreover, addition of biotin rescued the growth defect of the ΔbplA strain (see Fig. S5B). These data indicate that BplA plays a more important role than BirA in F. novicida growth in minimal medium. This is consistent with its superior biotin ligase activity in E. coli (Fig. 3) and in vitro (Fig. 4).
We hypothesized that the contribution of BplA to F. novicida growth in minimal medium would be reflected during macrophage infection, where the bacteria must traffic through the nutrient-limited host cell phagosome. To test this premise, we infected murine bone marrow-derived macrophages with the wild-type and deletion strains and quantified levels of intracellular bacteria at 5.5 h postinfection. In contrast to wild-type bacteria, which readily replicated within these macrophages, the ΔbplA mutant strain replicated poorly and was present at roughly 10-fold-lower levels (Fig. 6A). Complementation of the ΔbplA mutation with a plasmid expressing the wild-type gene restored wild-type levels of replication. The birA mutant, however, replicated similarly to the wild-type strain (Fig. 6A), indicating that BirA is not required for F. novicida intracellular replication. These data indicate that BplA makes a much more significant contribution to F. novicida replication than does BirA. Furthermore, bplA but not birA transcripts were up-regulated during macrophage infection (Fig. 6B), a further indication of a significant role for BplA.
FIG 6 .
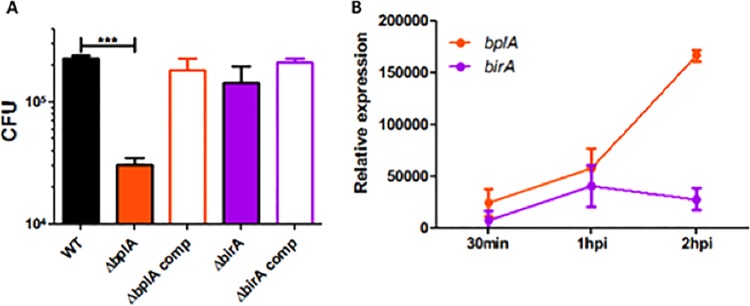
F. novicida BplA is required for replication in macrophages. (A) Murine bone marrow-derived macrophages were infected with a 20:1 MOI of wild-type F. novicida U112 (WT) and the ΔbplA, ΔbirA, bplA complemented (comp), and birA complemented (comp) strains. CFU were quantified at 5.5 h postinfection. (B) Murine BMDM macrophages were infected with wild-type F. novicida U112 (MOI 20:1), and expression of bplA and birA was quantified by qRT-PCR relative to expression of the housekeeping gene uvrD at 30 min, 1 h, and 2 h postinfection (hpi). ***, P < 0.0005.
BplA is required for F. novicida virulence in mice.
Since BplA was required for F. novicida replication in macrophages, a process thought to be required for replication in vivo, we tested if BplA was similarly required for virulence in mice. Mice were infected subcutaneously with the wild-type strain, the ΔbplA mutant strain, or the ΔbirA mutant strain. At 48 h postinfection, the ΔbplA mutant strain was present at significantly lower levels than the wild-type strain in the skin (6-fold), spleen (27-fold), and liver (39-fold) (Fig. 7). In contrast, the ΔbirA mutant strain was not significantly attenuated compared to the wild-type strain. Taken together, these data demonstrate the strong contribution of BplA to F. novicida virulence, as well as its inability to be replaced by BirA. Hence, the two enzymes play distinct roles in F. novicida physiology.
FIG 7 .
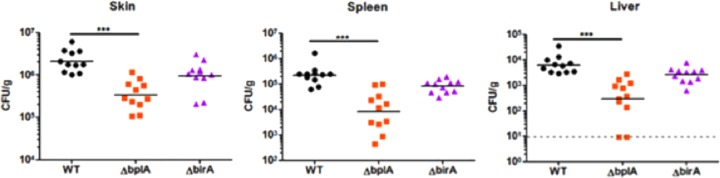
F. novicida BplA is required for replication in mice. Mice were infected subcutaneously with 1 × 105 CFU of wild-type F. novicida U112 (WT) or the ΔbplA or ΔbirA strain. At 48 h postinfection, skin samples obtained at the site of infection as well as the spleen and liver were harvested, and CFU were enumerated after plating. ***, P < 0.0005.
DISCUSSION
We investigated why F. novicida encodes two genes encoding putative biotin protein ligases. This seemed a wasteful extravagance given its small genome (ca. 40% that of E. coli K-12). The low sequence conservation between the two ligases (27% identical residues) argues that neither protein evolved from the other. Indeed, virtually all of the residues conserved between the two proteins are those common to all biotin protein ligases. Both F. novicida proteins align almost as well with Bacillus subtilis BirA as they do with one another (data not shown). The location of the birA gene at the end of the bioBFCD operon (see Fig. S1 in the supplemental material) argues that birA would have been the first biotin ligase to appear in a Francisella ancestor, given that there is no regulatory or enzymatic reason that the protein must be encoded at that location. If so, then BplA would have been a later acquisition, perhaps to facilitate pathogenesis. However, the sequences encoding the two proteins give no evidence for recent acquisition; both genes have the low G+C content characteristic of Francisella genomes.
BplA, the main F. novicida biotinylation enzyme, is required for growth in nutrient-limiting environments (see Fig. S5A in the supplemental material), replication in mouse macrophages (Fig. 6A), where its expression is up-regulated (Fig. 6B), and in vivo virulence (Fig. 7). These data provide further evidence of the link between bacterial metabolism and F. novicida virulence. Given that BplA does the “heavy lifting” in AccB biotinylation, what is the physiological role of BirA? BirA plays no obvious role in pathogenesis and is a remarkably poor ligase, as is evident from its inability to support growth of the E. coli BirA ligase mutant strain at low biotin concentrations (Fig. 3) and its poor enzymatic activity in vitro (Fig. 4). Given the presence of the robust BplA ligase, the retention of BirA in F. novicida and several other Francisella species argues that its physiological role is likely to regulate biotin synthesis and thereby prevent wasteful synthesis of this coenzyme. Synthesis of a biotin molecule by the E. coli pathway consumes 15 ATP equivalents, and thus, unconstrained biotin synthesis could exert a significant metabolic cost in Francisella (which seems very likely to use the same pathway as E. coli). In bacteria that have only a BirA ligase (e.g., E. coli), bio operon transcription responds to both biotin limitation and increased supply of apo (unbiotinylated)-AccB acceptor protein (Fig. 1). As noted above, the two derepression modes act to decrease the levels of the BirA–biotinoyl-AMP complex required to bind the bio operator. However, the presence of BplA argues that in F. novicida, regulation of bio operon transcription by the supply of apo-AccB would not take place, because the very active BplA would modify AccB and thereby short circuit this mode of regulation. Hence, F. novicida BirA likely primarily functions to monitor the intracellular concentration of biotin and would perform this task only at high intracellular biotin concentrations, because only then could it form the key regulatory ligand, biotinoyl-AMP. The poor affinity of BirA for biotin seems to be an advantage, in that it would prevent the regulatory system from starving the BplA ligase for biotin. The finding that one of the highly virulent F. tularensis strains (strain SchuS4) encodes a full-length BplA but an inactive truncated BirA indicates that BplA is sufficient to provide requisite biotin ligase activity to support growth. It also suggests that highly efficient biotin ligase activity in the absence of biotin operon repression during infection by this intracellular pathogen may optimally promote bacterial virulence. This may be a paradigm employed more broadly by diverse intracellular pathogens.
MATERIALS AND METHODS
Strains and growth conditions.
All E. coli strains were derivatives of the wild type K-12 strain (see Table S1 in the supplemental material) and were routinely maintained in LB medium (Luria-Bertani medium containing 10 g of tryptone, 5 g of yeast extract and 10 g of NaCl per liter) or on LB agar plates. The defined M9 minimal medium contained 0.1% vitamin-free Casamino Acids and either 0.4% glucose or 0.2% arabinose. Antibiotics were supplemented as needed (micrograms per milliliter): sodium ampicillin, 100; tetracycline HCl, 10; and kanamycin sulfate, 50. Due to deletion of the araD gene, strain BM4062 is sensitive to arabinose due to accumulation of toxic ribulose-5-phosphate. To allow full induction of the plasmid paraBAD promoter that drives expression of the F. novicida ligases we repaired the ΔaraD mutation by phage P1 transduction of BM4062 with a lysate of strain CAG12095 with selection for tetracycline resistance, followed by screening for growth on arabinose as the sole carbon source.
To generate the F. novicida bplA and birA deletion mutants, PCR was used to amplify flanking DNA regions upstream and downstream of the gene of interest. A kanamycin resistance cassette was inserted between these flanking regions using Gibson assembly (New England Biolabs) and transformed into chemically competent wild-type strain U112 as previously described. The primers used to create the kanamycin-resistant deletion mutants contained Flp recombinase target (FRT) sites flanking the kanamycin resistance cassette, which allowed a clean deletion of each mutant to be made using the plasmid pFFlp, encoding the Flp recombinase, as previously described (9). The bplA and birA strains were complemented in trans by ligation of the genes into the EcoRI and BamHI sites of the broad-host-range vector pBAV1K-T5-GFP. The resulting plasmids were transformed into the clean bplA and birA deletion mutant strains, respectively.
Ligase plasmids and DNA manipulations.
The two putative biotin protein ligases-encoding genes bplA (FTN_0568) and birA (FTN_0811) were amplified by standard PCR genomic DNA of F. novicida U112 using Phusion high-fidelity DNA polymerase (New England Biolabs). Following gel purification of the bplA and birA PCR products, they were each inserted into the medium-copy-number, arabinose-inducible expression vector pBAD322 (18) using XmaI and SphI digestions to give plasmids pBAD322-bplA and pBAD322-birA, respectively (see Table S1 in the supplemental material). Similarly, the genes were also inserted into the T7 promoter expression vector pET28(a) by use of BamHI and XhoI digestion (see Table S2 in the supplemental material), resulting in pET28-bplA and pET28-birA (see Table S1). All constructs were verified by DNA sequencing.
Bio-5′-AMP synthesis reactions.
The assay for ligase-catalyzed in vitro protein biotinylation activity was performed as described previously (4), with some modifications. Protein concentrations were determined using the extinction coefficients calculated from the protein sequence using the ExPASY Tools website. The assays contained 50 mM Tris-HCl (pH 8), 5 mM tris-(2-carboxyethyl)phosphine, 5 mM MgCl2, 20 µM biotin, and 5 µM ATP plus 16.5 nM [α-32P]ATP, 100 mM KCl, and 2 µM ligase. Each of the reaction mixtures was incubated at 37°C for 30 min. For each ligase protein tested, two identical tubes were incubated in parallel, and after the 30-min incubation, AccB-87 (50 µM) was added to one of each pair of tubes, while the other tube was left untreated. The tubes were incubated for an additional 15 min at 37°C. One microliter of each reaction mixture was applied to an Analtech Avicel microcrystalline cellulose thin-layer-chromatography plate, and the plates were developed in isobutyric acid-NH4OH-water (66:1:33 by volume) (19). The chromatograms were dried for 10 h, exposed to a phosphorimaging plate, and visualized using a Fujifilm FLA-3000 PhosphorImager and Fujifilm Image Gauge software (version 3.4 for Mac OS).
Analyses of in vitro BirA biotin attachment activity.
Low-resolution matrix-assisted laser desorption/ionization (MALDI) was used to measure the level of ligase-catalyzed biotinylation of AccB-87 as previously reported (20). Reaction mixtures contained 100 µM AccB-87, 3 µM ligase, 100 µM biotin, 1 mM ATP, 10 mM MgCl2, 100 mM KCl, 5 mM tris-(2-carboxyethyl)phosphine in 50 mM Tris-HCl (pH 8.0) were incubated at 37°C for 16 h, dialyzed against 25 mM ammonium acetate, lyophilized to dryness, and submitted for MALDI analyses.
Growth of F. novicida.
Bacteria were subcultured to an optical density at 600 nm (OD600) of 0.03 in Chamberlain’s medium (CHB). Subcultures were read hourly using a SynergyMx BioTek plate reader (Applied Biosystems) for 24 h. Biotin (50 nM) (Merck KGaA) was added when appropriate.
Quantitative real-time PCR.
RNA was isolated from mid-log-phase broth cultures or macrophages infected with wild-type F. novicida U112 (multiplicity of infection [MOI], 20:1) at various time points by TRI reagent and column purification with a Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA). Quantitative real-time PCR (qRT-PCR) was performed using the Power Sybr green RNA-to-CT one-step kit (Applied Biosystems). Relative transcript levels were calculated by normalizing CT values to DNA helicase II (uvrD and FTN_1594) and plotted as 2−ΔΔCT.
Macrophage infections.
Murine bone marrow-derived macrophages (BMDM) were prepared as described previously (22, 23). Briefly, bone marrow was collected from the femurs of mice. Bone marrow cells were plated in sterile petri dishes and incubated in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS)- and 15% macrophage colony-stimulating factor (M-CSF)-conditioned medium. Bone marrow cells were incubated at 37°C with 5% CO2 and harvested after 6 days. All BMDM were incubated before and during infection in 24 well plates at 37°C with 5% CO2. For infection, BMDM were seeded at 5 × 105 cells per well and incubated overnight at 37°C with 5% CO2. BMDM were infected at a multiplicity of infection (MOI) of 20:1. At 5.5 h postinfection, macrophages were lysed with 1% saponin. Lysates were serially diluted in phosphate-buffered saline (PBS) and plated onto TSA containing 0.1% cysteine to enumerate CFU.
Mouse infections.
For mouse infections, female C57BL/6 mice (6 to 8 weeks old) (Jackson Laboratory, Bar Harbor, ME) were housed under specific-pathogen-free conditions at Emory University. Experimental studies were performed in accordance with the Institutional Animal Care and Use Committee guidelines. Mice were infected subcutaneously with 2 × 105 CFU in 50 µl sterile PBS. After 48 h, the mice were sacrificed, and the spleens, livers, and skin at the site of infection were harvested, homogenized, and plated for CFU on Mueller-Hinton (MH) plates supplemented with 0.1% l-cysteine.
Statistical analysis.
Macrophage replication and qRT-PCR were analyzed for significance using unpaired, two-tailed, Student’s t tests. The mouse infection data were analyzed for significance using the Mann-Whitney test.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Genome organization of the bio operon and putative biotin protein ligase genes of Francisella species. The bplA and birA loci are highlighted in orange and purple, respectively, whereas the purple circle denotes the BirA-binding site predicted by analogy with E. coli. The asterisk indicates that the F. tularensis SchuS4 birA gene is a pseudogene that encodes a protein of 44 residues rather than the intact 320-residue protein. Download
Purification, identification, and characterization of the F. novicida BplA and BirA proteins. (A and B) SDS-PAGE profiles of purified BplA and BirA; (C and D) mass spectral identification of the tryptic peptides of the two proteins; (E and F) chemical cross-linking assays of the solution structures of the F. novicida ligases; (G) test of the possibility that F. novicida BplA and BirA are able to form mixed multimer species. Download
F. novicida contains a single biotinylated protein, the AccB subunit of acetyl-CoA carboxylase. (A) Alignments of the AccB proteins of Francisella species with that of E. coli. (B) F. novicida AccB was modeled on the known structure of the E. coli protein. (C and D) Streptavidin-probed Western blots of SDS-PAGE gel separation of extracts of E. coli and three F. novicida strains, respectively. Download
F. novicida BirA binds its cognate operator and weakly binds the E. coli bioO operator. (A) Multiple alignment of putative Francisella BirA binding sites with that of E. coli. (B) F. novicida BplA binding to the F. novicida bioO operator. (C) F. novicida BirA binding to the E. coli bioO operator. (D) F. novicida BirA binding to its cognate bioO operator. (E and F) E. coli BirA binding to its cognate bioO operator and that of F. novicida, respectively. Download
F. novicida BplA contributes to growth in minimal medium. Wild-type F. novicida U112 and the ΔbirA, ΔbplA, birA complemented (comp), and bplA complemented (comp) strains were grown in Chamberlain’s minimal medium (CHB) in the absence or presence of exogenous biotin (50 nM) and the OD600 was measured every hour. The left panel indicated that F. novicida BplA is required for wild-type growth in CHB, and the right panel shows rescue of the growth defect by addition of biotin. Download
Bacterial strains and plasmids used in this study.
DNA primers used in this study.
ACKNOWLEDGMENTS
This work was supported by the start-up package from Zhejiang University (YF) and Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (grant no. LR15H190001) (YF), National Institutes of Health (NIH) grant AI15650 from National Institute of Allergy and Infectious Diseases (JEC). DSW was supported by NIH/NIAID grant U54-AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award. Y.F. is a recipient of the “Young 1000 Talents” Award.
We thank Peter Yau (Biotechnology Center) and Kelvin Tucker (Mass Spectrometry Laboratory), University of Illinois at Urbana-Champaign for assistance in mass spectrometry.
Footnotes
Citation Feng Y, Chin CY, Chakravartty V, Gao R, Crispell EK, Weiss DS, Cronan JE. 2015. The atypical occurrence of two biotin protein ligases in Francisella novicida is due to distinct roles in virulence and biotin metabolism. mBio 6(3):e00591-15. doi:10.1128/mBio.00591-15.
REFERENCES
- 1.Chapman-Smith A, Cronan JE Jr. 1999. The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci 24:359–363. doi: 10.1016/S0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- 2.Cronan JE. 2014. Biotin and lipoic acid: synthesis, attachment, and regulation. Ecosal Plus. doi: 10.1128/ecosalplus.ESP-0001-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henke SK, Cronan JE. 2014. Successful conversion of the Bacillus subtilis BirA Group II biotin protein ligase into a group I ligase. PLoS One 9:e96757. doi: 10.1371/journal.pone.0096757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravartty V, Cronan JE. 2013. The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase. J Biol Chem 288:36029–36039. doi: 10.1074/jbc.M113.525618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Beckett D. 1996. Evidence for interdomain interaction in the Escherichia coli repressor of biotin biosynthesis from studies of an N-terminal domain deletion mutant. Biochemistry 35:1783–1792. doi: 10.1021/bi952269e. [DOI] [PubMed] [Google Scholar]
- 6.Cronan JE., Jr 1989. The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell 58:427–429. doi: 10.1016/0092-8674(89)90421-2. [DOI] [PubMed] [Google Scholar]
- 7.Solbiati J, Cronan JE. 2010. The switch regulating transcription of the Escherichia coli biotin operon does not require extensive protein-protein interactions. Chem Biol 17:11–17. doi: 10.1016/j.chembiol.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leelaporn A, Yongyod S, Limsrivanichakorn S, Yungyuen T, Kiratisin P. 2008. Francisella novicida bacteremia, Thailand. Emerg Infect Dis 14:1935–1937. doi: 10.3201/eid1412.080435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y, Napier BA, Manandhar M, Henke SK, Weiss DS, Cronan JE. 2014. A Francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol Microbiol 91:300–314. doi: 10.1111/mmi.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker DF, Campbell AM. 1980. Use of bio-lac fusion strains to study regulation of biotin biosynthesis in Escherichia coli. J Bacteriol 143:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard PK, Shaw J, Otsuka AJ. 1985. Nucleotide sequence of the birA gene encoding the biotin operon repressor and biotin holoenzyme synthetase functions of Escherichia coli. Gene 35:321–331. doi: 10.1016/0378-1119(85)90011-3. [DOI] [PubMed] [Google Scholar]
- 12.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li SJ, Cronan JE Jr. 1992. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267:855–863. [PubMed] [Google Scholar]
- 14.Li SJ, Cronan JE Jr. 1993. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol 175:332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cronan JE., Jr 2001. The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the “thumb” structure id essential and that the domain functions as a dimer. J Biol Chem 276:37355–37364. doi: 10.1074/jbc.M106353200. [DOI] [PubMed] [Google Scholar]
- 16.Napier BA, Meyer L, Bina JE, Miller MA, Sjöstedt A, Weiss DS. 2012. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc Natl Acad Sci U S A 109:18084–18089. doi: 10.1073/pnas.1206411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streaker ED, Beckett D. 1998. A map of the biotin repressor-biotin operator interface: binding of a winged helix-turn-helix protein dimer to a forty base-pair site. J Mol Biol 278:787–800. doi: 10.1006/jmbi.1998.1733. [DOI] [PubMed] [Google Scholar]
- 18.Cronan JE. 2006. A family of arabinose-inducible Escherichia coli expression vectors having pBR322 copy control. Plasmid 55:152–157. doi: 10.1016/j.plasmid.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Prakash O, Eisenberg MA. 1979. Biotinyl 5′-adenylate: corepressor role in the regulation of the biotin genes of Escherichia coli K-12. Proc Natl Acad Sci U S A 76:5592–5595. doi: 10.1073/pnas.76.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Zhang H, Cronan JE. 2013. Profligate biotin synthesis in α-proteobacteria—a developing or degenerating regulatory system? Mol Microbiol 88:77–92. doi: 10.1111/mmi.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchida KM, Otsuka AJ. 1987. Isolation and characterization of Escherichia coli birA intragenic suppressors. Mol Gen Genet 210:234–240. doi: 10.1007/BF00325688. [DOI] [PubMed] [Google Scholar]
- 22.Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS. 2011. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS One 6:e24201. doi: 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A 104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Genome organization of the bio operon and putative biotin protein ligase genes of Francisella species. The bplA and birA loci are highlighted in orange and purple, respectively, whereas the purple circle denotes the BirA-binding site predicted by analogy with E. coli. The asterisk indicates that the F. tularensis SchuS4 birA gene is a pseudogene that encodes a protein of 44 residues rather than the intact 320-residue protein. Download
Purification, identification, and characterization of the F. novicida BplA and BirA proteins. (A and B) SDS-PAGE profiles of purified BplA and BirA; (C and D) mass spectral identification of the tryptic peptides of the two proteins; (E and F) chemical cross-linking assays of the solution structures of the F. novicida ligases; (G) test of the possibility that F. novicida BplA and BirA are able to form mixed multimer species. Download
F. novicida contains a single biotinylated protein, the AccB subunit of acetyl-CoA carboxylase. (A) Alignments of the AccB proteins of Francisella species with that of E. coli. (B) F. novicida AccB was modeled on the known structure of the E. coli protein. (C and D) Streptavidin-probed Western blots of SDS-PAGE gel separation of extracts of E. coli and three F. novicida strains, respectively. Download
F. novicida BirA binds its cognate operator and weakly binds the E. coli bioO operator. (A) Multiple alignment of putative Francisella BirA binding sites with that of E. coli. (B) F. novicida BplA binding to the F. novicida bioO operator. (C) F. novicida BirA binding to the E. coli bioO operator. (D) F. novicida BirA binding to its cognate bioO operator. (E and F) E. coli BirA binding to its cognate bioO operator and that of F. novicida, respectively. Download
F. novicida BplA contributes to growth in minimal medium. Wild-type F. novicida U112 and the ΔbirA, ΔbplA, birA complemented (comp), and bplA complemented (comp) strains were grown in Chamberlain’s minimal medium (CHB) in the absence or presence of exogenous biotin (50 nM) and the OD600 was measured every hour. The left panel indicated that F. novicida BplA is required for wild-type growth in CHB, and the right panel shows rescue of the growth defect by addition of biotin. Download
Bacterial strains and plasmids used in this study.
DNA primers used in this study.