Abstract
A PCR assay was developed for the detection of Streptococcus suis serotypes 2 and 1/2. This multiplex PCR is based on the amplification of the gene coding for 16S rRNA of S. suis and on the amplification of the cps2J gene coding for the capsule of S. suis serotypes 2 and 1/2. An internal control was constructed and added in this test to monitor the efficiency of amplification in each reaction. To evaluate the specificity of the test, 31 strains of other bacterial species related to S. suis or isolated from pigs and 42 strains of S. suis serotypes 1 and 3 to 34 were analyzed. The detection threshold of the test was 28 S. suis CFU/ml. The specificity and the sensitivity of the multiplex PCR test and the presence of an internal control allowed the analysis of biological samples without a culture step. The PCR assay was then applied to the detection of 14 S. suis serotype 1/2 strains, 88 S. suis serotype 2 strains isolated from pigs, and 25 S. suis serotype 2 strains isolated from humans. This test was also applied to analyze tonsil samples of pigs experimentally infected and carrier pigs without any symptoms.
Streptococcus suis is an important pathogen of swine, causing meningitis, arthritis, pericarditis, polyserositis, septicemia, and sudden death of weaning piglets as well as growing pigs (15). Moreover, S. suis may be isolated from healthy pigs, and these animals are a source of S. suis transmission in pig herds. This bacterium is also a zoonotic agent responsible for meningitis, septicemia, arthritis, and endocarditis in humans. Most cases have involved individuals who had occupational exposure to pigs, like butchers, slaughterhouse workers, veterinarians, and pig farmers (12, 18, 28, 29). More recently, several cases of human S. suis infection acquired from wild boars have been reported (2, 10, 21, 22).
Thirty-five capsular serotypes of S. suis have been described (types 1/2 and 1 through 34) (27). Serotypes 2, 1/2, 9, 7, and 3 are usually isolated in France from diseased or dead pigs, mainly in cases of meningitis, arthritis, and septicemia (5). Although serotype 2 is considered to be the most-virulent serotype in most countries, strains belonging to other serotypes can also cause disease in pigs (9, 15).
Currently, bacteriological techniques are routinely used to detect S. suis. Recently, PCR tests were developed. Monoplex PCR tests, based on sequences of type-specific capsular genes of S. suis, were developed to detect specifically serotypes 2 and 1/2, 1 and 14, 7 and 9 (25, 26). Then, these methods were changed into multiplex PCR tests (31). A test based on amplification of the epf gene encoding the extracellular factor proteins of virulent serotype 2 was also described previously (30). In 1999, Okwumabua et al. described a PCR assay based on the gene encoding the suilysin of S. suis type 2, but this target was not conserved across capsular types or pathogenic strains (19). In 2000, Boye et al. described a method to detect S. suis by in situ hybridization with a species-specific probe targeting 16S rRNA (6). This method does not permit the detection of serotypes 32 through 34. In 2001, tRNA intergenic length polymorphism analysis (tDNA-PCR) combined with capillary electrophoresis was described by Baele et al. to identify streptococci, including three S. suis strains (1). In 2003, a multiplex PCR test based on S. suis cps genes specific to serotypes 2 (and 1/2), 1 (and 14), 7, and 9 and on the gdh gene encoding the glutamate dehydrogenase of S. suis serotype 2 was developed by Okwumabua et al. (20). This PCR assay allowed the amplification of all serotypes of S. suis with the target based on the gdh gene. However, this method was only applied to detect or characterize S. suis from pure cultures.
In the present study, we report the development of a multiplex PCR test to detect S. suis species and serotypes 2 and 1/2 from tonsillar specimens, sampled from live or dead animals, without a culture step. An internal control was constructed and added in the multiplex PCR to monitor the efficiency of amplification in each reaction. Compared to bacteriological methods, the PCR assay was fast and sensitive. This PCR assay was used to study S. suis infections using two S. suis serotype 2 strains and one S. suis serotype 1/2 strain in experimentally infected specific-pathogen-free (SPF) piglets.
MATERIALS AND METHODS
Bacterial strains.
The specificity of the PCR assay was tested with a collection of 203 strains representing 172 S. suis strains belonging to one of the 35 capsular types described as well as 25 bacterial species other than S. suis (Table 1). For some experiments, the reference strain S735 was also used. In addition, three French porcine field strains of S. suis were used for experimental infections: S. suis capsular serotype 2 strains 332 and 347, isolated from septicemia and from palatine tonsils of clinically healthy pigs, respectively, and S. suis capsular serotype 1/2 (strain 353) isolated from tonsils of a clinically healthy pig.
TABLE 1.
Bacterial strains used in the PCR specificity test
| Species | Strain(s) | No. of strains tested (n = 203) |
|---|---|---|
| Streptococcus suis serotype 1 | Reference strain 5428a | 1 |
| Streptococcus suis serotype 1/2 | Reference strain 2651a and field strainsb | 14 |
| Streptococcus suis serotype 2 | ATCC 43765, ATCC 700794, ATCC 700796c | 3 |
| Field strains isolated from pigsd | 88 | |
| Field strains isolated from humanse | 25 | |
| Streptococcus suis serotypes 3 to 34 | Reference strainsa and field strainsf | 41 |
| Streptococcus agalactiae | ATCC 13813 | 1 |
| Streptococcus acidominimus | NCDO 2025g | 1 |
| Streptococcus alactolyticus | ATCC 43077 | 1 |
| Streptococcus anginosus | ATCC 33397 | 1 |
| Streptococcus bovis | ATCC 33317 | 1 |
| Streptococcus constellatus | ATCC 27823 | 1 |
| Streptococcus difficilis | ATCC 51487 | 1 |
| Streptococcus gordonii | ATCC 10558 | 1 |
| Streptococcus hyointestinalis | CCUG 27888h | 1 |
| Streptococcus intestinalis | ATCC 43492 | 1 |
| Streptococcus pneumoniae | ATCC 33400 | 1 |
| Streptococcus porcinus | ATCC 43138 and field strain | 2 |
| Streptococcus pyogenes | ATCC 12344 | 1 |
| Escherichia coli | Field strains | 2 |
| Campylobacter jejuni | Field strain | 1 |
| Campylobacter coli | Field strain | 1 |
| Mycoplasma hyopneumoniae | ATCC 25934 and field strain | 2 |
| Mycoplasma hyosynoviae | ATCC 25591 and field strain | 2 |
| Mycoplasma hyorhinis | ATCC 17981 and field strain | 2 |
| Mycoplasma flocculare | ATCC 27399 | 1 |
| Actinobacillus pleuropneumoniaei | ATCC 27088 and field strain | 2 |
| Actinobacillus lignieresii | ATCC 49236 | 1 |
| Actinobacillus rossii | ATCC 27072 | 1 |
| Pasteurella multocida | Field strain | 1 |
| Staphylococcus aureus | Field strain | 1 |
Groupe de Recherche sur les Maladies Infectieuses du Porc, Faculté de Médecine Vétérinaire, Université de Montréal, St. Hyacinthe, Québec, Canada (8).
Field strains isolated in France, including strain 353 used in the experimental infection.
ATCC, American Type Culture Collection, Manassas, Va.
Field strains isolated in France, including strains 332 and 347 used in the experimental infection.
Field strains isolated in France, The Netherlands, Canada, and the United Kingdom.
Field strains isolated in France.
NCDO, National Collection of Dairy Organisms, Shinfield, Reading, United Kingdom.
CCUG, Culture Collection University of Göteborg, Göteborg, Sweden.
A. pleuropneumoniae serotype 1.
Streptococcus sp. and Actinobacillus lignieresii were cultivated on Columbia agar base supplemented with 5% sheep blood (AES Laboratories, Combourg, France). Mycoplasma hyosynoviae, Mycoplasma hyopneumoniae, and Mycoplasma hyorhinis were cultivated on Friis medium (13). Campylobacter coli and Campylobacter jejuni were cultivated as previously described (17). The other strains were cultivated on pleuropneumoniae-like organism agar (PPLO agar; Difco, Cergy Pontoise, France) supplemented with nicotinamide dinucleotide (10 μg/ml), glucose (1 mg/ml), and 5% decomplemented horse serum. All strains were incubated at 37°C in 5% CO2.
DNA preparation.
Samples were prepared for PCR as described by Kellog and Kwok (16). Briefly, 1 ml of each initial suspension (IS) were centrifuged (12,000 × g, 4°C, 20 min) and the pellets were resuspended in a mixture of 250 μl of 10 mM Tris HCl (pH 8.3), 100 mM KCl, 2.5 mM MgCl2, and 250 μl of 10 mM Tris HCl (pH 8.3), 2.5 mM MgCl2, 1% (vol/vol) Tween 20 (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France), 1% (vol/vol) Triton X-100 (Sigma-Aldrich Chimie), 0.01% (vol/vol) Nonidet P-40 (Sigma-Aldrich Chimie), and proteinase K (120 μg/ml; Sigma-Aldrich Chimie). Samples were incubated for 1 h at 60°C prior to proteinase K heat inactivation at 95°C for 10 min, allowed to cool at room temperature, and kept at −20°C.
When inhibition of the PCR was observed, DNA was reextracted as follows. Four hundred microliters of lysate was placed with 400 μl of phenol-chloroform-isoamyl(ic) alcohol (25:24:1), vortexed, and centrifuged at 10,000 × g for 30 s. Then, the supernatant was mixed with 400 μl of chloroform-isoamyl(ic) alcohol (24:1), vortexed, and centrifuged, and the supernatant was mixed with 50 μl of 3 M sodium acetate buffer (pH 5.5) and 400 μl of isopropanol for 30 min at 4°C to precipitate the DNA. After centrifugation at 10,000 × g for 15 min, the DNA pellet was washed with 70% ethanol, dried, and resuspended in 50 μl of double-distilled water.
Construction of the PCR IPC.
To check for the presence of inhibitors within the PCR mixture, an internal positive control (IPC) was constructed. IPC was synthesized in one PCR. The primers used in this reaction (CI 6-s and CI 7-as) possessed 5′ overhanging ends which were identical to the primers used in the PCR specific for S. suis serotype 2 (cps2J-s and cps2J-as), whereas their 3′ ends were complementary to a predetermined DNA sequence (16S ribosomal DNA [rDNA]) of S. suis of defined length and sequence (Table 2) (AF009477). The IPC sequence was different from the 16S rDNA sequence amplified by the 16S-195(s) and 16S-489(as) primers used in the multiplex PCR test.
TABLE 2.
Primers used to construct the PCR internal control and for multiplex PCR
| Primer | Sequence (5′-3′) | PCR product size (bp) |
|---|---|---|
| 16S-195(s) | CAGTATTTACCGCATGGTAGATAT | 294 |
| 16S-489(as2) | GTAAGATACCGTCAAGTGAGAA | |
| cps2J-s | GTTGAGTCCTTATACACCTGTT | 459 |
| cps2J-as | CAGAAAATTCATATTGTCCACC | |
| CI 6-s | GTTGAGTCCTTATACACCTGTTACTCAGTGCCGCAGCTAACGCATT | 620 |
| CI 7-as | CAGAAAATTCATATTGTCCACCCGACTTCACCCCAATCATCTATCC |
The IPC was a 16S rDNA fragment of 620 bp from S. suis generated by PCR. The PCR mixture contained PCR buffer II (20 mM Tris-HCl [pH 8.4], 50 mM KCl, 1% glycerol, Thermostable AccuPrime protein, 1.5 mM MgCl2, 200 μM [each] deoxynucleoside triphosphate), 400 nM (each) CI 6-s and CI 7-as primers, 1 U of AccuPrime Taq DNA polymerase (Invitrogen, Cergy Pontoise, France), and 5 μl of a cell lysate of pure culture of S. suis S735 reference strain. Amplification was performed in a Perkin-Elmer Cetus (Courtaboeuf, France) GeneAmp PCR system 9600. The reaction procedure consisted of 40 cycles of denaturation at 94°C for 30 s, primer annealing at 65°C for 25 s, and extension at 72°C for 10 s. The PCR product was purified using a commercially available kit (Life Technologies, Cergy Pontoise, France). This IPC was stored in double-distilled water at −20°C. DNA concentration was determined spectrophotometrically.
Multiplex PCR conditions.
The multiplex PCR developed in this study permitted the simultaneous detection of the S. suis species and serotypes 2 and 1/2. The 294-bp PCR product, specific to S. suis, was obtained with the forward primer 16S-195(s) and the reverse primer 16S-489(as2) (Table 2) defined on the 16S rDNA sequence (AF009477) (8). The second primer set, detecting serotypes 2 and 1/2, was composed of a forward primer, cps2J-s, and a reverse primer, cps2J-as (Table 2), and enabled the amplification of 459-bp products. These primers were defined on the capsular gene cps (AF118389), and they also amplified the IPC (620 bp).
The multiplex PCR mixture contained PCR buffer [67 mM Tris-HCl, 16 mM (NH4)2SO4, 0.01% Tween 20, 2.5 mM MgCl2 (pH 8.8)], a 600 μM concentration of each deoxynucleoside triphosphate (Pharmacia Biotech, Orsay, France), 1.1 μM cps2J-s and cps2J-as primers, 600 nM 16S-195(s) and 16S-489(as2) primers, 2.5 U of Taq DNA polymerase (Eurobio, Les Ulis, France), 3 fg of IPC, and 5 μl of the DNA template. The DNA template was replaced by double-distilled water for the negative control. Amplification was performed in a Perkin-Elmer Cetus GeneAmp PCR system 9600. The reaction procedure consisted of 40 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 30 s, and extension at 72°C for 60 s. The amplified products were separated in a 2% agarose gel in TBE buffer (90 mM Tris, 90 mM borate, 2.5 mM EDTA [pH 8]) for 1 h at a constant voltage of 125 V. Amplified products were stained with ethidium bromide and detected by UV transillumination. The Smart Ladder was used as a molecular size standard (Eurogentec, Angers, France).
Sensitivity of the multiplex PCR.
The sensitivity of the PCR test was evaluated using 10-fold dilutions of a culture of S. suis reference strain serotype 2 (S735) at a titer of 2.8 × 105 CFU/ml. Then, each dilution (1 ml) was placed on tonsil biopsy specimens (6 mm3) obtained from S. suis-free animals and reduced into small pieces with a scalpel, and DNA was prepared as mentioned above (16).
Experimental infection.
All bacterial strains were prepared under the same conditions: one colony, isolated from an overnight culture on Columbia blood agar base, supplemented with 5% sheep blood, was resuspended in 5 ml of Todd-Hewitt broth (THB) (Difco) and incubated 18 h at 37°C, in 5% CO2. The bacterial cultures were then diluted in THB with 10% inactivated bovine serum, further incubated 6 h, and adjusted to 108 CFU/ml. For each inoculum, the bacterial concentration was confirmed by plating on Columbia blood agar base supplemented with 5% sheep blood.
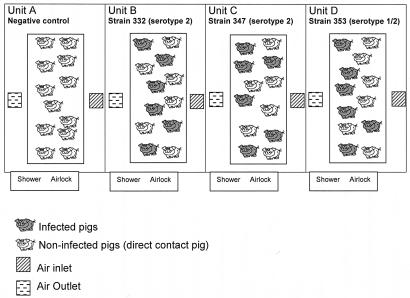
Forty-eight SPF Large White pigs, obtained from the experimental swine herd of the Agence Française de Sécurité Sanitaire des Aliments (Ploufragan, France) (7), 10 weeks of age, were divided into four experimental units (units A to D) (Fig. 1). . Very strict biosecurity measures were implemented in order to avoid contamination of the pigs: existence of an air filtration system and airlocks for each unit, unit-specific clothes, and compulsory showering after visiting the pigs. In unit A, six negative-control pigs received 2 ml of THB supplemented with 10% inactivated bovine serum by the intravenous route (group 1), and six animals (direct contact pigs) did not receive THB (group 2). In unit B, six pigs were infected intravenously with 1.81 × 108 CFU of S. suis strain 332 (group 3) and six contact pigs were not infected (group 4). In unit C, six pigs were inoculated intravenously with 3.23 × 108 CFU of S. suis strain 347 (group 5) and six contact pigs were not infected (group 6). In unit D, six pigs were inoculated intravenously with 1.29 × 108 CFU of S. suis strain 353 (group 7) and six contact pigs were not infected (group 8) (Fig. 1).
FIG. 1.
Experimental design. Negative control animals were in unit A and were divided into two groups: group 1 (six noninfected pigs that received sterile THB) and group 2 (six noninfected pigs that were in direct contact). In unit B, six pigs were infected with S. suis strain 332 (group 3) and six pigs were in direct contact (group 4). In unit C, six pigs were infected with S. suis strain 347 (group 5) and six pigs were in direct contact (group 6). In unit D, six pigs were infected with S. suis strain 353 (group 7) and six pigs were in direct contact (group 8).
Daily clinical examinations consisted of taking rectal temperature and looking for symptoms such as lameness, tremors, opisthotonos, nystagmus, or convulsions. Body weight was also recorded each week during the trial. Blood samples were collected 6 days before infection; on days 8, 16, and 21 postinfection (p.i.); and on sacrifice day (16 to 30 days p.i.) for serological and bacteriological analysis. Swabs from palatine tonsils were performed on day 16 p.i., placed in 2 ml of sterile water supplemented with NaCl (8.5 g/liter) (SW), and analyzed by classical bacteriological analysis and by PCR.
In each unit, 6 days before infection and on days 8, 16, and 21 p.i., 7 g of dehydrated granulated food, 15 g of feces, and 25 ml of drinking water were collected in four distant places. In each unit, four drag swabs (Sodibox, La Forêt Fouesnant, France), previously humidified with 5 ml of SW, were rubbed on the pen, on the air inlet system, and on the window. Food, feces, and dust samples were placed in 20 ml of SW, vortexed, and centrifuged at 4,000 × g for 15 min. The pellet was resuspended in 2 ml of SW (IS). All environmental samples were analyzed by bacteriological analysis and PCR.
Pigs were randomly sampled in each group, and necropsies were planned at intervals during 2 weeks (on days 16 to 30 p.i.). After euthanasia, the thoracic organs, peritoneum, brain, and joints were examined and swabs were collected from liver, heart, lung, joints, and muscle sampled beside two femur-ilium joints (swabs and biopsy). In addition, and to compare different tonsil samples, external swabs, biopsy specimens, and the whole tonsils were compared. Samples were taken even in the cases where no gross lesions were observed and placed in 2 ml of SW (IS). Biopsy specimens, of approximately 6 mm3, were reduced into small pieces with a scalpel and added to 2 ml of SW. All these ISs were analyzed by bacteriological analysis and by PCR.
Bacteriological analysis.
Ten microliters of each sample (or IS) was placed onto selective based Columbia medium supplemented with 5% sheep blood, 15 mg of nalidixic acid/liter, and 10 mg of colistin/liter. Then, the plates were incubated overnight at 37°C in 5% CO2. S. suis-like colonies were subcultivated on Columbia medium supplemented with 5% sheep blood, identified by PCR, and serotyped by slide agglutination using a type-specific hyperimmune serum (14).
Serological analysis.
All sera were tested with an indirect enzyme-linked immunosorbent assay (ELISA) using a protein extract from sonicated bacteria as antigen. This ELISA test was not attempted to be used as a diagnostic tool but rather was used to monitor the kinetics of the antibodies' response against proteins of the homologous strain. Three ELISA tests were developed, one for each strain (332, 347, and 353) as previously described by Cloutier et al. (9). Each serum was diluted at 1/160 before analysis. The positive control consisted of a serum from a pig experimentally infected with S. suis type 2, whereas the negative control corresponded to a serum from an uninfected SPF pig (tested in quadruplicate). o-Phenylenediamine at 0.4 mg/ml(Sigma-Aldrich Chimie) dissolved in 0.05 M citrate buffer (pH 5.5) with 0.5 M H2O2 was used as a substrate. After 15 min at 37°C in the dark, 25 μl of HCl was added in each well and optical density (OD) was measured at 492 nm on a kinetic microplate reader (Labsystems, Cergy-Pontoise, France). Results were reported as S/P ratios, which were defined as the OD obtained for each serum minus the mean OD of the negative control divided by the mean OD of the positive control.
Statistical analysis of data.
The data obtained in experimental study from pig groups were compared simultaneously by Kruskall-Wallis test and in two-by-two tables by Kolmogorov-Smirnov test. Associations between strain used for the assay, tonsillar carriership (as determined by external swab, biopsy sample, and whole tonsil), and method applied (PCR and bacteriological analysis) were assessed by Fisher exact test (n ≤ 5) or chi-square test (n > 5) on independence in two-by-two tables. These tests were carried out with Systat 9.0 program for Windows. Differences were estimated to be significant when probabilities (P) were lower than 0.05.
RESULTS
Specificity and sensitivity of the multiplex PCR.
In order to assess the specificity of the multiplex PCR, the different microorganisms listed in Table 1 were used as DNA templates. All S. suis strains, including reference strains as well as field strains, showed a fragment of 294 bp corresponding to a part of the 16S rDNA gene. None of the other bacterial species described in Table 1 showed any amplification product. A fragment of 489 bp corresponding to a part of the cps gene coding for the capsule was only obtained with S. suis serotypes 2 and 1/2 strains as expected.
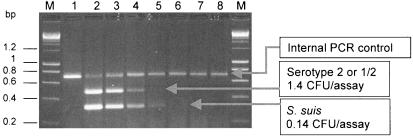
Sensitivity results are shown in Fig. 2. . The assay was carried out with 10-fold dilutions of S. suis DNA extracts obtained from a culture of reference strain S735 at a titer of 280 CFU/μl and mixed with a tonsil palatine biopsy specimen from an SPF pig (1 ml of dilution by biopsy). Under the conditions described in the trials (5 μl of DNA extract per assay), the detection limit of the PCR test was 1.4 CFU/assay, corresponding to 280 CFU/ml of tonsil sample for the product specific to serotypes 2 or 1/2 and 0.14 CFU/assay (28 CFU/ml of tonsil sample) for the product shared between all S. suis serotypes. Moreover, the IPC of 620 bp was also noticeable in Fig. 2.
FIG. 2.
Evaluation of detection limit of the multiplex PCR. Agarose electrophoresis of the PCR products obtained from serial dilutions of S. suis S735 strain culture at a titer of 2.8 × 105 CFU/ml (lane 2) and placed in tonsil biopsy specimens (1 ml of each dilution per biopsy specimen). The positions of the specific fragments of internal control, S. suis, and serotype 2 or 1/2 are indicated. Lanes: M, molecular mass marker (Smart Ladder; Eurogentec); 1, negative control of the PCR (only IPC was amplified); 3 to 8, PCR products at dilutions of 10, 102, 103, 104, 105, and 106.
Clinical signs and macroscopic lesions.
Data on clinical signs and macroscopic lesions are summarized in Table 3.
TABLE 3.
Clinical, pathological, bacteriological, PCR, and serological results observed for infected and contact pigs
| Unit | Group | Hyperthermia (days p.i.) | Lameness (days p.i.) | Macroscopic lesionsa | No. of pigsb in which the following was observed in indicated samples collected at necropsy:
|
ELISA results at necropsy (Sp mean) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. suis
|
S. suis DNA
|
||||||||||||||||
| Joints | Muscle | Liver | Heart | Lung | Tonsils | Joints | Muscle | Liver | Heart | Lung | Tonsils | ||||||
| A (THB) | 1 | No | No | No | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.015 ± 0.013 |
| 2 | No | No | No | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.027 ± 0.011 | |
| B (strain 332, type 2) | 3 | Yes (1-2) | Yes (1-22) | Arthritis (4), pneumonia (1) | 0 | 1 | 0 | 0 | 0 | 6 | 0 | 0 | 1 | 0 | 0 | 6 | 0.333 ± 0.099 |
| 4 | No | No | Arthritis (4) | 2 | 2 | 1 | 1 | 1 | 6 | 1 | 2 | 1 | 1 | 1 | 6 | 0.179 ± 0.035 | |
| C (strain 347, type 2) | 5 | Yes (1-2) | Yes (1-10) | Arthritis (4) | 1 | 2 | 0 | 3 | 2 | 5 | 0 | 2 | 0 | 2 | 2 | 6 | 0.644 ± 0.045 |
| 6 | No | No | No | 1 | 0 | 0 | 1 | 1 | 6 | 1 | 0 | 0 | 0 | 0 | 6 | 0.399 ± 0.086 | |
| D (strain 353, type 1/2) | 7 | No | No | Arthritis (3) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0.053 ± 0.012 |
| 8 | No | No | Arthritis (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0.047 ± 0.013 | |
Data in parentheses are numbers of pigs (out of a group of six) in which lesions were observed.
Data are numbers of pigs out of a group of six.
(i) Groups 1 and 2.
Negative control animals did not exhibit any clinical signs of S. suis infection. Postmortem examinations did not reveal any lesions in these pigs. The average daily weight gains (ADGs) were 981 g (group 1) and 993 g (group 2).
(ii) Groups 3 and 4 (S. suis type 2).
Rectal temperatures of three piglets infected with strain 332 (group 3) were moderate (40.2 ± 0.3°C) during 24 to 48 h p.i. Under the conditions of this trial (level three biosecurity), the normal temperature of SPF piglets is 39.5°C. All the piglets presented lameness, 24 h p.i. and during 1 to 22 days. Four of them developed arthritis associated with pneumonia in one animal. Contact pigs (group 4) did not develop clinical signs, but four animals showed arthritis at necropsy. The body growth of animals in group 3 was retarded. Differences in ADG between group 3 (395 g) and group 4 (921 g) were significant (P < 0.05), although no differences were noted between negative control pigs and contact pigs (P > 0.05).
(iii) Groups 5 and 6 (S. suis type 2).
Two piglets infected with strain 347 (group 5) developed hyperthermia (40 ± 0.3°C) during 24 to 48 h, and lameness was observed in four pigs (during 1 to 10 days). Postmortem examinations revealed arthritis in these animals, whereas contact pigs (group 6) had neither symptoms nor lesions. The body growth was affected by the experimental infection (group 5). The ADG was 550 g, significantly different (P < 0.05) from those observed for piglets in negative control groups and group 6 (ADG = 970 g).
(iv) Groups 7 and 8 (S. suis type 1/2).
No clinical signs were noticeable in groups 7 and 8, but moderate arthritis was observed at necropsy in three infected and one contact pigs. The ADG were, respectively, 927 and 875 g (groups 7 and 8), and no difference was obtained between these two groups and negative control groups (P > 0.05).
Detection of S. suis.
All samples collected from animals in negative control groups and analyzed 16 days p.i. (live pigs) and after euthanasia (16 to 30 days p.i.) were negative by PCR and bacteriological analysis. Data are summarized in Tables 3 and 4.
TABLE 4.
Detection of S. suis serotype 2 or 1/2 by bacteriological analysis and PCRa
| Group | No. of samples in which S. suis was detected at:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 16 days p.i.
|
Necropsy day
|
|||||||
| Bacteriology (external swab) | PCR (external swab) | Bacteriology
|
PCR
|
|||||
| External swab | Biopsy | Whole | External swab | Biopsy | Whole | |||
| 1 (THB) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 (strain 332, type 2) | 4 | 6 | 4 | 6 | 4 | 5 | 6 | 6 |
| 4 | 5 | 6 | 4 | 6 | 6 | 6 | 6 | 6 |
| 5 (strain 347, type 2) | 4 | 5 | 2 | 5 | 5 | 6 | 6 | 6 |
| 6 | 5 | 6 | 3 | 4 | 6 | 6 | 6 | 6 |
| 7 (strain 353, type 1/2) | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 4 |
| 8 | 0 | 0 | 0 | 0 | 0 | 3 | 5 | 5 |
Tonsil samples (from external swab, biopsy, and whole tonsil) from live pigs (16 days p.i.) and pigs after euthanasia (16 to 30 days p.i.) were studied.
(i) Environmental samples.
All environmental samples were negative by PCR and bacteriological analysis.
(ii) Results obtained with live pigs (16 days p.i.) (Table 4).
The comparison of PCR as well as bacteriological results, obtained from tonsils in infected or in contact pigs did not show any differences between groups 3, 4, 5, and 6 (P > 0.05). Results from groups 7 and 8 were negative.
(iii) Results obtained in euthanized piglets (16 to 30 days p.i.).
The results, obtained by PCR and bacteriological analysis from 24 dead animals (Tables 3 and 4), confirmed the presence of S. suis type 2 (strains 332 and 347) infection, respectively, in 14 and 19 samples from joints, muscle, liver, heart, and lungs, but results by the two techniques were not significantly different (P > 0.05). S. suis type 1/2 was isolated in only one pig (group 7) from the joints. Sixteen to thirty days p.i., S. suis type 2 was detected from the tonsils of 24 and 23 pigs by PCR and culture, respectively, whereas nine pigs were identified as positive by PCR and negative by culture in groups 7 and 8 (S. suis type 1/2). In these groups, the difference between the two tests was significant (P < 0.05). The results were not significantly different between the three samples of tonsils (external swab, biopsy sample, and whole tonsil) (Table 4). No significant difference was observed between infected and contact pigs and between groups 3, 4, 5, and 6 (P > 0.05).
Serological results.
The ELISA results obtained from negative controls but also from groups 7 and 8 were negative. Pigs infected with S. suis type 2 (strains 332 and 347) developed humoral antibodies detectable by ELISA from 16 days p.i. The level of antibodies increased until the end of the experiment. Contact pigs of both groups were also seropositive with a moderate level of antibodies.
DISCUSSION
The multiplex PCR assay described in this study was based on the amplification of a gene fragment coding for 16S rRNA of S. suis and on the amplification a cps2J gene fragment encoding S. suis serotype 2 and 1/2 capsular biosynthesis (24). Furthermore, our test contained an internal PCR control to eliminate false-negative samples due to inhibitors of polymerization. All S. suis strains tested in this study were detected by our multiplex PCR test, while none of the other bacterial species showed a positive reaction.
The 35 capsular types of S. suis were recognized by this PCR test, which was also able to detect S. suis in specimens from live pigs experimentally infected. S. suis isolates belonging to any serotype isolated from tonsil could be potentially virulent. Recently, in our laboratory, S. suis serotype 5 was isolated from a subject with septicemia without any other bacterial isolation. This serotype was also isolated from nursery pigs with serious cases of meningitis on a Canadian farm (9). Some S. suis strains isolated from healthy carrier pigs (particularly in tonsils) are able to induce transmission of S. suis infection between pigs or between herds. These pigs should be carefully checked, and the detection of S. suis species in tonsils is the first step of the diagnosis. Thus, the PCR assay described in this study, based on the 16S rDNA region conserved across capsular types, allowed the detection of S. suis species and would be very useful for epidemiological studies. Other PCR tests were previously developed to detect S. suis species (6, 19, 31). Okwumabua et al. and Wisselink et al. have also reported two PCR tests to detect two of the S. suis virulence factors, the suilysin and the extracellular factor, respectively (19, 30). However, the absence of these proteins in some virulent strains may preclude the routine use of these tests. Moreover, in a previous study we showed that these proteins were present neither in all S. suis strains nor in all virulent European strains (5, 19). In 2003, Okwumabua et al. developed a multiplex PCR based on the gdh gene, encoding the glutamate deshydrogenase of S. suis serotype 2, allowing the amplification of all S. suis serotypes (20). This method was very attractive, but it was only applied to detect or characterize S. suis from pure cultures.
Our multiplex PCR assay allowed also the detection of all S. suis serotypes and more specifically serotypes 2 and 1/2. A preliminary study was performed to develop a PCR test able to detect serotype 2 alone, because this serotype is the major cause of disease in France and because it is a zoonotic agent that causes septicemia, endocarditis, and meningitis in humans (2, 5, 10, 21, 22, 29). The first step was to show genomic specificities in serotype 2 strains. We sequenced the four potential target genes (cps2F, cps2H, cps2I, and cps2J) that code for S. suis capsule, in serotypes 2 and 1/2 reference strains. In the original work describing the sequencing of these genes, the serotype 1/2 reference strain was not analyzed (23). The alignment of these nucleotide sequences showed a perfect identity for the cps2H, cps2I, and cps2J genes of the two reference strains. However, a substitution (T to C) was detected in the cps2F gene of serotype 2. Since this mutation leads to the disappearance of an SspI restriction site in the serotype 1/2, a PCR-restriction fragment length polymorphism was developed. However, this substitution was not detected in all serotype 2 strains isolated from subjects in the field (data not shown). In this context, we decided to develop a PCR test able to detect simultaneously serotypes 2 and 1/2. These serotypes are the most frequently detected in France (77.3%) (5). Several PCR tests, targeted on the cps locus, were previously described to detect these two serotypes (20, 26, 31). We designed original primers from the cps locus in order to (i) have a hybridization temperature compatible with the primers based on 16S rDNA, (ii) avoid the appearance of dimers, and (iii) have a PCR product differing in size from the product of 16S-195s and 16S-489 as primer pair.
The sensitivity of our multiplex PCR test was evaluated in vitro with SPF pig tonsils experimentally infected with S. suis type 2 reference strain and was found to be 28 CFU of S. suis/ml of tonsil sample. In the in vitro conditions described in our study, the PCR test was evaluated to be 20 times more sensitive than culture-positive results (28 versus 500 CFU of S. suis/ml). Our assay is more sensitive than other PCR tests previously described: the detection threshold of the multiplex test developed by Wisselink et al. (31) was 10 fg of chromosomal DNA in 25 μl of clinical sample (approximately 200 CFU/ml according to our reckoning). This estimate probably lacks accuracy because the test was performed not with tonsils experimentally infected in vitro but with purified DNA. The sensitivity of the PCR tests described by Smith et al. and Okwumabua et al. were not evaluated (20, 25, 26).
The comparison of PCR and bacteriological isolation obtained from tonsils of live pigs (healthy carriers) as well as in other organs in dead pigs, with or without macroscopic lesions, did not show any significant differences, with the exception of groups 7 and 8 infected with serotype 1/2 (P < 0.05). It is possible that the number of organisms present in samples from groups 3 to 6 is above the detection limit for both assays. Our multiplex PCR assay, with an internal control and without a culture step, is easy to perform. Ninety-six samples could be analyzed simultaneously in 6 h, whereas bacteriological isolations require at least 4 days (including primary isolation, cloning, biochemical identification, and serotyping). The major difficulty of bacteriological isolation is to locate S. suis colonies from multi-infected samples such as tonsils. On the other hand, tonsil specimens appeared to be helpful to detect S. suis in live pigs. Our PCR assay used samples directly and was able to detect and identify S. suis serotype 2 and 1/2 among suspicious α-hemolytic colonies on blood agar medium. In practice, the main advantages of this test are its abilities (i) to detect S. suis from multi-infected samples, (ii) to reduce the time required to identify the bacteria, and (iii) to increase the number of colonies analyzed at the same time.
The comparison of tonsil biopsy specimens, external swabs, and whole tonsils did not show any significant differences. These results differ from those published by Fittipaldi et al. in 2003 (11), with Actinobacillus pleuropneumoniae infection. These authors showed that the PCR detection rate was higher with whole tonsils than with tonsil biopsy specimens. However, in that study, conventional naturally infected pigs were tested. External tonsil swabs will be chosen in the future because external tonsil swabs are less traumatizing for pigs.
In this experimental study, clinical signs and macroscopic lesions induced in SPF piglets were moderate except for arthritis (especially for pigs infected with S. suis serotype 1/2). These results are different from those of our previous trials carried out under these standardized conditions with different strains of S. suis type 2 (3, 4). There was a transmission from infected to contact pigs, since we detected S. suis in contact pigs during the trial (on day 16) and at necropsy. At the end of the assay, all contact pigs in units B and C were healthy carriers pigs with no clinical symptoms of the disease (P > 0.05).
In conclusion, the assay in the present work may be used routinely to identify pigs carrying S. suis serotypes 2 and 1/2 and all other serotypes. It may also be applicable for epidemiological studies and transmission studies of S. suis and can contribute to the control of S. suis infection.
Acknowledgments
We thank Roland Cariolet, Bernard Beaurepaire, Gérard Bénévent, Pierre Ecobichon, and Jean-Claude Rault (Service de production de porcs assainis et d'expérimentation, AFSSA-Ploufragan) for skilled technical assistance and Anne Gautier-Bouchardon for critical reading of the manuscript.
REFERENCES
- 1.Baele, M., V. Storms, F. Haesebrouck, L. A. Devriese, M. Gillis, G. Verschraegen, T. de Baere, and M. Vaneechoutte. 2001. Application and evaluation of the interlaboratory reproducibility of tRNA intergenic length polymorphism analysis (tDNA-PCR) for identification of Streptococcus species. J. Clin. Microbiol. 39:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bensaid, T., B. Bonnefoi-Kyriacou, C. Dupel-Pottier, O. Bellon, E. Lagier, and H. Chardon. 2003. Streptococcus suis meningitis following wild boar hunting. Presse Med. 32:1077-1078. [PubMed] [Google Scholar]
- 3.Berthelot-Hérault, F., R. Cariolet, A. Labbe, M. Gottschalk, J. Y. Cardinal, and M. Kobisch. 2001. Experimental infection of specific pathogen free piglets with French strains of Streptococcus suis capsular type 2. Can. J. Vet. Res. 65:196-200. [PMC free article] [PubMed] [Google Scholar]
- 4.Berthelot-Hérault, F., M. Gottschalk, A. Labbe, R. Cariolet, and M. Kobisch. 2001. Experimental airborne transmission of Streptococcus suis capsular type 2 in pigs. Vet. Microbiol. 82:69-80. [DOI] [PubMed] [Google Scholar]
- 5.Berthelot-Hérault, F., H. Morvan, A. M. Kéribin, M. Gottschalk, and M. Kobisch. 2000. Production of muramidase released protein (MRP), extracellular factor (EF) and haemolysin by field isolates of Streptococcus suis capsular type 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet. Res. 31:473-479. [DOI] [PubMed] [Google Scholar]
- 6.Boye, M., A. A. Feenstra, C. Tegtmeier, L. O. Andresen, S. R. Rasmussen, and V. Bille-Hansen. 2000. Detection of Streptococcus suis by in situ hybridization, indirect immunofluorescence, and peroxidase-antiperoxidase assays in formalin-fixed, paraffin-embedded tissue sections from pigs. J. Vet. Diagn. Investig. 12:224-232. [DOI] [PubMed] [Google Scholar]
- 7.Cariolet, R., P. Marie, G. Moreau, and H. Robert. 1994. Rappel des différentes méthodes d'obtention de porcelets assainis: conditions de maintien du statut sanitaire et valorisation de ces animaux. Journées Rech. Porcine France 26:1-12. [Google Scholar]
- 8.Chatellier, S., J. Harel, Y. Zhang, M. Gottschalk, R. Higgins, L. A. Devriese, and R. Brousseau. 1998. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int. J. Syst. Bacteriol. 48:581-589. [DOI] [PubMed] [Google Scholar]
- 9.Cloutier, G., S. D'Allaire, G. Martinez, C. Surprenant, S. Lacouture, and M. Gottschalk. 2003. Epidemiology of Streptococcus suis serotype 5 infection in a pig herd with and without clinical disease. Vet. Microbiol. 97:135-151. [DOI] [PubMed] [Google Scholar]
- 10.Durand, F., C. L. Périno, C. Recule, J. P. Brion, M. Kobisch, F. Guerber, and J. Croizé. 2001. Bacteriological diagnosis of Streptococcus suis meningitis. Eur. J. Clin. Microbiol. 20:519-521. [DOI] [PubMed] [Google Scholar]
- 11.Fittipaldi, N., A. Broes, J. Harel, M. Kobisch, and M. Gottschalk. 2003. Evaluation and field validation of PCR tests for detection of Actinobacillus pleuropneumoniae in subclinically infected pigs. J. Clin. Microbiol. 41:5085-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.François, B., V. Gissot, M. C. Ploy, and P. Vignon. 1998. Recurrent septic shock due to Streptococcus suis. J. Clin. Microbiol. 36:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friis, N. F. 1975. Some recommendations concerning primary isolation of Mycoplasma suispneumoniae and Mycoplasma flocculare: a survey. Nord. Vet. Med. 27:337-339. [PubMed] [Google Scholar]
- 14.Gottschalk, M., R. Higgins, M. Jacques, K. R. Mittal, and J. Henrichsen. 1989. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 27:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins, R., and M. Gottschalk. 1999. Streptococcal diseases, p. 563-578. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa University Press, Ames.
- 16.Kellog, D. E., and S. Kwok. 1990. Detection of human immunodeficiency virus, p. 339-343. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 17.Le Minor, L., and M. Véron. 1990. Bactériologie médicale. Médecine-Sciences, Flammarion, Paris, France.
- 18.Matsuo, H., and S. Sakamoto. 2003. Purulent meningitis caused by Streptococcus suis in a pig breeder. Kansenshogaku Zasshi 77:340-342. [DOI] [PubMed] [Google Scholar]
- 19.Okwumabua, O., O. Abdelmagid, and M. M. Chengappa. 1999. Hybridization analysis of the gene encoding a hemolysin (suilysin) of Streptococcus suis type 2: evidence for the absence of the gene in some isolates. FEMS Microbiol. Lett. 181:113-121. [DOI] [PubMed] [Google Scholar]
- 20.Okwumabua, O., M. O'Connor, and E. Shull. 2003. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 218:79-84. [DOI] [PubMed] [Google Scholar]
- 21.Pedroli, S., M. Kobisch, O. Beauchet, J. P. Chaussinand, and F. Lucht. 2003. Streptococcus suis bacteriemia. Presse Med. 32:599-601. [PubMed] [Google Scholar]
- 22.Rosenkranz, M, H. A. Elsner, H. J. Sturenburg, C. Weiller, J. Rother, and I. Sobottka. 2003. Streptococcus suis meningitis and septicemia contracted from a wild boar in Germany. J. Neurol. 250:869-870. [DOI] [PubMed] [Google Scholar]
- 23.Smith, H. E., M. Damman, J. van der Velde, F. Wagenaar, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, H. E., R. de Vries, R. van't Slot, and M. A. Smits. 2000. The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb. Pathog. 29:127-134. [DOI] [PubMed] [Google Scholar]
- 25.Smith, H. E., L. van Bruijnsvoort, H. Buijs, H. J. Wisselink, and M. A. Smits. 1999. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol. Lett. 178:265-270. [DOI] [PubMed] [Google Scholar]
- 26.Smith, H. E., V. Veenbergen, J. van der Velde, M. Damman, H. J. Wisselink, and M. A. Smits. 1999. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 37:3146-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-387. [DOI] [PubMed] [Google Scholar]
- 28.Strangmann, E., H. Froleke, and K. P. Kohse. 2002. Septic shock caused by Streptococcus suis: case report and investigation of a risk group. Int. J. Hyg. Environ. Health. 205:385-392. [DOI] [PubMed] [Google Scholar]
- 29.Trottier, S., R. Higgins, G. Brochu, and M. Gottschalk. 1991. A case of human endocarditis due to Streptococcus suis in North America. Rev. Infect. Dis. 13:1251-1252. [DOI] [PubMed] [Google Scholar]
- 30.Wisselink, H. J., F. H. Reek, U. Vecht, N. Stockhofe-Zurwieden, M. A. Smits, and H. E. Smith. 1999. Detection of virulent strains of Streptococcus suis type 2 and highly virulent strains of Streptococcus suis type 1 in tonsillar specimens of pigs by PCR. Vet. Microbiol. 67:143-157. [DOI] [PubMed] [Google Scholar]
- 31.Wisselink, H. J., J. J. Joosten, and H. E. Smith. 2002. Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J. Clin. Microbiol. 40:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]