ABSTRACT
Cryptococcus neoformans is a human opportunistic fungal pathogen causing severe disseminated meningoencephalitis, mostly in patients with cellular immune defects. This species is divided into three serotypes: A, D, and the AD hybrid. Our objectives were to compare population structures of serotype A and D clinical isolates and to assess whether infections with AD hybrids differ from infections with the other serotypes. For this purpose, we analyzed 483 isolates and the corresponding clinical data from 234 patients enrolled during the CryptoA/D study or the nationwide survey on cryptococcosis in France. Isolates were characterized in terms of ploidy, serotype, mating type, and genotype, utilizing flow cytometry, serotype- and mating type-specific PCR amplifications, and multilocus sequence typing (MLST) methods. Our results suggest that C. neoformans serotypes A and D have different routes of multiplication (primarily clonal expansion versus recombination events for serotype A and serotype D, respectively) and important genomic differences. Cryptococcosis includes a high proportion of proven or probable infections (21.5%) due to a mixture of genotypes, serotypes, and/or ploidies. Multivariate analysis showed that parameters independently associated with failure to achieve cerebrospinal fluid (CSF) sterilization by week 2 were a high serum antigen titer, the lack of flucytosine during induction therapy, and the occurrence of mixed infection, while infections caused by AD hybrids were more likely to be associated with CSF sterilization. Our study provides additional evidence for the possible speciation of C. neoformans var. neoformans and grubii and highlights the importance of careful characterization of causative isolates.
IMPORTANCE
Cryptococcus neoformans is an environmental fungus causing severe disease, estimated to be responsible for 600,000 deaths per year worldwide. This species is divided into serotypes A and D and an AD hybrid, and these could be considered two different species and an interspecies hybrid. The objectives of our study were to compare population structures of serotype A and serotype D and to assess whether infections with AD hybrids differ from infections with serotype A or D isolates in terms of clinical presentation and outcome. For this purpose, we used clinical data and strains from patients diagnosed with cryptococcosis in France. Our results suggest that, according to the serotype, isolates have different routes of multiplication and high genomic differences, confirming the possible speciation of serotypes A and D. Furthermore, we observed a better prognosis for infections caused by AD hybrid than those caused by serotype A or D, at least for those diagnosed in France.
INTRODUCTION
Cryptococcus neoformans is a life-threatening human fungal pathogen causing meningoencephalitis, mainly in patients with cellular immune defects, such as those with acquired immunodeficiency syndrome (AIDS). This yeast is estimated to cause 1 million annual cases globally and nearly 625,000 deaths/year (1). This species exists in two mating types (MATa and MATα) (2) and two varieties, C. neoformans var. grubii (serotype A) and C. neoformans var. neoformans (serotype D), which were recently proposed as distinct species (3); most serotype D isolates are found in Europe (4, 5). The third serotype (AD hybrid) results from the fusion of serotypes A and D and in some cases has an apparent African origin (6–8). The proportion of AD hybrids varies worldwide (1.8% in Thailand, 1.3 to 5.9% in the Americas, and 3.4 to 45% in Europe) (4, 9). The allelic profiles for the mating types are also heterogeneous: a majority of αADα, fewer aADα, and even fewer αADa strains in the United States, a majority of αADa strains in Spain, Portugal, and Germany, and a similar proportion of αADa and aADα strains in Italy (10). Of note, some AD hybrid isolates have only one mating type allele because of partial or complete chromosome deletion or chromosome loss and reduplication, suggesting genomic instability (11, 12). For serotype A, the vast majority of clinical and environmental isolates are MATα (0.1 to 2% MATa) (13–15), except in sub-Saharan Africa (10% MATa) (16). For serotype D, it is established that 15% of Dutch isolates are MATa (17). αADα hybrids, which have two α type mating alleles, also occur and result from unisexual reproduction (18).
The C. neoformans population structure has been studied mainly for serotype A using PCR fingerprinting, multilocus sequence typing (MLST), or variable-number tandem repeat (VNTR) methods (7, 17, 19–24). The majority of serotype A isolates exhibit clonal expansion with very few recombination events and low genetic diversity (2). These results show mitotic clonal expansion, inbreeding via unisexual reproduction of clonally related or identical isolates, or both. Geographic specificity has been observed so far only for isolates recovered in Botswana that exhibit higher genetic diversity and recombination events (25) and for environmental isolates in India that show evidence of recombination and extensive gene flow (26). The experimental design (sampling and geographical area) may contribute to differences in the extent of clonal expansion versus recombination events reported (6). For serotype D, a unique molecular type is usually described as VNIV or AFLP2 (27–30). Evidence of recombination was found after analysis of 58 environmental isolates from North Carolina (31) and 33 from other areas (11), which differs from results obtained with Dutch isolates (17).
In humans, serotype A is the more frequent serotype and is thought to be associated with more severe infections, at least in HIV-infected patients (5). In experimental animal models, most studies have established higher virulence for serotype A than serotype D and for MATα than MATa (32, 33). Contradictory results have been published on AD hybrids. Their virulence may vary depending on the inoculation route and also on the chromosomal composition, with higher virulence for strains containing a majority of the serotype A genome (12, 34, 35). Overall, there are few clinical data on patients infected with AD hybrids.
Our objectives were thus to (i) analyze the population structure of serotype A versus serotype D using isolates recovered from patients diagnosed with cryptococcosis in France and (ii) analyze clinical presentations and outcomes in patients infected with AD hybrids compared to serotype A or D.
RESULTS
Molecular characterization of clinical isolates.
Among the 400 CryptoA/D isolates recovered from the 181 patients, 61% (244) corresponded to serotype A, 19% (76) to serotype D, and 20% (80) to AD hybrids. Of note, the serotypes determined here by PCR matched those obtained previously using fluorescence microscopy (5) except for patients infected with AD hybrids that were previously considered infected with either serotype A (20/131, 15%) or D (11/50, 22%). Overall, diploid strains were found in 52 (29%) patients (37 hybrids, 6 serotype A, and 9 serotype D). None of the serotype A isolates were MATa, whereas 13% (13/97) of the serotype D isolates were MATa. Among the 37 (20%) patients infected with AD hybrids, 25 (67%) were infected with αADa, 3 (8%) with αADα, and 6 (16%) with aADα. For three hybrids, only one allele of the mating types was identified (1 of each aAD-, -ADa, and -ADα), probably due to partial or complete chromosome loss.
Among the 181 CryptoA/D patients, 26 (14.4%) had proven mixed infection with mixed serotypes (12 patients), ploidies (6), genotypes (2), or a combination of mixed serotypes/genotypes (2) or serotypes/ploidies (4) (Table 1), confirming data obtained for a subset of the patients (36). Patients with mixed genotypes of serotype A were infected with unrelated (patient 27) or related (patient 197) isolates. Patients with mixed ploidies were infected with haploid and diploid isolates sharing the same genetic profile, suggesting in vivo diploidization by endoreplication (36). Some patients were infected with unrelated A (or D) and AD isolates (differences in ≥3/5 loci), suggesting coinoculation. Finally, one patient was infected with potentially related D and AD isolates (differing at only 1 of 5 loci), suggesting possible in vivo hybridization even though the serotype A partner was never uncovered (36) (see Table S1 in the supplemental material). Mixed infections were suspected for 13 additional patients (PCR on the original haploid isolate suggested a mixture of A and D or the presence of AD, but the 10 single colonies analyzed for each patient were haploid and either A [9/13] or D [4/13]).
TABLE 1 .
Molecular information for isolates recovered in the 26 proven mixed infections
| Patient | Origin isolate | Serotype, mating type, and ploidy | Single-colony isolate | ST of serotype: |
Type of mixed infection | Probable origin of mixed infection | |
|---|---|---|---|---|---|---|---|
| A | D | ||||||
| 5 | AD5-87 | Aα, n | AD11-71 | 104 | Serotypes | Coinoculation | |
| Dα, n | AD11-72 | 129 | |||||
| 10 | AD5-85 | αADa, 2n | AD5-35 | Serotypes + genotypes | Coinoculation | ||
| AD4-64 | Aα, n | AD8-93 | 63 | ||||
| αADa, 2n | AD2-86 | ||||||
| 20a | AD3-28 | Aα, n | AD9-61 | 63 | Serotypes | Coinoculation | |
| Dα, 2n | AD1-75 | 114 | |||||
| 21 | AD3-88 | Aα, n | AD3-87 | 23 | Serotypes | Coinoculation | |
| Dα, n | AD3-89 | 119 | |||||
| 23a | AD4-5 | Aα, n | AD7-84 | 23 | Serotypes | Coinoculation | |
| Dα, 2n | AD4-16 | 122 | |||||
| 27 | AD4-44 | Aα, n | 23 | Genotypes | Coinoculation | ||
| AD4-45 | Aα, n | 77 | |||||
| 34 | AD3-74 | Aα, n | AD8-83 | 69 | Serotypes | Coinoculation | |
| Dα, n | AD7-28 | 134 | |||||
| 35 | AD3-37 | Dα, n | AD11-22 | 108 | Serotypes | Coinoculation | |
| Aα, n | AD5-71 | 45 | |||||
| 51 | AD5-26 | Dα, n | AD12-34 | 135 | Ploidies | In vivo endoreplication | |
| Dα, 2n | AD12-35 | 135 | |||||
| 62 | AD4-62 | aADα, 2n | AD3-35 | Serotypes | Possible in vivo hybridization | ||
| Dα, n | AD2-25 | 131 | |||||
| 71a | AD4-26 | Da, n | AD10-73 | 132 | Serotypes + ploidies | Coinoculation + in vivo endoreplication | |
| Da, 2n | AD10-72 | 132 | |||||
| AD4-27 | Aα, n | AD7-68 | 32 | ||||
| Da, n | AD10-72 | 132 | |||||
| 80 | AD3-91 | Dα, n | AD3-91 | 121 | Serotypes | Coinoculation | |
| aADα, 2n | AD7-25 | ||||||
| 82 | AD7-3 | Dα, n | AD11-77 | 120 | Ploidies | In vivo endoreplication | |
| Dα, 2n | AD11-79 | 120 | |||||
| 96 | AD3-23 | Aα, n | AD4-41 | 63 | Ploidies | In vivo endoreplication | |
| Aα, 2n | AD4-34 | 63 | |||||
| 100 | AD4-70 | αADa, 2n | AD5-70 | Serotypes | Coinoculation | ||
| Aα, n | AD12-8 | 106 | |||||
| 119a | AD1-60 | Da, n | AD6-82 | 130 | Ploidies | In vivo endoreplication | |
| Da, 2n | AD10-75 | 130 | |||||
| 130 | AD1-66 | Aα, n | AD8-36 | 46 | Serotypes | Coinoculation | |
| Dα, n | AD1-84 | 122 | |||||
| 139 | AD4-77 | Aα, n | AD3-64 | 71 | Ploidies | In vivo endoreplication | |
| Aα, 2n | AD3-76 | ||||||
| 140 | AD4-80 | Aα, n | AD2-77 | 63 | Ploidies | In vivo endoreplication | |
| Aα, 2n | AD2-55 | ||||||
| 161a | AD1-76 | Aα, n | AD7-53 | 46 | Serotypes + ploidies | Coinoculation + in vivo endoreplication | |
| Dα, n | AD1-70 | 121 | |||||
| AD1-77 | Aα, n | AD7-99 | 46 | ||||
| Aα, 2n | AD8-18 | 46 | |||||
| 177a | AD4-20 | Aα, n | AD9-73 | 63 | Serotypes + ploidies | Coinoculation + in vivo endoreplication | |
| Dα, n | AD10-32 | 114 | |||||
| AD4-21 | Dα, 2n | 114 | |||||
| 188a | AD1-12 | Aα, n | 63 | Serotypes + genotypes | Coinoculation | ||
| AD1-36 | Aα, n | AD8-34 | 32 | ||||
| Dα, n | AD2-78 | 121 | |||||
| 197 | AD3-57 | Aα, n | 63 | Genotypes | Coinoculation or microevolution | ||
| AD3-58 | Aα, n | 58 | |||||
| 198a | AD4-58 | αADa, 2n | AD10-66 | Serotypes | Coinoculation | ||
| AD5-14 | Aα, n | AD10-49 | 46 | ||||
| 199 | AD5-15 | Aα, n | AD11-98 | 46 | Serotypes + ploidies | Coinoculation + in vivo endoreplication | |
| Aα, 2n | AD11-99 | 46 | |||||
| AD5-16 | αADa, 2n | AD11-68 | |||||
| 217 | AD3-17 | Aα, n | AD11-42 | 40 | Serotypes | Coinoculation | |
| Dα, n | AD11-43 | 133 | |||||
Mixed infection that was described in our previous study (36).
Comparison of genetic diversity and population structures of C. neoformans.
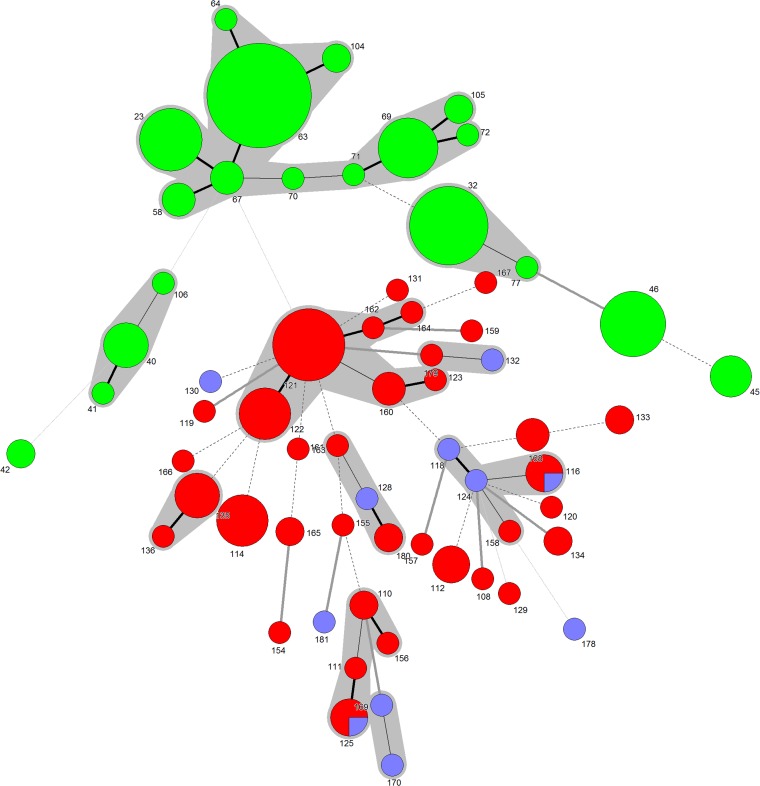
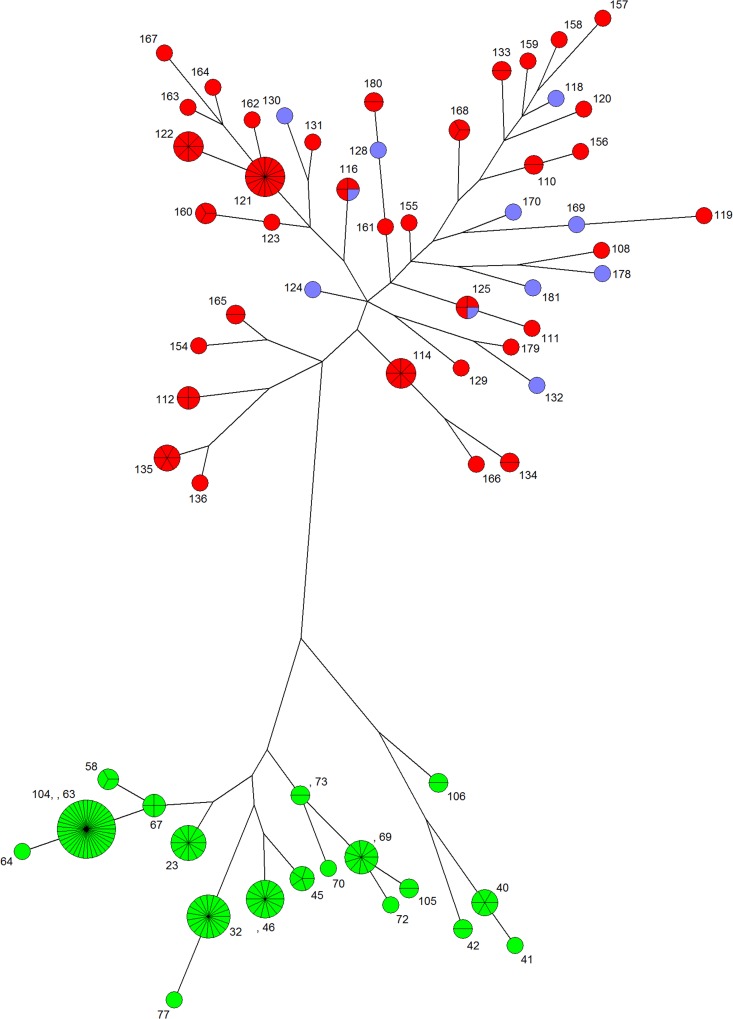
Serotypes A and D differed in the number of genetic profiles (19 sequence types [STs] grouped in 3 major clusters and 3 singletons for the 121 serotype A isolates, versus 44 STs grouped in 7 clusters and 20 singletons for the 97 serotype D isolates) (Fig. 1; also, see Tables S2 and S3 in the supplemental material). For serotype D, the discriminatory power was 0.95, confirming the robustness of the MLST method for the serotype D population. ST121 was the major profile (17/97, 17.5%), but most STs corresponded to a single isolate. Isolates harboring MATa belonged to 10 STs, including 2 STs shared with MATα isolates (ST116 and ST125) (Fig. 1). The genetic diversity was greater for serotype D than for serotype A, according to maximum-parsimony phylogenetic analysis (Fig. 2), gene diversity, and average number of alleles per locus (Table 2).
FIG 1 .
Minimum-spanning trees for isolates of serotype A and D. Minimum-spanning trees were constructed with the ST allelic profiles of the 7 MLST loci for the 97 isolates of serotype D and the 121 serotype A isolates. Green nodes, serotype A MATα; red nodes, serotype D MATα; blue nodes, serotype D MATa. The denomination of the sequence type (ST) is indicated for each node. The size of the node is proportional to the number of isolates sharing the same ST, whereas the lines between STs indicate inferred phylogenetic relationships and are in bold black, plain black, discontinuous black, bold grey, or plain grey depending on the number of allelic mismatches between profiles (1, 2, 3, 4, or more than 4, respectively). Clusters are in grey and correspond to partition of nodes that differ by a maximum of two loci.
FIG 2 .
Maximum-parsimony trees for isolates of serotypes A and D. Maximum-parsimony trees were constructed with concatenate sequences of the 7 MLST loci for the 97 serotype D isolates and the 121 serotype A isolates. Green nodes, serotype A MATα; red nodes, serotype D MATα; blue nodes, serotype D MATa. The denomination of the sequence type (ST) is indicated for each node. The size of the nodes increased with the number of isolates sharing similar sequences. The size of the lines between nodes increased with the number of differing nucleotides. Logarithmic scaling for branches was used.
TABLE 2 .
Comparison of population structure of Cryptococcus neoformans serotype D and serotype A
| Parameter | Serotype D (97 patients) | Serotype A (118 patients) |
|---|---|---|
| % (no.) of patients infected with MATa isolates | 13.4 (13) | 0 (0) |
| % (no.) of patients infected with diploid isolates | 9.3 (9) | 5.1 (6) |
| No. of STs | 44 | 19 |
| No. of combination with significant congruence/total of pairwise combinations | 10/21 | 8/21 |
| No. of graphs with allelic compatibility/total (Fig. 3 and Fig. S1) | 19/21 | 6/21 |
| Gene diversity (H) | 0.95 | 0.86 |
| Average no. of alleles per locus (N) | 11.57 | 5.57 |
| Index of association standardized (IA) | 0.00356 (44 STs); 0.1736 (97 isolates) | 0.2537 (19 STs); 0.2958 (121 isolates) |
| No. of segregating sites (S) | 76 | 81 |
| No. of recombination events (R) | 116 | 0.3 |
| Minimum no. of recombination events (Rm) | 16 | 8 |
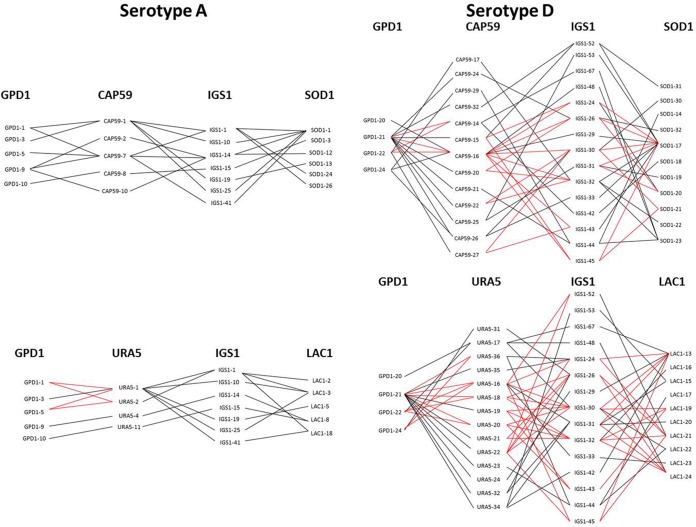
For both populations, the observed variance was significantly different from the expected variance (P < 0.0001), suggesting linkage disequilibrium among the 7 loci. Similarly, a significant test of congruence for 10/21 combinations for serotype D and 8/21 for serotype A suggested that some loci exhibited coevolution confirming linkage disequilibrium. However, based on the allelic compatibility test, compatibility was observed in 19/21 graphs for serotype D and in 6/21 graphs for serotype A (Fig. 3; also, see Fig. S1 in the supplemental material) serving as evidence of recombination in both but more recombination events among serotype D than serotype A populations. Furthermore, the index of association (IA) was not significantly different from zero, suggesting that some recombination events could occur in the serotype D population, which contrasted with results obtained for serotype A. Recombination parameters (S, R, and Rm) differed between serotypes D and A (Table 2). Altogether, these results suggested recombination events in the serotype D and clonal expansion for the serotype A populations.
FIG 3 .
Comparison of allelic compatibility tests generated for serotype A and serotype D. An hourglass shape (red lines) indicates the presence of all four possible pairs of alleles and serves as evidence of recombination. A selection of 6 out of the 21 tests is shown, demonstrating allelic compatibility in 1/6 tests for serotype A and 6/6 tests for serotype D. All graphs are provided in Fig. S1 in the supplemental material.
Finally, multiple alignment of the concatenated sequences of the 7 loci (>4,000 bp in length) showed that similarity ranged from 98.7% to 100% among serotype A isolates and from 99.2% to 100% among serotype D isolates. In contrast, 81 to 92% similarity was observed between sequences of A and D isolates. Multiple alignments revealed 7 gap positions corresponding to 20 (range, 1-7) bp for serotype A isolates, and 22 gap positions corresponding to 81 (range, 1-19) bp for serotype D isolates. These indels were distributed among all isolates and localized mainly in the IGS1 locus (5/7 positions for serotype A and 18/22 positions for serotype D), which is an untranslated region.
Influence of AD hybrids on clinical presentation and outcome of cryptococcosis.
We compared the characteristics of the patients depending on the infecting serotype (A, D, and AD) for the 155 patients infected with a unique strain (Tables 3 and 4). Overall, the three populations did not differ in terms of sex or age. The proportion of patients infected with AD hybrids did not differ according to the continent of birth, whereas there were no serotype D isolates in patients born in Africa. Of note, no association between geographical origin and cluster or ST was observed for serotype A or D isolates. There was a trend toward fewer serotype D and more AD hybrids among HIV-infected patients, whereas it was the opposite among HIV-negative patients. When four risk categories among the 33 HIV-negative patients (“malignancy,” “solid organ transplantation,” “others” [disease/treatment], and “none” [no known risk factor]) were considered, “malignancy” and “others” were the major risk factors for the 19 patients infected with A, “malignancy” and “none” were the major risk factors for the 10 infected with D, and all four categories were recorded for the 4 patients infected with AD. Clinical presentation and disease severity were similar despite a trend toward less disseminated infection and less frequent abnormal lung imaging with AD hybrids, lower serum antigen titers with serotype D and AD hybrids, and less frequent abnormal brain imaging with serotype D. Intracranial pressure was recorded for only 21 patients and was increased in 16 (14 infected with serotype A, 1 with D, and 1 with AD). Induction therapy with a combination of amphotericin B and flucytosine was prescribed with similar frequencies to patients infected with serotypes A and AD and significantly less frequently to patients infected with serotype D. Fluconazole induction therapy mirrored that of the amphotericin B-flucytosine combination. The proportion of mycological failure (nonsterilization at week 2 despite antifungal therapy) was lower in the case of the AD hybrid than in that of serotype A or D. This was significant when patients with meningoencephalitis were considered and near significant when all cases (meningeal and nonmeningeal cryptococcosis) were considered.
TABLE 3 .
Characteristics of the 155 patients with single infection due to one of the three serotypes of C. neoformans
| Characteristic | Serotype A (n = 98.0) | Serotype D (n = 26.0) | Serotype AD (n = 31.0) | P |
|---|---|---|---|---|
| No. of males/total (%)a | 79/98 (80.6) | 21/26 (80.8) | 24/31 (77.4) | 0.957 |
| Age (yr) (mean ± SD) | 41.2 ± 11.9 | 45.8 ± 12.5 | 42.7 ± 12.5 | 0.217 |
| No. born in Africa/total (%) | 31/98 (31.6) | 0/26 (0) | 6/31 (19.4) | 0.001 |
| No. HIV infected/total (%) | 79/98 (80.6) | 16/26 (61.5) | 27/31 (87.1) | 0.063 |
| Mean CD4/mm3 ± SD for HIV-infected patients | 50 ± 82 | 44 ± 56 | 41 ± 54 | 0.866 |
| No. with abnormal neurology/total (%) | 40/98 (40.8) | 10/26 (38.7) | 15/31 (48.4) | 0.733 |
| No. with meningoencephalitis/total (%) | 82/95 (86.3) | 19/23 (82.6) | 26/28 (92.9) | 0.545 |
| No. with fungemia/total (%) | 42/94 (44.7) | 11/25 (44.0) | 11/28 (39.3) | 0.888 |
| No. with dissemination/total (%) | 60/98 (61.2) | 16/26 (61.5) | 12/31 (38.7) | 0.081 |
| No. with high serum antigen titer (≥512)/total (%) | 47/89 (52.8) | 8/23 (34.8) | 9/28 (32.1) | 0.083 |
| No. with high CSF antigen titer (≥512)/total (%) | 40/85 (47.1) | 6/17 (35.3) | 8/24 (33.3) | 0.398 |
| No. with abnormal brain imaging/total (%) | 28/80 (35.0) | 2/18 (11.1) | 11/27 (40.7) | 0.084 |
| No. with abnormal lung imaging/total (%) | 48/97 (49.5) | 11/23 (47.8) | 8/30 (26.7) | 0.088 |
| No. with AMB + 5FC as induction therapy/total (%)b | 54/98 (55.1) | 7/26 (26.9) | 14/31 (45.2) | 0.031 |
| No. with fluconazole as induction therapy/total (%) | 24/92 (26.1) | 11/24 (45.8) | 12/29 (41.4) | 0.086 |
| No. with mycological failure at day 15/total (%)c | 33/77 (42.9) | 7/22 (31.8) | 4/24 (16.7) | 0.059 |
| No. with CSF mycological failure at day 15/total (%)d | 31/66 (47.0) | 6/17 (35.3) | 3/23 (13.0) | 0.013 |
| No. who died within 90 days after diagnosis/total (%) | 21/82 (25.6) | 7/22 (31.8) | 7/25 (28.0) | 0.812 |
Total number of patients evaluated or for whom the information was available.
AMB, amphotericin B; 5FC, flucytosine.
Persistence of viable cryptococci in cultured samples.
Persistence of viable cryptococci in cerebrospinal fluid samples.
TABLE 4 .
Independent parameters associated with mycological failure at week 2 for 123 patients
| Parameter | Univariate analysis |
Multivariate analysisa |
|||
|---|---|---|---|---|---|
| % with mycological failure (no./total) | % with mycological cure (no./total) | P value | OR (95% CI) | P | |
| Male sex | 92.2 (47/51) | 77.8 (56/72) | 0.046 | ||
| Disseminated infection | 74.5 (38/51) | 59.7 (43/72) | 0.122 | ||
| Mixed infections | 21.6 (11/51) | 8.3 (6/72) | 0.061 | 5.6 (1.4–22.6) | 0.015 |
| Lack of 5FC | 54.9 (28/51) | 34.7 (25/72) | 0.029 | 5.8 (2.0–17.2) | 0.001 |
| Infection by AD hybrid | 7.8 (4/51) | 31.9 (23/72) | 0.002 | 0.1 (0.02–0.46) | 0.003 |
| Serum antigen titer > 512 | 63.8 (30/47) | 40.9 (27/66) | 0.022 | 5.0 (1.7–14.4) | 0.003 |
OR, odds ratio; CI, confidence interval.
In a multivariate analysis on the entire database (unique and mixed infections), parameters independently associated with mycological failure (based on cerebrospinal fluid [CSF] sterilization) were a high serum antigen titer, lack of flucytosine during induction therapy, and mixed infection, while infection with AD hybrids was more often associated with mycological cure (Table 4). Overall survival at 3 months was not different in patients infected by hybrids compared to the others. Ten patients (1 with AD hybrids and 9 with serotype A) had neurological sequelae.
DISCUSSION
We used clinical data and isolates collected during the nationwide surveillance on cryptococcosis and the CryptoA/D study to further analyze Cryptococcus biology and the disease it causes. Parts of the data sets have been used in other studies (5, 36, 37). However, the discovery of AD hybrid isolates among those previously classified as serotype A or D allowed us to analyze here how the disease caused by AD hybrids in humans differed from that caused by A or D (5). Likewise, expanding our search for mixed infections to the entire data set of the CryptoA/D study allowed us to confirm the high incidence of mixed infections during cryptococcosis but also to include it with the data for AD hybrids in the model of multivariate analysis studying the parameters influencing outcome of infection. We thus confirmed previous findings but also extended our understanding of the biology of an important fungal pathogen and of parameters potentially useful for the management of cryptococcosis.
The diversity of the French C. neoformans clinical isolates was higher than previously reported (5), with a proportion of AD hybrids (20%) and a distribution of mating type profiles similar to European data (9). Cogliati et al. and Li et al. found that 71% of AD hybrids harbored the MATa allele (73% here), suggesting that AD hybrids could be a reservoir preserving the MATa mating type, rarely found outside Africa for serotype A (9, 11). The hypothetical African origin of AD hybrids (7) was further supported by the similar proportion of AD hybrids among patients born in Africa and in other continents in our study.
The difference in the population structures of serotypes A and D was striking. Serotype A is considered more virulent than serotype D, and its clonal expansion (at least for MATα) could contribute to maintain genomic markers associated with virulence. Our data (low genetic diversity, lack of MATa, linkage disequilibrium, values of recombination parameters, and low allelic compatibility) confirmed the hypothesis of clonal expansion for serotype A in France. The presence of both mating types can facilitate recombination in a population, and high genetic diversity within a population is often associated with sexual reproduction (11, 38). It is known that sexual reproduction of serotype D is robust and not strain specific (39), whereas 50% of clinical and environmental isolates of serotype A are fertile (40). Despite linkage disequilibrium, our results (high proportion of MATa and diploid isolates, high genetic diversity, higher proportions of indels in coding regions, high allelic compatibility, and values of recombination tests) suggest that recombination events can occur among serotypes D isolates. Despite the possibility of recombination (31), no geographic specificity was observed, with the limitation that only clinical isolates were analyzed.
The possible speciation of serotypes A and D has been the subject of debate for many years (3, 6). Known differences between the two serotypes include geographical distribution (41), host-related susceptibility and type of disease produced (5), skin tropism (41), maximal growth temperature (42), proportion of MATa (16), monokaryotic fruiting for MATa (43), and overall genome differences (85 to 90% nucleotide sequence identity between JEC21 and H99 genomes [44, 45]). Here, we found major differences in population structure and 8 to 19% nucleotide difference over 4,000 bp. Reproductive isolation has been considered the main mechanism of speciation (46). Even if serotype A diverged from the serotype D lineage about 18.5 million years ago (8), incomplete intervarietal sexual cycles occurred at different times (0 to 3.2 million years ago), leading to introgression of C. neoformans var. grubii into C. neoformans var. neoformans and to the origins of AD hybrids (9, 44). AD hybrids seem to be locked in the diploid state because of genomic differences preventing progression through meiosis. Furthermore, basidiospores generated from intervarietal matings have a low viability and low propensity to germinate (39). On the basis of reproductive isolation already described and phylogenetic distance confirmed by our results, both varieties could be classified into different species (6, 47). In their new study, Hagen et al. (3) propose recognition of both varieties of C. neoformans as new species (C. neoformans for serotype A and the newly proposed Cryptococcus deneoformans for serotype D). This is based on the phylogenetic analysis of 11 genetic loci and on biochemical, physiological, and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry profiles. Our results are thus in agreement with this new taxonomic classification (3).
A fascinating discovery of the systematic screening of original isolates was the high proportion of mixed infections (21.5%), confirming and extending with 181 patients the results obtained with 49 (36). As reported, these mixed infection were the results of either coinoculation or in vivo evolution (transitions in ploidy or microevolution). However, we did not uncover hybridization that could explain the simultaneous presence of A, D, and AD hybrids in the same patient. It was important to expand the sample size to the entire population of the CryptoA/D study to ensure that our original finding was not due to some selection bias. This study also allowed us to determine by univariate analysis that patients infected with AD hybrids differed from those infected with serotype A or D (Table 4), especially in terms of type of infection (less frequent dissemination based on culture and serum antigen titers and less frequent lung involvement) and in terms of response to induction treatment (more CSF sterilization). Multivariate analysis showed that parameters independently associated with a lack of CSF sterilization at week 2 after initiation of treatment were a high serum antigen titer (>512), the lack of flucytosine during induction therapy (as reported before [5]), and the occurrence of mixed infection, while infection with AD hybrids was more often associated with CSF sterilization. Nevertheless, differences in CSF sterilization do not prove differences in virulence in humans. We should keep in mind that the AD hybrids identified in patients diagnosed in France are different from those found in Africa, and indeed, Wiesner et al. found no survivors among the 8 Ugandan patients infected by AD hybrids (48), whereas survival was similar whatever the infecting serotype for the patients enrolled in the CryptoA/D study. Virulence of AD hybrids has been assessed in experimental infections using relatively few strains (12, 32, 34). The mating type, the inoculation route, and the presence of the Aα allele have been associated with enhanced virulence. A recent study, however, demonstrated a significantly lower virulence only for the aADa hybrids, providing evidence for negative epistatic interactions between Aa and Da alleles (49). This was not assessed here because of the small number of patients involved for some of the allele combinations. A higher DNA content does not explain differences in virulence for AD hybrids because genetically related haploid and diploid isolates exhibit similar virulence (36, 49). However, diploid cells are thought to be larger than haploid cells, and cell size can have an impact on phagocytosis (described for titan cells [50]). Chromosomal composition (proportion of serotype A or D genomes) in AD hybrids varies with partial and/or complete chromosome deletion and/or duplication (11, 12, 35), with an impact on capsule structure, as shown here by the difference in serotype assignment achieved with a monoclonal antibody (51).
Our study again highlights the importance of combining clinical and molecular data on original isolates for the study of cryptococcosis and most probably other fungal infections. This approach could be essential to identify putative species- and clade-specific risk factors and possible associations between particular strain types and host microenvironment and could lead to optimized recommendations for the management of the patients.
MATERIALS AND METHODS
Ethics statement.
The CryptoA/D study is a prospective multicenter observational study that enrolled 230 patients with cryptococcosis in France between 1997 and 2001 (5). The CryptoA/D study was approved by the ethical committee and reported to the French Ministry of Health (registration number DGS970089). Patients enrolled in the CryptoA/D study gave their written informed consent for a systematic workup. Data were analyzed anonymously. Clinical data and isolates were collected. The cryptococcosis surveillance program is approved by the Institut Pasteur Internal Review Board (2009–14/IRB).
Cryptococcus neoformans isolates.
Molecular characterization was performed for 400 isolates obtained from 380 original isolates recovered from 181 patients (CryptoA/D isolates) and for 53 serotype D isolates collected from 53 patients during the surveillance program implemented at the National Reference Center for Invasive Mycoses and antifungals (CNRMA) (52). Reference strains JEC21 (serotype D, MATα), JEC20 (serotype D, MATa), H99 (serotype A, MATα), KN99a (serotype, A MATa), and KN99α (serotype A, MATα) were used.
Molecular characterization. (i) Determination of ploidy.
Cells were prepared for flow cytometry (53). Data were acquired from 30,000 cells using the FL2 channel of a BD FACScan flow cytometer (Becton, Dickinson Company, Franklin Lakes, NJ, USA). Analysis was performed using CellQuest software version 3.3 (BD Biosciences, San Jose, CA, USA) using the profile of H99α as a reference for haploidy.
(ii) Determination of mating type and serotype.
Yeasts were grown for 24 h at 28°C on solid YNB medium. DNA was extracted using the High-Pure PCR template preparation kit (Roche Applied Science, Indianapolis, IN). PCR were performed on an iCycler thermocycler (Bio-Rad, Hercules, CA) using primers specific for the serotype and the mating type (SXI1α/SXI2a and STE20α/a) (12). Serotype-specific primers for the PAK1 and GPA1 genes were also used for all original cultures and selected single colonies (12). For some isolates, other STE20 primers were used (Table 5) because of a deletion in the STE20 sequences.
TABLE 5 .
New primers specific of the serotype and mating type used for amplification of STE20
| Serotype/mating type | Primer | 5′–3′ sequence | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|
| Dα | JOHE21312 | AGCACCAGCCTATGGAGTCCGTCT | 668 | 60 |
| JOHE21322 | TCAAAAGGTTGTCAGACTTGATGT | |||
| Da | JOHE21313 | CACATCTCAGATGCCATTTTACCA | 526 | 60 |
| JOHE21323 | TCATCACAATGATCTCATTCACAA | |||
| Aa | JOHE21314 | CTAACTCTACTACACCTCACGGCA | 457 | 11 |
| JOHE21324 | CGCACTGCAAAATAGATAAGTCTG | |||
| Aα | JOHE21691 | AGCATCAGCTTTTGGAGTCTAC | 413 | Wenjun Li (Duke University) |
| JOHE21692 | AGCATCAGCTTTTGGAGTCTAC |
(iii) MLST.
Multilocus sequence typing (MLST) was performed on all serotype A and D isolates using the published scheme (CAP59, URA5, GPD1, SOD1, LAC1, IGS1, PLB1) (19) with slightly different conditions for serotype D (36). Of note, LAC1 and URA5 loci are both localized on chromosomes 8 and 7 for serotypes A and D, respectively, whereas the other five loci are on different chromosomes. Sequences were edited with Chromas Pro version 1.41 (Technelysium Pty. Ltd., Helensvale, Queensland, Australia) and Mega version 5.1 (54). Concatenate sequences of the 7 MLST loci were aligned to construct a similarity matrix using BioNumerics version 6.6 (Applied Maths, NV). The MLST allelic sequences and sequence type (ST) are available online (http://mlst.mycologylab.org/defaultinfo.aspx?Page=CN).
Discriminatory power was determined by using Hunter coefficient (55). Frequency of each ST was calculated by using START software version 2 and LIAN3.5 software (56). Gene diversity (H) was calculated as [n/(n − 1)](1 − Σp2i), where n is the number of samples and pi is the relative frequency of the ith allele. Average number of alleles per locus was determined as (1/k)Σni, where k is the total number of loci and ni is the number of alleles for one locus.
Genotypes for AD hybrid were determined for 5 loci (PLB1, GPD1, SOD1, IGS1, and LAC1) by using serotype-specific primers (11, 36). We were not able to design serotype-specific primers for the CAP59 and URA5 loci due to high sequence similarity between A and D alleles.
Phylogenetic analysis.
For the population genetics analysis, only one isolate per patient was used except when different genotypes were observed in a given patient. A total of 97 isolates from 97 patients was studied for serotype D and 121 isolates from 118 patients for serotype A. Minimum-spanning trees were constructed with MLST alleles using BioNumerics. Clusters were defined as partitions of nodes having a maximum distance of two loci. Isolates located in the same cluster were considered related. Maximum-parsimony trees were constructed using BioNumerics, with concatenate sequences of the 7 MLST loci aligned using ClustalW.
An allele compatibility test was performed from ST global isolates by generation of 21 graphs. This test calculates the proportion of loci that show phylogenetic compatibility when compared in pairwise combinations. In the simplest case of phylogenetic compatibility, for two loci with two alleles each, if all four possible genotypes are found in the population, these two loci are called phylogenetically incompatible. An hourglass shape thus indicates the presence of all four possible pairs of alleles and serves as evidence for recombination (57).
The index of congruence (Icong) was calculated using online calculation (58) (http://mobyle.pasteur.fr/cgi-bin/portal.py#welcome) for testing topological similarity between haplotypes trees by comparing pairwise combination (here, 21 combinations for 7 MLST loci) and calculating the P value for each combination. Congruence between trees can suggest coevolution of genes, i.e., no recombination events.
The index of association (IA) and linkage disequilibrium were determined by using START software version 2 and LIAN3.5 software (56). IA is the ratio of the observed variance in the association of alleles among loci to the corresponding expected variance based on random associations. Significant associations among alleles at different loci are inconsistent with random recombination but consistent with clonality.
DnaSP software version 5.10 was used to calculate recombination parameters, with S representing the number of segregating sites, R the number of recombination events, and Rm the number of recombination events that can be parsimoniously inferred from the sequences (59).
Statistical analysis.
Correlations between clinical and molecular data were analyzed for the CryptoA/D patients (Stata 10.0; Stata Corporation, College Station, TX). Comparisons between groups were done using chi square or Fisher exact tests for categorical variables, and Student’s t test or one-way analysis of variance for continuous variables. For the multivariate analysis, logistic regression was used to determine factors associated with mycological failure (lack of CSF sterilization) at week 2. Odds ratios (OR) and 95% confidence intervals (CI) were determined by means of logistic regression analysis. Variables that were clinically relevant with P values of 0.25 were entered simultaneously into the initial model. Variables were removed following a backward-stepwise selection procedure, leaving only variables with P values of 0.05 in the final model. We estimated overall survival (cumulative survival probabilities and their 95% CIs) by the Kaplan-Meier method, and comparison of survival between groups was performed by log rank tests.
SUPPLEMENTAL MATERIAL
Allele compatibility test performed from 19 STs for serotype A population (A) and 44 STs serotype D population (B), by generation of 21 graphs. This test calculates the proportion of loci that show phylogenetic compatibility when compared in pairwise combinations. An hourglass shape (red lines) thus indicates the presence of all four possible pairs of alleles and serves as evidence of recombination. Download
MLST profiles of the isolates recovered from patients infected simultaneously by AD hybrids and serotypes A or D isolates.
MLST profiles and frequencies of the 44 sequence types (ST) for the 97 isolates of serotype D.
MLST profiles and frequencies of the 19 sequence types (ST) for the 121 isolates of serotype A.
ACKNOWLEDGMENTS
We gratefully acknowledge the Genotyping of Pathogens and Public Health sequencing facility (PF-8, Institut Pasteur). We also thank Sitali Simwami and Matthew Fisher (Imperial College London, London), Luciana Trilles and Wieland Meyer (University of Sydney, Australia) for C. neoformans MLST allele designation. We thank Wenjun Li, Sheng Sun (Duke University, USA), Sylvain Brisse (Institut Pasteur, Paris, France), and Alexandre Alanio (Hôpital Saint Louis, Paris, France) for helpful discussions.
We have no conflicts of interest.
The financial contribution of Institut Pasteur (through the “programme de recherche clinique”), Institut de Veille Sanitaire (through the National Reference Center for Invasive Mycoses and Antifungals) and of Ensemble contre le Sida-SIDACTION (through a grant to Françoise Dromer) is gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The French Cryptococcosis Study Group is composed of the following individuals, who actively participated in the data collection: J. Achard, D. Chabasse, J. P. Bouchara (Angers); S. Bland, J. P. Bru (Annecy); M. Pulik, F. Leturdu (Argenteuil); X. Lepeu, H. Lefrand, C. Jensen (Avignon); M. Ferrand, M. Larrouy (Bayonne); M. Bentata, C. Bouges-Michel, J. Camuset, L. Guillevin, B. Jarrousse, M. Robineau, J. J. Rousset (Bobigny); B. Couprie, M. Dupon, H. Dutronc, J. Y. Lacut, J. L. Pellegrin, J. M. Ragnaud, J. F. Viallard, F. X. Weill (Bordeaux); M. E. Bougnoux, X. Montreal, S. Morelon, E. Rouveix, (Boulogne); P. Granier, H. de Montclos (Bourg-en-Bresse); A. Desveaux, M. Gavignet, A. S. Labussiere, M. Mornet (Bourges); L. De Saint-Martin, E. Moalic (Brest); J. Roucoules, J. F. Loriferne, G. Otterbein (Bry-sur-Marne); J. F. Desson, M. Leporrier, C. Duhamel, J. Bonhomme (Caen); J. M. Korach (Chalons en Champagne); B. Salles, C. Sire (Chalon/Saône); V. Herve, B. Souleau, (Clamart); J. Beytout, M. Cambon, D. Pons, P. Poirier (Clermont-Ferrand); Y. Boussougant, D. Dreyfuss, X. Michon, P. Vinceneux (Colombes); G. Belkacem-Belkaki, S. Bretagne, M. Chousterman, P. Grimberg, A. S. Lascaux, A. Schaeffer, A. Sobel (Créteil); J. L. Bacri, G. Berthelot (Dieppe); A. Bonnin, M. Duong, J. Lopez, H. Portier, F. Dalle (Dijon); M. Gauthier, O. Salmon (Évry); J. Bizet (Fresnes); J. L. Gaillard, C. Perronne (Garches); M. A. Desailly, H. Maisonneuve (La Roche-sur-Yon); J. P. Bedos, J. Doll, O. Eloy, J. C. Ghnassia, S. Roussin-Bretagne (Le Chesnay); C. Brocard, P. Guiffault, A. Layet, A. Morel (Le Havre); F. Botterel, P. Bouree, J. F. Delfraissy, Y. Kertaimont, A. Angoulvant, P. Lozeron, K. Rérat, G. Saïd (Le Kremlin-Bicêtre); X. Cricks (Les Mureaux); M. L. Darde, A. Jaccard, B. Bouteille, D. Azjenberg (Limoges); D. Bouhour, E. Dannaoui, X. Mallet, D. Peyramond, M. A. Piens, C. Trepo (Lyon); L. Berardi, F. Tremolieres (Mantes-la-Jolie); Y. Berland, A. Blancard, L. Collet, J. Delmont, H. Gallais, X. Gamby, A. Michel Nguyen, J. Moreau, N. Petit, J. M. Sainty, J. Sampol-Roubicek (Marseille); M. Bietrix, M. Nezri (Martigues); A. Fiacre, S. Levy (Meaux); C. Chandesris, X. La Torre (Montargis); P. Andres, E. Billaud, F. Boiffin, M. Hamidou, O. Morin, B. Planchon, P. Poirier, F. Raffi, D. Villers, F. Morio (Nantes); P. Clevenbergh, F. De Salvador, P. Dellamonica, X. Durand, M. Gari-Toussaint (Nice); A. Romaru, M. Texereau (Niort); L. Bret, T. Prazuk, D. Poisson (Orléans); X. Bernard, Y. Pacheco (Pierre-Bénite); B. Becq-Giraudon, C. Kauffmann-Lacroix, J. C. Meurice, T. Pasdeloup (Poitiers); J. Deville, D. Toubas (Reims); C. Arvieux, F. Cartier, S. Chevrier, B. Degeilh, T. Frouget, C. Guiguen, P. Le Cavorzin, J. P. Gangneux, C. Michelet, V. Noyon (Rennes); P. Abboud, P. Brasseur, J. Leroy, J. F. Muir, L. Favennec (Rouen); P. Babinet, F. Fraisse, N. Godineau, S. Hamane, P. Margent, D. Mechali, M. Thuong (Saint-Denis); C. Soler, (Saint-Mandé); B. Hery, J. Y. Leberre (Saint-Nazaire); A. Gregory, O. Prevot (Saint-Julien-en-Genevois); D. Christmann, J. Waller, V. Letscher-Bru (Strasbourg); O. Bletry, P. Cahen, D. Zucman (Suresnes); B. Fortier (Toul); X. Aubert, S. Chadapaud, X. Delbeck, A. Lafeuillade, X. Raoult (Toulon); E. Bonnet, S. Cassaing, A. Gadroy, M. D. Linas, J. F. Magnaval, P. Massip, L. Prudhomme, L. Sailler, (Toulouse); V. Baclet, C. Coignard, Y. Mouton, I. Ravaux (Tourcoing); C. Eloy, A. Fur, L. Rezzouk (Troyes); C. Fontier, E. Mazards (Valenciennes); M. F. Biava, P. Canton, L. Kures, C. Rabaud, M. Machouart (Vandœuvre-lès-Nancy); D. Vittecocq, E. Chachaty (Villejuif); S. Dellion, O. Patey (Villeneuve-St.-Georges); J. P. Bedos, O. Benveniste, C. Bouchard, S. Belaich, C. Carbon, C. Chochillon, J. P. Coulaud, V. Descamps, X. Duval, C. Leport, F. Lheriteau, P. Longuet, H. Mouas, F. Vachon, J. L. Vilde, P. Yeni (Hôpital Bichat-Claude Bernard, Paris); V. Lavarde, C. Piketty (Hôpital Broussais, Paris); B. Christoforov, J. Dupouy-Camet, J. P. Luton, A. Paugam, M. T. Baixench (Hôpital Cochin, Paris); N. Desplaces, G. Raguin (Hôpital de La Croix-Saint-Simon, Paris); P. Chevalier, M. Kazatchkine, V. Lavarde, A. Meyrier (Hôpital Européen Georges Pompidou, Paris); A. Bernadou, M. Cornet, J. P. Marie, S. Oudart (Hôpital de l’Hôtel-Dieu, Paris); M. Gayraud, Y. Pean (Institut Mutualiste Montsouris, Paris); C. Aznar, B. Dupont, H. Poncelet, Amaury de Gouvelho, Clarisse Loyer, Odile Launay (Hôpital de l’Institut Pasteur); P. Berche, B. Dupont, V. Mathé, (Hôpital Necker-Enfants Malades, Paris); L. Baril, P. Bossi, F. Bricaire, J. Carrière, A. Datry, S. Herson, M. Jouan, M. Levy-Soussan, C. Mouquet, B. Orcel, M. M. Thiebaut (Hôpital Pitié-Salpétrière, Paris); J. Frottier, J. B. Guiard-Schmidt, B. Lebeau, J. L. Meynard, M. C. Meyohas, J. L. Poirot, P. Roux, X. Urban (Hôpital Saint-Antoine, Paris); F. Daniel, J. Gilquin, J. F. Timsit (Hôpital Saint-Joseph, Paris); C. Lacroix, JC Brouet, J. M. Decazes, F. Derouin, B. Eurin, J. R. Legall, C. Legendre, S. Neuville (Hôpital Saint-Louis, Paris); J. P. Escande (Hôpital Tarnier, Paris); G. Delzant, G. Kac, C. Trivalle (Hôpital Tenon, Paris). Also representing the French National Reference Center for Invasive Mycoses & Antifungals were Anne Boullié, Karine Boukris-Sitbon, Olivier Lortholary, Dea Garcia-Hermoso, and Damien Hoinard.
Footnotes
Citation Desnos-Ollivier M, Patel S, Raoux-Barbot D, Heitman J, Dromer F, The French Cryptococcosis Study Group. 2015. Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. mBio 6(3):e00311-15. doi:10.1128/mBio.00311-15.
Contributor Information
Liise-anne Pirofski, Albert Einstein College of Medicine.
Collaborators: The French Cryptococcosis Study Group, J. Achard, D. Chabasse, J. P. Bouchara, S. Bland, J. P. Bru, M. Pulik, F. Leturdu, X. Lepeu, H. Lefrand, C. Jensen, M. Ferrand, M. Larrouy, M. Bentata, C. Bouges-Michel, J. Camuset, L. Guillevin, B. Jarrousse, M. Robineau, J. J. Rousset, B. Couprie, M. Dupon, H. Dutronc, J. Y. Lacut, J. L. Pellegrin, J. M. Ragnaud, J. F. Viallard, F. X. Weill, M. E. Bougnoux, X. Montreal, S. Morelon, E. Rouveix, P. Granier, H. de Montclos, A. Desveaux, M. Gavignet, A. S. Labussiere, M. Mornet, L. De Saint-Martin, E. Moalic, J. Roucoules, J. F. Loriferne, G. Otterbein, J. F. Desson, M. Leporrier, C. Duhamel, J. Bonhomme, J. M. Korach, B. Salles, C. Sire, V. Herve, B. Souleau, J. Beytout, M. Cambon, D. Pons, P. Poirier, Y. Boussougant, D. Dreyfuss, X. Michon, P. Vinceneux, G. Belkacem-Belkaki, S. Bretagne, M. Chousterman, P. Grimberg, A. S. Lascaux, A. Schaeffer, A. Sobel, J. L. Bacri, G. Berthelot, A. Bonnin, M. Duong, J. Lopez, H. Portier, F. Dalle, M. Gauthier, O. Salmon, J. Bizet, J. L. Gaillard, C. Perronne, M. A. Desailly, H. Maisonneuve, J. P. Bedos, J. Doll, O. Eloy, J. C. Ghnassia, S. Roussin-Bretagne, C. Brocard, P. Guiffault, A. Layet, A. Morel, F. Botterel, P. Bouree, J. F. Delfraissy, Y. Kertaimont, A. Angoulvant, P. Lozeron, K. Rérat, G. Saïd, X. Cricks, M. L. Darde, A. Jaccard, B. Bouteille, D. Azjenberg, D. Bouhour, E. Dannaoui, X. Mallet, D. Peyramond, M. A. Piens, C. Trepo, L. Berardi, F. Tremolieres, Y. Berland, A. Blancard, L. Collet, J. Delmont, H. Gallais, X. Gamby, A. Michel Nguyen, J. Moreau, N. Petit, J. M. Sainty, J. Sampol-Roubicek, M. Bietrix, M. Nezri, A. Fiacre, S. Levy, C. Chandesris, X. La Torre, P. Andres, E. Billaud, F. Boiffin, M. Hamidou, O. Morin, B. Planchon, P. Poirier, F. Raffi, D. Villers, F. Morio, P. Clevenbergh, F. De Salvador, P. Dellamonica, X. Durand, M. Gari-Toussaint, A. Romaru, M. Texereau, L. Bret, T. Prazuk, D. Poisson, X. Bernard, Y. Pacheco, B. Becq-Giraudon, C. Kauffmann-Lacroix, J. C. Meurice, T. Pasdeloup, J. Deville, D. Toubas, C. Arvieux, F. Cartier, S. Chevrier, B. Degeilh, T. Frouget, C. Guiguen, P. Le Cavorzin, J. P. Gangneux, C. Michelet, V. Noyon, P. Abboud, P. Brasseur, J. Leroy, J. F. Muir, L. Favennec, P. Babinet, F. Fraisse, N. Godineau, S. Hamane, P. Margent, D. Mechali, M. Thuong, C. Soler, B. Hery, J. Y. Leberre, A. Gregory, O. Prevot, D. Christmann, J. Waller, V. Letscher-Bru, O. Bletry, P. Cahen, D. Zucman, B. Fortier, X. Aubert, S. Chadapaud, X. Delbeck, A. Lafeuillade, X. Raoult, E. Bonnet, S. Cassaing, A. Gadroy, M. D. Linas, J. F. Magnaval, P. Massip, L. Prudhomme, L. Sailler, V. Baclet, C. Coignard, Y. Mouton, I. Ravaux, C. Eloy, A. Fur, L. Rezzouk, C. Fontier, E. Mazards, M. F. Biava, P. Canton, L. Kures, C. Rabaud, M. Machouart, D. Vittecocq, E. Chachaty, S. Dellion, O. Patey, J. P. Bedos, O. Benveniste, C. Bouchard, S. Belaich, C. Carbon, C. Chochillon, J. P. Coulaud, V. Descamps, X. Duval, C. Leport, F. Lheriteau, P. Longuet, H. Mouas, F. Vachon, J. L. Vilde, P. Yeni, V. Lavarde, C. Piketty, B. Christoforov, J. Dupouy-Camet, J. P. Luton, A. Paugam, M. T. Baixench, N. Desplaces, G. Raguin, P. Chevalier, M. Kazatchkine, V. Lavarde, A. Meyrier, A. Bernadou, M. Cornet, J. P. Marie, S. Oudart, M. Gayraud, Y. Pean, C. Aznar, B. Dupont, H. Poncelet, Amaury de Gouvelho, Clarisse Loyer, Odile Launay, P. Berche, B. Dupont, V. Mathé, L. Baril, P. Bossi, F. Bricaire, J. Carrière, A. Datry, S. Herson, M. Jouan, M. Levy-Soussan, C. Mouquet, B. Orcel, M. M. Thiebaut, J. Frottier, J. B. Guiard-Schmidt, B. Lebeau, J. L. Meynard, M. C. Meyohas, J. L. Poirot, P. Roux, X. Urban, F. Daniel, J. Gilquin, J. F. Timsit, C. Lacroix, JC Brouet, J. M. Decazes, F. Derouin, B. Eurin, J. R. Legall, C. Legendre, S. Neuville, J. P. Escande, G. Delzant, G. Kac, C. Trivalle, Anne Boullié, Karine Boukris-Sitbon, Olivier Lortholary, Dea Garcia-Hermoso, and Damien Hoinard
REFERENCES
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Litvintseva AP, Xu J, Mitchell TG. 2011. Population structure and ecology of Cryptococcus neoformans and Cryptococcus gattii, p 97–111. In Heitman J, Kozel TR, Kwon-Chung J, Perfect JR, Casadevall A (ed), Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC. [Google Scholar]
- 3.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T. 23 February 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Viviani MA, Cogliati M, Esposto MC, Lemmer K, Tintelnot K, Colom Valiente MF, Swinne D, Velegraki A, Velho R, European Confederation of Medical Mycology (ECMM) Cryptococcosis Working Group . 2006. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res 6:614–619. doi: 10.1111/j.1567-1364.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- 5.Dromer F, Mathoulin-Pélissier S, Launay O, Lortholary O, French Cryptococcosis Study Group . 2007. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med 4:e21. doi: 10.1371/journal.pmed.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol 60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 7.Litvintseva AP, Lin X, Templeton I, Heitman J, Mitchell TG. 2007. Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathog 3:e114. doi: 10.1371/journal.ppat.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Vilgalys R, Mitchell TG. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol 9:1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- 9.Cogliati M, Lin X, Viviani MA. 2011. Hybridization and its importance in the Cryptococcus species complex, p 359–370. In Heitman J, Kozel TR, Kwon-Chung J, Perfect JR, Casadevall A (ed), Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC. [Google Scholar]
- 10.Cogliati M, Zamfirova R, Tortorano AM, Viviani MA, FIMUA Cryptococcosis network . 2011. Population structure of Italian Cryptococcus neoformans variety grubii clinical isolates, oral session #1. 8th International Conference on Cryptococcus and Cryptococcosis, Charleston, South Carolina, USA. [Google Scholar]
- 11.Li W, Averette AF, Desnos-Ollivier M, Ni M, Dromer F, Heitman J. 2012. Genetic diversity and genomic plasticity of Cryptococcus neoformans AD hybrid strains. G3 (Bethesda) 2:83–97. doi: 10.1534/g3.111.001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengeler KB, Cox GM, Heitman J. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun 69:115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzot SP, Hamdan JS, Currie BP, Casadevall A. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol 35:2243–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitman J, Filler SG, Edwards JE, Mitchell AP. 2006. Molecular principles of fungal pathogenesis. ASM Press, Washington, DC. [Google Scholar]
- 15.Kwon-Chung KJ, Bennett JE. 1978. Distribution of a and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol 108:337–340. [DOI] [PubMed] [Google Scholar]
- 16.Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell 2:1162–1168. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen F, Illnait-Zaragozí MT, Meis JF, Chew WH, Curfs-Breuker I, Mouton JW, Hoepelman AI, Spanjaard L, Verweij PE, Kampinga GA, Kuijper EJ, Boekhout T, Klaassen CH. 2012. Extensive genetic diversity within the Dutch clinical Cryptococcus neoformans population. J Clin Microbiol 50:1918–1926. doi: 10.1128/JCM.06750-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. 2007. Alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet 3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanafy A, Kaocharoen S, Jover-Botella A, Katsu M, Iida S, Kogure T, Gonoi T, Mikami Y, Meyer W. 2008. Multilocus microsatellite typing for Cryptococcus neoformans var. grubii. Med Mycol 46:685–696. doi: 10.1080/13693780802027062. [DOI] [PubMed] [Google Scholar]
- 22.Karaoglu H, Lee CM, Carter D, Meyer W. 2008. Development of polymorphic microsatellite markers for Cryptococcus neoformans. Mol Ecol Resour 8:1136–1138. doi: 10.1111/j.1755-0998.2008.02196.x. [DOI] [PubMed] [Google Scholar]
- 23.Karaoglu H, Lee CM, Meyer W. 2005. Survey of simple sequence repeats in completed fungal genomes. Mol Biol Evol 22:639–649. doi: 10.1093/molbev/msi057. [DOI] [PubMed] [Google Scholar]
- 24.Illnait-Zaragozi MT, Martínez-Machín GF, Fernández-Andreu CM, Boekhout T, Meis JF, Klaassen CH. 2010. Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PLoS One 5:e9124. doi: 10.1371/journal.pone.0009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litvintseva AP, Mitchell TG. 2012. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathog 8:e1002495. doi: 10.1371/journal.ppat.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, Xu J. 2008. Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology 154:1513–1524. doi: 10.1099/mic.0.2007/015594-0. [DOI] [PubMed] [Google Scholar]
- 27.Boekhout T, van Belkum A, Leenders AC, Verbrugh HA, Mukamurangwa P, Swinne D, Scheffers WA. 1997. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol 47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 28.Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E, IberoAmerican Cryptococcal Study Group . 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis 9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, Viviani MA, Chindamporn A, Sukroongreung S, John MA, Ellis DH, Sorrell TC. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790–1799. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto Y, Kohno S, Koga H, Kakeya H, Tomono K, Kaku M, Yamazaki T, Arisawa M, Hara K. 1995. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J Clin Microbiol 33:3328–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol 43:556–564. doi: 10.1128/JCM.43.2.556-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barchiesi F, Cogliati M, Esposto MC, Spreghini E, Schimizzi AM, Wickes BL, Scalise G, Viviani MA. 2005. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J Infect 51:10–16. doi: 10.1016/j.jinf.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun 71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturvedi V, Fan J, Stein B, Behr MJ, Samsonoff WA, Wickes BL, Chaturvedi S. 2002. Molecular genetic analyses of mating pheromones reveal intervariety mating or hybridization in Cryptococcus neoformans. Infect Immun 70:5225–5235. doi: 10.1128/IAI.70.9.5225-5235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu G, Liu I, Sham A, Stajich JE, Dietrich FS, Kronstad JW. 2008. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans. Genome Biol 9:R41. doi: 10.1186/gb-2008-9-2-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desnos-Ollivier M, Patel S, Spaulding AR, Charlier C, Garcia-Hermoso D, Nielsen K, Dromer F. 2010. Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. mBio 1:e00091-10. doi: 10.1128/mBio.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alanio A, Desnos-Ollivier M, Dromer F. 2011. Dynamics of Cryptococcus neoformans-macrophage interactions reveal that fungal background influences outcome during cryptococcal meningoencephalitis in humans. mBio 2:e00158-11. doi: 10.1128/mBio.00158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell LT, Carter DA. 2006. Looking for sex in the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res 6:588–598. doi: 10.1111/j.1567-1364.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- 39.Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol 3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- 40.Fraser JA, Heitman J. 2006. Sex, MAT, and the evolution of fungal virulence, p 13–33. In Heitman J, Filler GF, Edwards JEJ, Mitchell AP (ed), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC. [Google Scholar]
- 41.Dromer F, Mathoulin S, Dupont B, Laporte A. 1996. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993). French Cryptococcosis Study Group. Clin Infect Dis 23:82–90. doi: 10.1093/clinids/23.1.82. [DOI] [PubMed] [Google Scholar]
- 42.Tortorano AM, Viviani MA, Rigoni AL, Cogliati M, Roverselli A, Pagano A. 1997. Prevalence of serotype D in Cryptococcus neoformans isolates from HIV positive and HIV negative patients in Italy. Mycoses 40:297–302. doi: 10.1111/j.1439-0507.1997.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 43.Tscharke RL, Lazera M, Chang YC, Wickes BL, Kwon-Chung KJ. 2003. Haploid fruiting in Cryptococcus neoformans is not mating type alpha-specific. Fungal Genet Biol 39:230–237. doi: 10.1016/S1087-1845(03)00046-X. [DOI] [PubMed] [Google Scholar]
- 44.Kavanaugh LA, Fraser JA, Dietrich FS. 2006. Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol Biol Evol 23:1879–1890. doi: 10.1093/molbev/msl070. [DOI] [PubMed] [Google Scholar]
- 45.Kronstad JW, Loftus BJ, Lodge JK. 2011. The Cryptococcus genomes: tools for comparative genomics and expression analysis, p 115–126. In Heitman J, Kozel TR, Kwon-Chung J, Perfect JR, Casadevall A (ed), Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC. [Google Scholar]
- 46.Kohn LM. 2005. Mechanisms of fungal speciation. Annu Rev Phytopathol 43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- 47.Diaz MR, Boekhout T, Kiesling T, Fell JW. 2005. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res 5:1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Wiesner DL, Moskalenko O, Corcoran JM, McDonald T, Rolfes MA, Meya DB, Kajumbula H, Kambugu A, Bohjanen PR, Knight JF, Boulware DR, Nielsen K. 2012. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 3:e00196-12. doi: 10.1128/mBio.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin X, Nielsen K, Patel S, Heitman J. 2008. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect Immun 76:2923–2938. doi: 10.1128/IAI.00168-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okagaki LH, Nielsen K. 2012. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell 11:820–826. doi: 10.1128/EC.00121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charlier C, Chrétien F, Baudrimont M, Mordelet E, Lortholary O, Dromer F. 2005. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am J Pathol 166:421–432. doi: 10.1016/S0002-9440(10)62265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dromer F, Mathoulin-Pélissier S, Fontanet A, Ronin O, Dupont B, Lortholary O, French Cryptococcosis Study Group . 2004. Epidemiology of HIV-associated cryptococcosis in France (1985–2001): comparison of the pre- and post-HAART eras. AIDS 18:555–562. doi: 10.1097/00002030-200402200-00024. [DOI] [PubMed] [Google Scholar]
- 53.Sia RA, Lengeler KB, Heitman J. 2000. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol 29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter PR. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol 28:1903–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jolley KA, Feil EJ, Chan MS, Maiden MC. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- 57.Agapow P, Burt A. 2001. Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1:101–102. doi: 10.1046/j.1471-8278.2000.00014.x. [DOI] [Google Scholar]
- 58.De Vienne DM, Giraud T, Martin OC. 2007. A congruence index for testing topological similarity between trees. Bioinformatics 23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- 59.Librado P, Rozas J. 2009. DnaSP V5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 60.Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Allele compatibility test performed from 19 STs for serotype A population (A) and 44 STs serotype D population (B), by generation of 21 graphs. This test calculates the proportion of loci that show phylogenetic compatibility when compared in pairwise combinations. An hourglass shape (red lines) thus indicates the presence of all four possible pairs of alleles and serves as evidence of recombination. Download
MLST profiles of the isolates recovered from patients infected simultaneously by AD hybrids and serotypes A or D isolates.
MLST profiles and frequencies of the 44 sequence types (ST) for the 97 isolates of serotype D.
MLST profiles and frequencies of the 19 sequence types (ST) for the 121 isolates of serotype A.