Abstract
Pulsed-field gel electrophoresis (PFGE) has been used as an epidemiological tool for surveillance studies of Bordetella pertussis since the early 1990s. To date there is no standardized procedure for comparison of results, and therefore it has been difficult to directly compare PFGE results between laboratories. We propose a profile-based reference system for PFGE characterization of B. pertussis strain variation and to establish traceability of B. pertussis PFGE results. We initially suggest 35 Swedish reference strains as reference material for PFGE traceability. This reference material is deposited at the Culture Collection of the University of Gothenburg, Gothenburg, Sweden. Altogether, 1,810 Swedish clinical isolates from between 1970 and 2003 were studied, together with the Swedish Pw vaccine strain, six reference strains, and two U.S. isolates. Our system provides evidence that profiles obtained by using only one enzyme, i.e., XbaI, give enough data to analyze the epidemiological relationship between them. Characterization with one enzyme is far less labor intensive, yielding results in half the time than when a two-enzyme procedure is used. Also, we can see that there is a correlation between PFGE profile and pertactin type. One common PFGE profile, BpSR11 (n = 455), showed 100% prn2 and 100% Fim3 when analyzed for pertactin type and serotype. On the other hand, strains with the same profile may express various serotypes when isolated over longer periods of time. Subculturing of the same isolate eight times or lyophilization caused no change in PFGE profile.
Whooping cough caused by Bordetella pertussis is a worldwide disease. The organism continues to circulate even in populations where high vaccine coverage of infants and children is achieved (8, 16). Although the mortality and morbidity are substantially reduced, peaks of disease appear every 3 to 5 years (2, 6, 10). Waning immunity and changed epidemiology are often referred to as explanations of this phenomenon (10, 28).
In 1998 Mooi et al. (19), focusing on the polymorphism of pertactin and pertussis toxin, suggested a vaccine-driven evolution as the reason for the emergence of new subtypes in The Netherlands. Recent Dutch isolates were shown to be distinguishable from strains that had been used for the production of vaccines. These same findings were reproduced in many other countries (10, 12, 14, 22, 29).
Acellular pertussis vaccines were introduced nationwide in Sweden in 1996, 17 years after the withdrawal of whole-cell pertussis vaccine from the childhood immunization schedule (20). In October 1997 a whooping cough surveillance program started, including collection of strains from Swedish laboratories for clinical microbiology.
Pulsed-field gel electrophoresis (PFGE) is used as an epidemiological tool for surveillance studies of B. pertussis strain variation over time and for identification of outbreak-associated isolates (4, 5, 7, 14, 22, 24, 29). It is of particular interest to monitor the bacterial population and its possible influence on vaccine effectiveness after the introduction of acellular vaccines. With the aid of PFGE, it has been possible to identify isolates that are epidemiologically related which were previously indistinguishable from each other by other typing methods (13). It is also possible to type an endemic flora of closely related organisms frequently recovered from infected patients for which no direct or epidemiological linkage can be demonstrated (25).
One problem with PFGE results is that the direct and precise comparison of profiles from different laboratories is difficult because of variation in technique and terminology. Currently there is no standardized protocol for performing PFGE or criteria for analyzing the fragment patterns. Protocol harmonization on a European level and even an international level is necessary for interlaboratory comparison of prevalent and emerging new variants.
In 1999 European research groups working together proposed a reference methodology for epidemiological typing of B. pertussis (18). The PFGE protocol proposed in that methodology suggested a two-enzyme system, i.e., XbaI and SpeI, for chromosomal DNA cleavage. Based on the profiles obtained with each of these two enzymes, Weber et al. (29) identified six main groups of B. pertussis, i.e., groups I, II, III, IVα, IVβ, and V. Reference strains were recommended for strain characterization at the group level (18, 29). Many recent studies have put forward the idea of a one-enzyme system, primarily using XbaI (4, 5, 7, 14, 22, 24). In many of these studies, references to internationally available type strains were lacking (4, 5, 7, 14).
The aims of this study were to establish traceability of B. pertussis PFGE results, as follows: (i) to describe a profile-based reference system, i.e., a reference method and reference materials with Swedish clinical material; (ii) to examine other characteristics such as toxin, pertactin, and serotype for these profiles; (iii) to study the stability of the relationship between isolates and PFGE profile; and (iv) to make key isolates available by depositing them in a culture collection bank.
MATERIALS AND METHODS
Clinical isolates.
Altogether, 1,810 Swedish clinical isolates were studied (Table 1). Clinical isolates were collected, either by our laboratory or by hospital laboratories participating in the whooping cough surveillance network (20). The isolates included here were collected between 1970 and 2003, covering various periods of vaccine status in the population (Table 1). There was (i) a whole-cell vaccine period from the 1950s to 1978 (26); (ii) a vaccine-free period from 1979 to 1996, represented by two acellular vaccine trials, the Biken study from 1986 to 1987 (1) and trial I from 1992 to 1995 (11); and (iii) a surveillance period from 1997 to 2003 after the reintroduction of general vaccination with acellular pertussis vaccines in Sweden (20).
TABLE 1.
Swedish isolates used for establishment of a PFGE profile-based reference system for epidemiological typing of B. pertussis
| Period | Duration | Isolate collection | No. of isolates |
|---|---|---|---|
| Whole-cell vaccintion (Pw) | 1950-1978 | 1970-1977 | 70 |
| No general vaccination | 1979-1996 | 1986-1987 | 74 |
| 1992-1995 | 148 | ||
| Acellular vaccination (Pa) | 1997-present | 1997-2003 | 1,518 |
| Total | 1,810 |
Reference strains.
The following reference strains were used.
(i) Reference strains representing the different groups of B. pertussis put forward by Weber et al. in 2001 (29) (FR287, B902, FR743, Bp134, Tohama I, and strain 18323) were used for comparison. Tohama I is a well characterized and completely sequenced vaccine strain (21). Strain 18323 is a mouse-virulent challenge strain recommended by the World Health Organization as a reference strain (17).
(ii) Strain A639, recommended as a standard by Hardwick et al. in 2002 (15), was also included for comparison, together with A556; both were kindly provided by G. Sanden, Centers for Disease Control and Prevention, Atlanta, Ga.
(iii) The vaccine strain 44122, which was used for the production of a Swedish whole-cell vaccine up until 1979, was studied. Lyophilized material from 11 different vaccine strain vials from the years 1956 to 1964 was taken and analyzed. The material in the 11 vials represented harvest control material and thus was thought to be the same strain.
(iv) Reference strains for 35 key Swedish PFGE profiles have been deposited at the Culture Collection of the University of Gothenburg (CCUG), Gothenburg, Sweden (Table 2).
TABLE 2.
Reference strains for the 35 key Swedish profilesa
| Rank no. | Profile | n | Serotype (%) | prn type (%) | CCUG no. |
|---|---|---|---|---|---|
| 1 | BpSR11 | 455 | 3 (100) | 2 (100) | 48371 |
| 2 | BpSR1 | 214 | 2 (94) | 2 (99) | 48372 |
| 3 | BpSR16 | 144 | 2 (97) | 2 (99) | 48373 |
| 4 | BpSR31 | 71 | 3 (59) | 3 (100) | 48374 |
| 5 | BpSR5 | 64 | 3 (100) | 2 (100) | 48375 |
| 6 | BpSR18 | 54 | 2 (44) | 2 (98) | 48376 |
| 7 | BpSR28 | 42 | 3 (97) | 2 (100) | 48378 |
| 8 | BpSR10 | 39 | 3 (97) | 2 (100) | 48377 |
| 9 | BpSR19 | 38 | 2 (100) | 2 (100) | 48379 |
| 10 | BpSR26 | 35 | 2 (89) | 2 (97) | 48380 |
| 11 | BpSR25 | 30 | 3 (73) | 1/7 (97) | 48423 |
| 12 | BpSR32 | 30 | 3 (80) | 3 (100) | 48422 |
| 13 | BpSR63 | 28 | 2 (96) | 2 (100) | 48381 |
| 14 | BpSR9 | 25 | 3 (100) | 2 (100) | 48424 |
| 15 | BpSR29 | 23 | 3 (74) | 3 (96) | 48425 |
| 16 | BpSR68 | 23 | 2 (96) | 2 (100) | 48427 |
| 17 | BpSR12 | 20 | 3 (75) | 2 (100) | 48382 |
| 18 | BpSR15 | 20 | 2 (95) | 2 (100) | 48426 |
| 19 | BpSR7 | 20 | 2 (90) | 2 (100) | 48428 |
| 20 | BpSR2 | 19 | 2 (89) | 2 (100) | 48430 |
| 21 | BpSR3 | 18 | 3 (100) | 2 (100) | 48429 |
| 22 | BpSR30 | 13 | 3 (100) | 3 (100) | 48531 |
| 23 | BpSR98 | 12 | 2 (92) | 1/7 (100) | 48432 |
| 24 | BpSR106 | 12 | 2 (100) | 2 (100) | 48433 |
| 25 | BpSR45 | 10 | 2 (100) | 2 (100) | 48528 |
| 26 | BpSR13 | 9 | 3 (89) | 2 (100) | 48431 |
| 27 | BpSR6 | 9 | 3 (67) | 1/7 (100) | 48532 |
| 28 | BpSR23 | 9 | 2/3 (44) | 1/7 (92) | 48529 |
| 29 | BpSR24 | 8 | 3 (88) | 1/7 (100) | 48530 |
| 30 | BpSR4 | 8 | 3 (100) | 2 (100) | 48534 |
| 31 | BpSR64 | 8 | 2 (100) | 2 (100) | 48535 |
| 32 | BpSR97 | 8 | 2 (100) | 1/7 (100) | 48536 |
| 33 | BpSR147 | 7 | 2 (86) | 2 (100) | 48533 |
| 34 | BpSR159 | 7 | 2 (100) | 3 (100) | 48537 |
| 35 | BpSR17 | 7 | 2 (71) | 2 (100) | 48538 |
The reference strains have been deposited at the CCUG. The percentages for serotype and pertactin types represent the most prevalent types within the profile.
Strain characterization.
At the Swedish Institute for Infectious Disease Control, lyophilized and frozen isolates of B. pertussis were recultured on charcoal horse blood agar at 36°C for up to 3 days. These subcultures were stored frozen at −70°C according to the reference method (18).
A recommended standard methodology (18) based on fimbrial serotyping and gene typing of pertactin and pertussis toxin was used for epidemiological typing of B. pertussis. DNA fingerprinting by means of PFGE was slightly modified as described below.
PFGE.
PFGE was performed as described previously (18) with the exceptions of the following modifications. Only restriction enzyme XbaI was routinely used for digestion of chromosomal DNA. The running conditions were largely the same. Bacterial colonies in 1× TE (10 mM Tris-HCl [pH 8], 1 mM EDTA) suspension were adjusted to an optical density of 0.8 at 650 nm. Each plug (plugs were prepared in 1% low-melting-agarose [SeaPlaque; FMC Bioproducts]) was incubated in a 500-μl solution of lysis buffer (0.5 M EDTA [pH 8.0], 1% Sarkosyl) containing 1 mg of proteinase K (Invitrogen) (fungal; 1g [>20 U/mg]) per ml and incubated at 56°C overnight in a shaking incubator.
Washing of the plugs was carried out in three steps. For the first wash, 7.5 ml of prewarmed 1× TE was added to the plugs in 50-ml Falcon tubes. The plugs were then incubated at 56°C for 1 h in a shaking incubator. The plugs were cooled on ice, buffer was removed, and fresh 1× TE buffer was added. The plugs were again incubated at 56°C for 1 h on a shaking incubator. The third and final wash was carried out at room temperature for 1 h. The plug slices were incubated overnight at 37°C in 150 μl of buffer solution containing 40 U of XbaI (Amersham Pharmacia).
A DR III contour-clamped homogeneous electric field apparatus with a cooling module and variable speed pump (Bio-Rad) was used, with 2.2 liters of 0.5× Tris-borate-EDTA and 120 ml of 1% agarose (SeaKem GTG; FMC BioProducts) in 0.5× Tris-borate-EDTA.
The pulsed-field program was set to correspond to a migration period of 40 h. The procedural specifications were as follows: (i) pulse times of 5 to 6 s, 5.5 V/cm, and running time of 16 h, and (ii) pulse times of 8 to 35 s, 5.5 V/cm, and running time of 24 h. The cooling system was set to 14°C.
To obtain PFGE profiles with SpeI restriction enzyme, the same procedure as for XbaI was used. The only difference was the pulsing program, which is described by Mooi et al. (18). Thirty units of SpeI enzyme (New England Biolabs) was used to digest one plug slice. Digestion with XbaI or SpeI was carried out as an independent experiment.
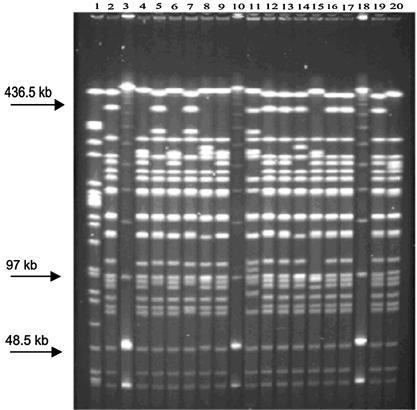
Strain T41 (Swedish reference 23 [BpSR23]) was used in every gel as an external quality control (Fig. 1). This strain was chosen because it showed evenly distanced DNA fragments encompassing all fragment sizes exhibited by our test strains on repeated pulsed-field gel analysis. T41 was run in the same lane alongside test strains in every gel. The reproducibility of the banding pattern of T41 was consistent between runs.
FIG. 1.
PFGE gel. Lane 1, Swedish vaccine strain. Lanes 3, 10, and 18, low-range marker. Lane 11, strain T41, which was applied in every gel as an intragel quality control. All other lanes show clinical test strains. High band resolution can be seen in the area also between 48.5 and 97.5 kb.
BioNumerics (Applied Maths, Inc.) version 3 software was used to normalize the DNA fragment migration distances relative to those of the Low Range PFGE DNA marker (New England BioLabs). Isolates restricted with the enzyme XbaI yielded 16 to 21 DNA bands of 45 to 435 kb in length.
Isolates with a DNA band pattern differing by ≥1 band were defined to be a distinct PFGE profile. High resolution was also shown in the area of 48.5 to 97.5 kb (Fig. 1).
The unweighted pair group method using arithmetic averages (UPGMA) was used as the clustering method, with a 1% band tolerance and 1% optimization settings with the Dice coefficient. All profiles were verified by visual comparison.
Serotyping.
Serotype analysis was performed by using a microplate agglutination assay with monoclonal antibodies against fimbria type 2 (Fim2) and Fim3. Anti-Fim2 was from hybridoma F2B2G8, and anti-Fim3 was from hybridoma C10C2D5 (9). The monoclonal antibodies used in this study were prepared at the Bordetella unit at the Institut Pasteur, Paris, France, and were kindly provided by N. Guiso. They were originally provided by the Pertussis Laboratory at the Food and Drug Administration by M. Brennan. The procedure followed the reference methodology, with the modification that the incubation was performed at 36°C. The positive controls used were B. pertussis FDA460, expressing Fim2 and Fim3; P25/98, expressing Fim2 only; and P611/98, expressing Fim3 only.
Gene typing.
A multiplex PCR was used for the amplification of the prn and ptxS1 genes. For determination of pertactin and toxin types, the PCR product was always sequenced on both DNA strands. Pyrosequencing was also used for characterization of the ptxS1 gene after validation of the technique (Application Note 210, Typing of bacteria: B. pertussis and parapertussis [Pyrosequencing AB, Uppsala, Sweden]).
(i) Pertactin.
Since variation in pertactin is essentially limited to region 1, only this region was sequenced for the majority of strains. Therefore, prn1 could not be identified separately from prn7, which has a single point mutation upstream of region 2.
The PCR conditions were according to the reference methodology with the following modifications: the prn forward primer AF was replaced by the primer 1PNU (5′-CGG CGC CAA TGT CAC GGT CCA-3′), and the reverse primer AR was replaced by 1PNL (5′-ATC GAC AGG GGC GCG GCT TGA-3′). The new primers overlap with the earlier ones.
(ii) Subunit 1 of pertussis toxin (ptxS1).
A total of 616 isolates were sequenced according to the reference methodology by dideoxy sequencing. For the remaining strains (n = 662), pyrosequencing was used. The two toxin primers for PCR, S1F2 and S1R2, were used as described earlier (18), together with the two primers used for sequencing, S1-MF and S1-MR.
RESULTS
PFGE analysis. (i) Number of PFGE profiles.
A total of 176 separate PFGE profiles obtained by using only XbaI digestion were identified in the Swedish B. pertussis collection. The profiles were designated BpSR1, BpSR2, BpSR3, etc., and for each profile one representative isolate was chosen as a reference strain. Thirty-five profiles were represented by more than seven isolates, i.e., 85% of all 1,810 isolates. In Table 2 the profiles are ordered by frequency.
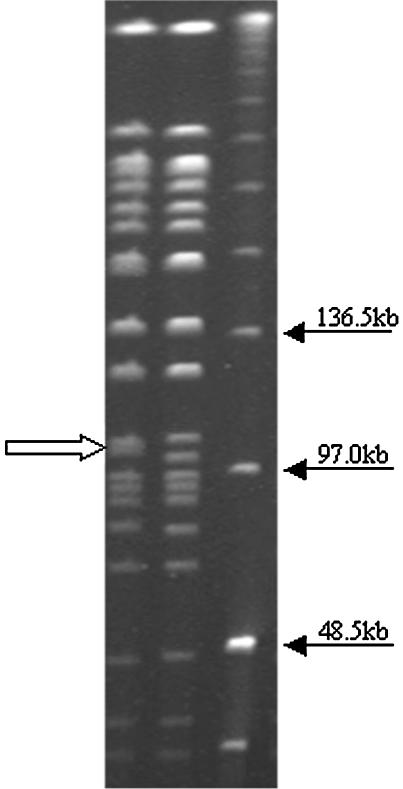
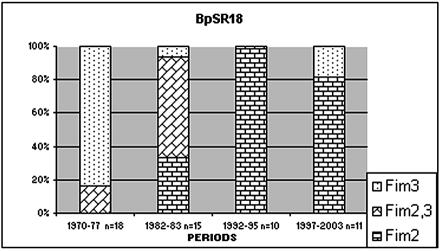
As described in Materials and Methods, profiles were defined as patterns differing by one or more bands; the profiles BpSR18 and BpSR16, for example, differed by only two fragment sizes (Fig. 2). Band 13 (108 kb) and band 14 (101 kb) in BpSR16 were located slightly closer to each when compared to band 13 and band 14 in BpSR18 (106 and 104kb). Phenotypically, however, the serotypes of these two closely matched profiles differed considerably. Ninety-seven percent of BpSR16 isolates (n = 114) expressed Fim2, but BpSR18 isolates (n = 54) expressed various fimbriae over a 30-year period (see Fig. 4).
FIG. 2.
PFGE gel. From left to right, profiles of BpSR16, BpSR18, and the low-range PFGE DNA marker (New England BioLabs). The only band difference between the two profiles is near the 97.0-kb region indicated by the white arrow.
FIG. 4.
Isolates with the BpSR18 profile express different fimbria serotypes during periods with different vaccination schedules.
(ii) Two enzymes versus one enzyme.
PFGE analysis based on profiles generated with two different enzymes is a laborious procedure. The most common profile during the acellular vaccine period, BpSR11, together with isolates representing the two profiles BpSR16 and BpSR18 (8 isolates of each), were selected to investigate whether profiles obtained by using SpeI instead of XbaI result in more discriminatory power. Sixteen isolates of BpSR11 chosen at random from a total of 455 BpSR11 profiles produced no different PFGE profiles.
For profiles BpSR16 and BpSR18, which showed small differences in profiles with the XbaI restriction enzyme, no difference could be shown after cleavage with SpeI (data not shown). In conclusion, the discriminatory power with one enzyme seemed to be adequate for surveillance and epidemiological purposes.
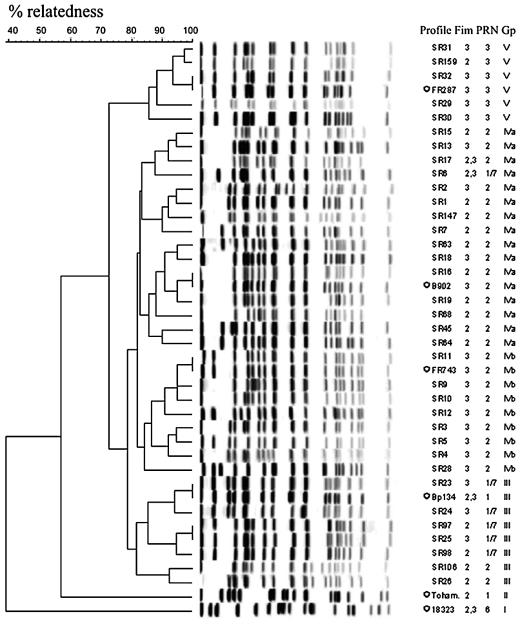
(iii) Dendrogram analysis of PFGE profiles.
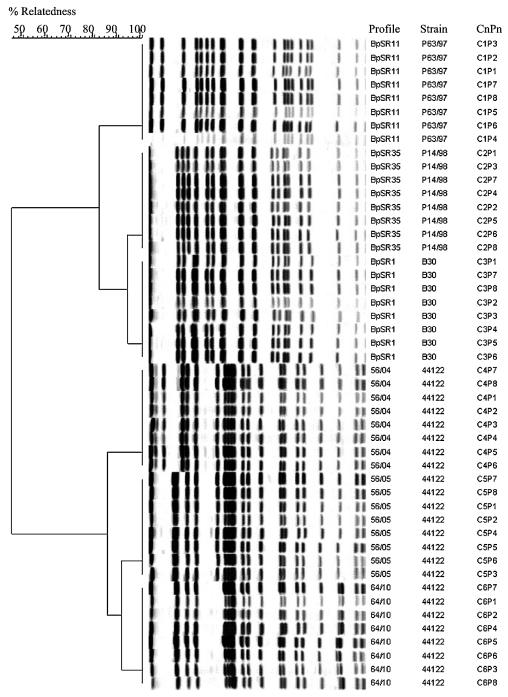
Dendrogram analysis performed on the 35 most prevalent Swedish profiles showed a minimum of 70% relatedness (Fig. 3).
FIG. 3.
Classification of the 35 prevalent Swedish profiles, including the six reference strains (indicated by asterisks). UPGMA with 1% band tolerance and 1% optimization settings was used as the clustering method. Only DNA profiles obtained after restriction with XbaI are shown. The serotype (Fim) and pertactin type (PRN) represent the most prevalent types within the profile. Gp, grouping system described by Weber et al. (29).
For comparison with the previously described grouping system (18, 29), the six reference strains representing six distinct groups were included in the dendrogram. The Swedish clinical isolates were organized in four clusters at a level of relatedness of 82%, corresponding to*FR287 (group V), *B902 (group IVα), *FR743 (group IVβ), and *Bp134 (group III) (Fig. 3). At this level there was also a close association of the cluster with prn. Increasing the cutoff slightly, however, made it possible to identify several more clusters; in particular, the B902 cluster IVα is heterogeneous.
The profile of the reference strain *FR743 is identical to the Swedish profile BpSR11. The reference strains *B902, *Bp134, and *FR287 have profiles identical to BpSR18, BpSR23, and BpSR32, respectively. The two clusters corresponding to Tohama I (group II or III) and strain 18323 (group I) had a much lower similarity, with relatednesses of 56 and 40%, respectively, and were not represented at all among the 176 profiles representing Swedish clinical isolates.
(iv) Reference strains A639 and A556.
The PFGE profile for the A639 isolate of U.S. origin, which was proposed to be a global reference, was not identical to that of any of the Swedish isolates but was very close to BpSR6 (Fig. 3), a profile that was prevalent during the 1970s (data not shown). The A556 profile was identical to BpSR5, the second most prevalent profile after 1997 (data not shown).
Other characteristics relating to PFGE profiles or clusters (Table 2). (i) Serotype.
The 1,810 isolates expressed Fim3, Fim2, or both Fim2 and Fim3. There was not a 100% relationship between profile and serotype. The association was between 80 and 100% in 27 of the 35 profiles (Table 2). Fewer than 1% of the isolates (n = 16) were not typeable. Interestingly, a stable PFGE profile of BpSR18 (n = 54) in the *B902 cluster was shown to shift serotype over the years (Fig. 4). The last isolate was collected in 1999. During the whole-cell vaccine period, the serotype was mainly Fim3 and to a lesser extent Fim2,3. Isolates from the vaccine trial from 1992 to 1995 they expressed Fim2. After reintroduction of vaccination, Fim3 reappeared.
(ii) Pertactin.
A total of 1,810 isolates were genotyped to determine the prn type. Three major types were detected among the isolates, prn1 (or prn7), prn2, and prn3. For 24 isolates the part discriminating for prn7 was also sequenced, but all remained prn1. The relationship between pertactin and profile was better than that between serotype and profile, with 34 of the 35 profiles showing a correlation of 96% to 100% between prn and profile (Table 2).
Four unusual prn types were identified in addition to the three common types: one isolate with prn4 in BpSR26; two isolates with prn9 (showing seven repeats in region 1) represented in BpSR16 and BpSR69; one isolate with prn11 (with five repeats with point mutations) in BpSR25; and finally one isolate with a prn type with only three repeats, which may be a new prn type and is not represented among the 35 most common profiles.
(iii) Toxin.
Toxin typing was restricted to 1,278 isolates. Toxin type ptxS1A has been by far the most prevalent subtype in all the sample materials over all periods studied. Toxin type B was found in 11 isolates, representing fewer than 1% of the isolates, all from the surveillance period. They represented six different profiles, i.e., BpSR87 (n = 3), BpSR111 (n = 3), BpSR94 (n = 2), and BpSR91, BpSR1, and BpSR26 (n = 1 each). Only BpSR1 and BpSR26 were ranked in the 35 most common profiles.
Relationship between the 35 most prevalent PFGE profiles and serotype or prn type (Table 2 and Fig. 3).
PFGE profiles within the four main clusters referring to the reference strains were correlated to a specific pertactin type (prn3, prn2, prn2, and prn1, respectively) (Fig. 3). The first three clusters are also closely associated with a specific serotype (Fim3, Fim2, and Fim3). The one with prn1 showed various serotype combinations (Fig. 3).
In conclusion, the cluster (Fig. 3) relating to *FR287 (group V) was characterized by a combination of Fim3 and prn3 (67% [96 of 144]). The next cluster, relating to*B902 (group IVα), was characterized by Fim2 and prn2 (87% [533 of 610]). In this cluster the most prevalent profile was BpSR1, with Fim2 in 94% and prn2 in 99% of the isolates. BpSR19 (n = 38), and BpSR45 (n = 10) showed this same combination in 100% of the isolates. On the other hand, the second most frequent profile during the Pw period, BpSR18 (Fim3/prn2), was also seen in this cluster.
The third cluster, identified by *Fr743 (IVβ), included isolates mainly with Fim3 and prn2 combinations (99% [662 of 671]). Here the most prevalent PFGE type was BpSR11 (n = 455) showing the same PFGE profile as the reference strain in 100% of the isolates. There was also a 100% association between serotype and prn type for BpSR5 (n = 64), BpSR9 (n = 25), and BpSR3 (n = 18).
In the fourth cluster, which was associated with *Bp134 (group III), prn1 variants were predominant (57%), in combination with various serotypes, most often Fim3 (73%). BpSR25 was the most prevalent profile in this group, which was recovered mainly in whole-cell vaccine period.
Stability and reproducibility of PFGE types. (i) Stability of profiles in long-term laboratory work.
PFGE of the strain used for production of the Swedish whole-cell vaccine, covering the 8-year period from 1956 to 1964, showed heterogeneity of profiles. At least four different profiles could be documented (Fig. 5); none of them were found in the environment. The profiles shifted over time, and different patterns could also be found within the same vial. This strain also was adapted to grow on conventional blood agar, as was 18323, both showing ptxS1E. In conclusion, reanalysis as part of quality assurance is advisable after numerous subcultures.
FIG. 5.
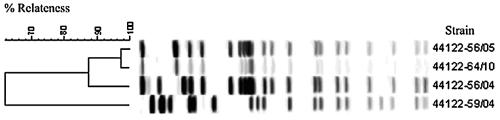
Strain 44122 was used for production of the Swedish whole-cell vaccine from the 1950s to 1978. PFGE of several clones from 11 different vials containing control material for vaccine production during the 8-year period from 1956 to 1964 showed heterogeneity of profiles. At least four different profiles could be documented (the profiles are identified by suffixes indicating year/month).
(ii) Stability of profiles in short-term laboratory work.
A test was designed to evaluate the stability and reproducibility of the generated profiles. Three clinical strains, P63/97 (C1) (profile BpSR11), P14/98 (C2) (profile BpSR35), and B30 (C3) (profile BpSR1) were chosen, together with the whole-cell vaccine strain 44122. The profiles BpSR11 and BpSR1 were the most prevalent, and in contrast, BpSR35 has been found only once so far.
For the vaccine strain, three of the four clones with different PFGE profiles were chosen (designated with a suffix showing year/month), i.e., 44122-56/04 (C4), 44122-56/05 (C5), and 44122-64/10 (C6). These clones were recovered from 11 vials of harvest control material from the period from 1956 to 1964. The selection of the vaccine strain variants was based on the hypothesis that heterogeneity in this strain could be a result of instability.
Together the six strains were serially subcultured three times a week for 3 weeks giving eight subcultures in all for each strain (P1 to P8). Material from each plate was stored frozen and used for each successive subculture. The 48 isolates were subjected to PFGE, serotyping, and prn gene typing.
The results of PFGE analysis, serotyping, and prn typing showed that all of the profiles were fully reproducible and consistent (Fig. 6). One exception is that P14/98 switched from Fim2 to Fim3 in the eighth subculture. It is worth noting that the three vaccine profiles were stable by themselves and did not differentiate into new profiles after subculture.
FIG. 6.
Dendrogram showing six strains (C1 to C6) subcultured for eight passages (P1 to P8), with Cn and Pn indicating the strain and passage number, respectively. No change in the pulsed-field profile could be identified. UPGMA with 1% band tolerance and 1% optimization settings was used as the clustering method. Only DNA profiles obtained after restriction with XbaI are shown.
The 35 Swedish reference strains deposited at CCUG were reanalyzed after lyophilization. To verify the reproducibility, PFGE was performed on the lyophilized vials received back from CCUG. All 35 PFGE profiles were found to be 100% identical to the reference material which had been sent to the culture collection bank.
Implementation of the PFGE profile system. (i) Prevalence of PFGE profiles over time.
When the most prevalent profiles among those in Table 2 were analyzed over time, it was shown that BpSR25 and BpSR18 occurred most frequently (27 of 70 [39%] and 18 of 70 [26%], respectively) during the whole-cell vaccine period. BpSR1 replaced these profiles during the vaccine-free period (80 of 222 [36%]). After reintroduction of general vaccination, BpSR11 has been the most frequent profile (455 of 1,518 [30%]). Some profiles were seen mainly as a peak during a limited period of time (e.g., BpSR31, showing prn3 during 1998 to 1999).
(ii) Outbreaks.
The profile-oriented approach can be used for documentation of international and national surveillance programs and for follow-up of minor local outbreaks. The major profiles were identified from isolates coming from all over Sweden. On the other hand, a less common profile, BpSR4 (Table 2), with eight isolates could be traced to Torsås only, a small county in the Kalmar region in southeast Sweden. All eight isolates were collected over a span of 6 weeks and then disappeared.
(iii) PFGE profiles for quality assurance.
PFGE of the strain used for production of the Swedish whole-cell vaccine covering the 8-year period from 1956 to 1964 showed heterogeneity of profiles. At least four different profiles were documented. The profiles shifted over time, and different patterns were also found within the same vial. This strain also was adapted to grow on conventional blood agar, as was 18323, both showing ptxS1E.
DISCUSSION
Chromosomal fingerprinting by means of PFGE is to date the most discriminative technique in use to reveal changes in the circulating population of B. pertussis. This method has been frequently used worldwide to provide laboratory data for characterization and tracing of B. pertussis isolates (4, 5, 7, 14, 24, 29). We have been able to successfully use PFGE as a tool to monitor the bacterial population in vaccine surveillance. PFGE profiles can also be related to clinical outcome. Isolates specifically associated with mortality, hospitalization, or persistent cough may be selected for further examination.
So far, however, it has been difficult to directly compare PFGE results between laboratories due to crucial technical differences. For comparability of results between laboratories, it is essential to identify key technical factors. The choice of endonucleases is one such factor. Mooi et al. recommended a method based on profiles obtained from each of two different enzymes (XbaI and SpeI) under high-resolution electrophoresis conditions (18). Beall et al., (3) studied three endonucleases (XbaI, DraI, and SpeI) and found that XbaI gave satisfactory results when pertussis epidemics were analyzed. The other two enzymes might give further resolution of slight differences but were not determined to be of general need. Extensive studies have also shown satisfactory discriminatory power for patterns generated with one restriction enzyme only (4, 5, 7, 15, 22). For analysis of the B. pertussis isolates collected at the Swedish Institute for Infectious Disease Control, we used only the XbaI restriction enzyme to obtain PFGE profiles. Generating PFGE profiles with one enzyme is cost efficient and provides adequate discrimination for epidemiological purposes.
Other technical factors influencing resolution capacity are electrophoresis conditions, e.g., migration period, pulse times, and voltage conditions (see Materials and Methods). The electrophoresis conditions described by Mooi et al. (18) generate good discriminatory power over the entire size range and also in the 45- to 97-kb size range. The control strain T41 was very useful for quality control of intragel results in these ranges.
A standardized procedure, however, is not sufficient for interlaboratory comparability. Reference material is also needed for full traceability. Therefore, representative isolates for the 35 most frequent profiles were deposited in the CCUG (www.ccug.gu.se). The collection can readily be extended with new interesting and significant isolates. It was shown that the profile-based system presented in this work was also applicable to the majority of isolates in a European culture collection (www.eupertstrain.org).
Stability is important for reproducibility. A long-term profile change was shown for the Swedish vaccine strain used for Pw production, indicating a need for reanalysis after numerous subcultures. On the other hand, the high natural frequencies of BpSR11 and BpSR1, the most prevalent profiles in Sweden, support the evidence for profile stability. Also, in a shorter term the profiles were quite stable. In our experiments, no changes of the profiles themselves were seen after subculture or lyophilization.
In the literature, small variations in PFGE patterns have been reported to occur in certain isolates of B. pertussis when they were cultured repeatedly or over a long time period (3, 25). PFGE profiles of closely related isolates may differ by changes consistent with a single genetic event, i.e., a point mutation or an insertion or deletion of DNA. Such changes typically result in two or three band differences (25). Chromosomal rearrangements may also contribute to genome variability (23). This is in agreement with the findings of Beall et al. (3). On the other hand, those authors showed that differences in PFGE profiles for single epidemic strains occurred occasionally upon repeated passages for 1.5 months.
The correlation between profile and prn type is very stable. In most cases, one profile correlates to one prn type, although there are also exceptions. This correlation between PFGE profile and prn genotype is of help in cluster analysis, as also shown by Weber et al. (29).
With this approach, four clusters were identified at 82% relatedness. These four clusters corresponded to the prn genotype and relate to the classification obtained with two enzymes as proposed by Weber et al. (29). In that system, the main Swedish clusters corresponded to groups V, IVα, IVβ, and III. In the Swedish material, groups II and I were not found. They were more related to noncirculating vaccine and reference strains. The reference strain A639 proposed by Hardwick et al. (15) did not represent any of the 35 Swedish isolates.
It should also be mentioned that the description of the B. pertussis groups (29) based on PFGE profiles is dependent on the choice of cutoff and that a rather small increase resulted in new clusters.
Considering the high association between every profile and the prn genotype, it is more likely that these clusters are true and that the profiles included in each cluster are genetically related. The use of one or two enzymes does not seem to be crucial for grouping of isolates, also indicating a strong link to the previously described reference system. Profile identification based on cleavage with one enzyme is more discriminatory than grouping alone, as it provides more adequate data to analyze the relationship between isolates. It is also more preferable from a practical point of view.
The situation for serotype may be different, as the bacteria are known to harbor genetic information for both Fim2 and Fim3. The profile and serotype combination was, however, most often shown to be stable, although the serotype was shown to shift over longer periods of time for some profiles e.g., BpSR18.
Other systems for genotyping have also been used for subtyping of B. pertussis. Multilocus sequence typing (MLST) is sequence based and therefore has the advantage of direct traceability and association to protein polymorphism. B. pertussis is a very homogeneous species, and consequently, finding polymorphic sites for MLST has proved to be difficult. In all, nine MLSTs types could be distinguished (27). The discriminatory capacity of PFGE was said to be greater than that of other techniques.
In conclusion, this report favors a focus on PFGE profile analysis based on cleavage with a single enzyme, i.e., XbaI, for basic traceability of new variants in surveillance studies and outbreak-associated transmission studies and was shown to have a potential usefulness as an instrument of quality control e.g., for the production of vaccines. It also offers a rational basis to select single isolates for extended analysis. There is a high correlation between PFGE profiles and different prn types. There is also a correlation between PFGE profiles and serotype, but this relationship seems to be temporary and shifts between periods. Some profiles are more dominant than others and may express different phenotypes in different periods. Some of the Swedish profiles were identified from samples collected over a period of 30 years; others were collected during local short outbreaks. Grouping of profiles may be rational for analysis of strain lineages.
Acknowledgments
This work was supported by GlaxoSmithKline and Aventis Pasteur and by the European Commission, contract no. QLK2-CT-2001-01819, Eupertstrain.
We thank all Swedish laboratories in clinical bacteriology for their generous contributions of B. pertussis isolates.
REFERENCES
- 1.Ad Hoc Group for the Study of Pertussis Vaccines. 1988. Placebo-controlled trial of two acellular vaccines in Sweden: protective efficacy and adverse events. Lancet i:955-960. [PubMed] [Google Scholar]
- 2.Andrews, R., A. Herceg, and C. Roberts. 1997. Pertussis notifications in Australia, 1991 to 1997. Commun. Dis. Intell. 21:145-148. [DOI] [PubMed] [Google Scholar]
- 3.Beall, B., P. K. Cassiday, and G. N. Sanden. 1995. Analysis of Bordetella pertussis isolates from an epidemic by pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:3083-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisgard, K. M., C. D. C. Christie, S. F. Reising, G. N. Sanden, P. K. Cassiday, C. Gomersall, W. A. Wattigney, N. E. Roberts, and P. M. Strebel. 2001. Molecular epidemiology of B. pertussis by pulsed-field gel electrophoresis profile: Cincinnati, 1989-1996. J. Infect. Dis. 183:1360-1367. [DOI] [PubMed] [Google Scholar]
- 5.Cassiday, P., G. Sanden, K. Heuvelman, F. Mooi, K. M. Bisgard, and T. Popovic. 2000. Polymorphism in B. pertussis pertactin and pertussis toxin virulence factors in the United States, 1935-1999. J. Infect. Dis. 182:1402-1408. [DOI] [PubMed] [Google Scholar]
- 6.de Melker, H. E., M. A. E. Conyn-van Spaendonck, H. C. Rümke, J. K. van Wijngaarden, F. R. Mooi, and J. F. P. Schellekens. 1997. Pertussis in The Netherlands: an outbreak despite high levels of immunization with whole-cell vaccine. Emerg. Infect. Dis. 3:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Moissac, Y. R., S. L. Ronald, and M. S. Peppler. 1994. Use of pulsed-field gel electrophoresis for epidemiological study of Bordetella pertussis in a whooping cough outbreak. J. Clin. Microbiol. 32:398-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Corbeira, P., R. Dal-Re, L. Aguilar, and J. Garcia-de-Lomas. 2000. Seroepidemiology of B. pertussis infections in the Spanish population: a cross-sectional study. Vaccine 21:2173-2176. [DOI] [PubMed] [Google Scholar]
- 9.Guiso, N., C. H. Wirsing von König, C. Becker, and H. Hallander. 2001. Fimbrial typing of Bordetella pertussis isolates: agglutination with polyclonal and monoclonal antisera. J. Clin. Microbiol. 39:1684-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guris, D., P. M. Strebel, B. Bardenheier, M. Brennan, R. Tachdjian, E. Finch, M. Wharton, and J. R. Livengood. 1999. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990-1996. Clin. Infect. Dis. 28:1230-1237. [DOI] [PubMed] [Google Scholar]
- 11.Gustafsson, L., H. O. Hallander, P. Olin, E. Reizenstein, and J. Storsaeter. 1996. A controlled trial of a two-component acellular, a five-component acellular, and a whole-cell pertussis vaccine. N. Engl. J. Med. 334:349-355. [DOI] [PubMed] [Google Scholar]
- 12.Gzyl, A., E. Augustynowicz, I. van Loo, and J. Slusarczyk. 2001. Temporal nucleotide changes in pertactin and pertussis toxin genes in B. pertussis strains isolated from clinical cases in Poland. Vaccine 20:299-303. [DOI] [PubMed] [Google Scholar]
- 13.Hahm, B. K., Y. Maldonado, E. Schreiber, A. K. Bhunia, and C. H. Nakatsu. 2003. Subtyping of foodborne and environmental isolates of Escherichia coli by multiplex-PCR, rep-PCR, PFGE, ribotyping and AFLP. J. Microbiol. Methods 53:387-399. [DOI] [PubMed] [Google Scholar]
- 14.Hardwick, T. H., P. K. Cassiday, R. S. Weyant, K. M. Bisgard, and G. N. Sanden. 2002. Changes in predominance and diversity of genomic subtypes of B. pertussis isolated in the United States, 1935 to 1999. Emerg. Infect. Dis. 8:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwick, T. H., B. Plikaytis, P. K. Cassiday, G. Cage, M. S. Peppler, D. Shea, D. Boxrud, and G. N. Sanden. 2002. Reproducibility of Bordetella pertussis genomic DNA fragments generated by XbaI restriction and resolved by pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juretzko, P., R. von Kries, M. Hermann, C. H. Wirsing von König, J. Weil, and G. Giani. 2002. Effectiveness of acellular pertussis vaccine assessed by hospital-based active surveillance in Germany. Clin. Infect. Dis. 35:162-167. [DOI] [PubMed] [Google Scholar]
- 17.Khattak, M. N., and R. C. Matthews. 1993. A comparison of the DNA fragment patterns of the mouse-virulent challenge strains and clinical isolates of B. pertussis. J. Infect. 27:119-124. [DOI] [PubMed] [Google Scholar]
- 18.Mooi, F. R., H. Hallander, C. H. Wirsing von König, B. Hoet, and N. Guiso. 2000. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur. J. Clin. Microbiol. Infect. Dis. 19:174-181. [DOI] [PubMed] [Google Scholar]
- 19.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. J. van der Heide, W. Gaastra, and R. J. L. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olin, P., L. Gustafsson, L. Barreto, L. Hessel, T. C. Mast, A. van Rie, H. Bogaerts, and J. Storsaeter. 2003. Declining pertussis incidence in Sweden following the introduction of acellular pertussis vaccine. Vaccine 21:2015-2021. [DOI] [PubMed] [Google Scholar]
- 21.Parkhill, J., et al. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 22.Peppler, M. S., S. Kuny, A. Nevesinjac, C. Rogers, Y. R. de Moissac, K. Knowles, M. Lorange, G. de Serres, and J. Talbot. 2003. Strain variation among Bordetella pertussis isolates from Quebec and Alberta provinces of Canada from 1985 to 1994. J. Clin. Microbiol. 41:3344-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stibitz, S., and M.-S. Yang. 1999. Genomic plasticity in natural populations of Bordetella pertussis. J. Bacteriol. 181:5512-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syedabubakar, S. N., R. S. Matthews, N. W. Preston, D. Owen, and V. Hillier. 1995. Application of pulsed field gel electrophoresis to the 1993 epidemic of whooping cough in the UK. Epidemiol. Infect. 115:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiru, M., P. Askelöf, M. Granström, and H. Hallander. 1997. Bordetella pertussis serotype of clinical isolates in Sweden during 1970-1995 and influence of vaccine efficacy studies. Dev. Biol. Stand. 89:239-245. [PubMed] [Google Scholar]
- 27.van Loo, I. H. M., J. Kees, A. Heuvelman, J. King, and F. R. Mooi. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J. Clin. Microbiol. 40:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Von König, C. H. W., S. Halperin, M. Riffelmann, and N. Guiso. 2002. Pertussis of adults and infants. Lancet Infect. Dis. 2:544-750. [DOI] [PubMed] [Google Scholar]
- 29.Weber, C., C. Boursaux-Eude, G. Coralie, V. Caro, and N. Guiso. 2001. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J. Clin. Microbiol. 39:4396-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]