Abstract
To investigate whether distinct populations have differing human immunodeficiency virus type 1 (HIV) neutralizing antibody responses, we compared 20 women from Tanzania's HIV Superinfection Study (HISIS) cohort, who were infected multiple HIV subtypes, and 22 women from the Centre for the AIDS Programme of Research in South Africa (CAPRISA) cohort, who were infected exclusively with HIV subtype C. By 2 years after infection, 35% of HISIS subjects developed neutralization breadth, compared with 9% of CAPRISA subjects (P = .0131). Cumulative viral loads between 3 and 12 months were higher in the HISIS group (P = .046) and strongly associated with breadth (P < .0001). While viral load was the strongest predictor, other factors may play a role, as the odds of developing breadth remained higher in HISIS even after correction for viral load.
Keywords: broadly neutralizing antibodies, Africa, cohorts, HIV genetic subtypes, viral load
Broadly neutralizing antibody (bNAb) responses are important for vaccines. Although most infected people develop some degree of neutralization cross-reactivity [1], approximately 10%–30% of individuals develop Abs that are very broadly neutralizing. These usually develop after 2–4 years and are associated with higher early viral loads [2]. Different populations may vary in their bNAb responses, owing to differences in host genetics, infecting subtype, and disease progression profiles. Subtype has been implicated in the potency and breadth of bNAbs, but most studies did not control for viral load and duration of infection [3]. Other viral factors influencing the development of breadth include early viral diversity, shorter variable loops, differences in glycosylation motifs, and, according to some studies, dual infection [4, 5]. The common targets between subtypes of human immunodeficiency virus (HIV)-specific bNAbs have been mapped to 5 sites on the envelope, the CD4 binding site, the V1V2 and V3/C3 regions of gp120, the membrane proximal external region in gp41, and the gp41-gp120 interface. However, some studies have shown that there may be subtype-specific epitope variants [3]. It remains unclear to what extent intersubtype recombinants impact the development of the bNAb response in infection.
To understand the evolution of bNAb responses in different populations in Africa, we compared the kinetics and breadth of neutralization responses in 2 cohorts: one in Tanzania (the HIV Superinfection Study [HISIS]), in which participants were infected with multiple HIV subtypes or recombinant forms [6]; and one in South Africa (the Centre for the AIDS Programme of Research in South Africa [CAPRISA]), in which all participants were exclusively infected with HIV subtype C [2]. This study showed that participants in the Tanzanian cohort had higher viral loads and significantly more bNAb responses than the South African cohort at 2 years after infection.
MATERIALS AND METHODS
Ethics Statement
Ethics approval was obtained from the Tanzania Ministry of Health, Ludwig Maximilians University (Munich, Germany), and the University of Cape Town, for HISIS; and the universities of KwaZulu-Natal (Cape Town) and Witwatersrand, for CAPRISA. All participants provided written informed consent.
Participants
For the HISIS cohort, high-risk, HIV-seronegative women from Mbeya, Tanzania, were recruited and followed up quarterly for 2 years [7]. The first 20 seropositive participants were included in this study.
For the CAPRISA cohort, high-risk, HIV-negative women from KwaZulu-Natal were monitored for recent HIV infection, as described previously [8], and the first 22 women infected in the CAPRISA 002 study were included in this study. Women were monitored at least monthly for the first year and quarterly thereafter.
Plasma HIV RNA loads in both cohorts were measured in a quantitation range of ≥400 to ≤750 000 virus copies/mm3, using the Amplicor HIV Monitor Test, version 1.5 (Roche, Basel, Switzerland). All participants were antiretroviral (ART) naive.
Estimation of Duration of Infection
The duration of Fiebig stage I/II infection was estimated as 14 days and that of Fiebig stages III/IV was estimated as 30 days. For participants beyond Fiebig stage IV infection, duration of infection was estimated as the midpoint between the last seronegative and first seropositive visits, or 45 days preceding the first seropositive visit.
Reverse Transcription Polymerase Chain Reaction Amplification and Sequencing
RNA was extracted and reverse transcribed as described elsewhere [9]. Single-genome amplification or end-point dilution was performed on complementary DNA derived from a plasma specimen obtained during the first seropositive visit, for HISIS subjects, and from the first visit of HIV detection in a plasma or serum specimen, for CAPRISA subjects, and amplicons were directly sequenced [9]. Sequence accession numbers are available in the supplementary material. Pairwise DNA distances and maximum likelihood trees were computed using Mega 4 software.
Characterization of Infecting Virus
Infecting virus sequences were subtyped using the REGA subtyping tool (http://bioafrica.mrc.ac.za/rega-genotype/html/subtypinghiv.html).
Panel Viral Isolates
The panel of molecularly cloned full-length env genes for HIV-1 Env pseudovirus production consisted of 4 representative clones from the standard panel of HIV strains for subtype A (Q168ENVa2, Q461ENVe2, Q842ENVd12, and Q23ENV17), subtype B (AC10.0.29, CAAN5342.A2, QH0692.42, and PVO.4), and subtype C (Du156.12, Du422.1, ZM214M.PL15, and ZM109F.PB4), obtained from the National Institutes of Health AIDS Research and Reference Reagent Program.
Cloning of Envelope Genes and Neutralization Assays
Functional env clones representing transmitted founder (T/F) virus were generated for 16 HISIS participants and inserted into mammalian expression vectors. HEK293T and TZM-bl cell lines were prepared, and Env-pseudotyped viruses were generated. Neutralization was measured and calculated as the median infectious dose (ID50) [2].
The following consensus C mutants were used for mapping HISIS_605 neutralization activity: N160A and K169E, for V2; I307A, H330Y, and N332A, for V3; K360V, E362N, L369P, T372V/T373M, and S375M, for CD4 binding site; T408A and T415I, for V4; R416A, for C4; and F468V, for V5.
Calculation of Breadth and Potency Scores and Statistical Analyses
Breadth and potency scores were calculated for each plasma sample according to the method of Blish et al [10]. Wilcoxon rank sum and Mann–Whitney U tests were performed using Graphpad Prism (GraphPad Software, La Jolla, California). Linear mixed model analyses fitted to viral load and corrections for repeated measures were performed using SAS, version 9.3 (SAS Institute, Cary, North Carolina).
RESULTS
Characteristics of the 2 Cohorts and Their Infecting Viruses
All participants in CAPRISA and HISIS were high-risk females who acquired HIV heterosexually and were recruited within a mean of 34 days from the estimated time of infection (range, 14–45 days), followed for 2 years (range, 24–27 months), and remained ART naive. Infections reflected the HIV diversity of their respective local epidemics, with HISIS participants infected with subtypes A (4 participants), C (8), D (1), and recombinant viruses (7 [2 with AC, 2 with ACD, 1 with AD, and 2 with CD]) [6], while all CAPRISA participants had subtype C infections [8] (Figure 1A and 1B).
Figure 1.
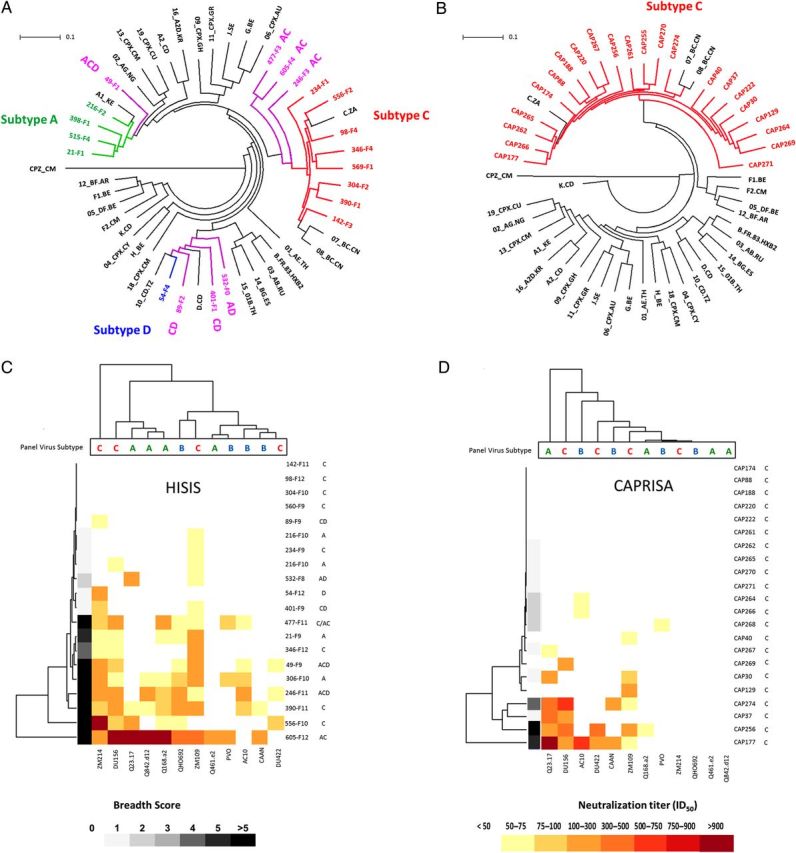
A and B, Circular maximum likelihood trees showing consensus-derived infecting viral sequences from the HISIS cohort (A) and the CAPRISA cohort (B). Thirty reference sequences, including 18 circulating recombinant form sequences and a SIVcpz outgroup are included (black). Branch tips are color coded as follows: subtype A, green; subtype B, blue; subtype C, red; and recombinants, purple. C and D, Hierarchical clustering of neutralization titers (displayed as the median infectious dose [ID50]) for plasma specimens from 20 HISIS donors (C) and 22 CAPRISA donors (D) and 12 panel viruses. The dendrograms on the left cluster plasma samples according to their neutralization capacity, while the dendrograms at the top cluster the panel viruses according to their neutralization susceptibilities. Donor plasma names and the subtype of their infecting viruses are at the right hand margins of panels C and D. Abbreviations: CAPRISA, Centre for the AIDS Programme of Research in South Africa; HISIS, HIV Superinfection Study.
HISIS Participants Had Greater Neutralization Breadth
Neutralization assays were conducted using a 12-pseudovirus panel consisting of subtypes A, B, and C (Supplementary Figure 1), and each plasma sample was assigned breadth and potency scores. Scores were calculated on the basis of overall neutralization titers and panel virus susceptibility. Two years after infection, there was significantly higher breadth among the Tanzanian participants, of whom 35% (7 of 20) developed bNAbs (defined as Abs that could neutralize >50% of the panel viruses at titers of over 1:50 [ID50 >50]), compared with only 9% (2 of 22) in the CAPRISA cohort (P = .0139; Figure 1C and 1D and Supplementary Figure 1). Accordingly, the HISIS cohort had a significantly higher mean breadth score (3.35) than the CAPRISA participants (0.95; P = .0131). Potency scores did not differ between the 2 cohorts. HISIS participants had higher mean breadth scores against subtype C panel viruses (1.65), followed by subtype B (0.9), and subtype A (0.8) (Supplementary Figure 1 and Figure 1C and 1D). No such trend was observed in CAPRISA participants.
Early Viral Loads Were Predictive of Breadth at 2 Years and Were Higher in the HISIS Cohort
We found a strong association with breadth at 2 years and plasma viral load area under the curve (AUC) between 3 and 12 months (P < .0001; r2 = 0.4747). Furthermore, the median plasma viral load AUC for individuals in HISIS was significantly higher than for those in CAPRISA (P = .046, by the Mann–Whitney U test; Supplementary Figure 2A). To determine whether there was an effect independent of viral load, a multinomial regression model was fit to breadth as a 3-level categorical outcome (no breadth [breadth score, 0]; moderate breath [breadth score, 1–4]; and broadly neutralizing (breadth score, ≥5]) with an adjustment for the influence of plasma viral load AUC. We found that the odds of developing bNAb responses among HISIS participants was 5.88 times that among CAPRISA participants (P = .0644). The odds remained higher after correcting for plasma viral load AUC (odds ratio, 3.1), although this was no longer statistically significant (P = .2920). Nevertheless, these data are consistent with contributions from other factors to the development of greater breadth in the HISIS group.
Viral Characteristics and Development of Neutralization Breadth
There was a high incidence of subtype C infection (40%) and intersubtype recombinant virus infection (35%) in HISIS participants (Supplementary Figure 1). However, neither finding was associated with significant differences in mean breadth scores both before and after accounting for viral load. We were thus unable to establish subtype as a major factor in the development of breadth in the HISIS cohort.
High diversity in early infection and superinfection with a second strain following seroconversion has been associated with neutralization breadth [4, 5]. In HISIS, 7 of 20 subjects (35%) were reported to be dually infected (incorporating both coinfection and superinfection) [6, 11], compared with 3 of 22 subjects (14%) in the CAPRISA group [12] (Supplementary Figure 1). However, we found no significant association between dual infection and the development of breadth in either cohort or in a combined analysis, using a Wilcoxon rank sum test and a multinomial regression model.
A comparison of variable loops between the cohorts found no significant difference in V1-V5 length (Supplementary Figure 2B). We also found no significant association between the loop length of the infecting virus and the development of breadth.
Development of Autologous NAb Responses
We compared the kinetics of autologous NAb responses in the HISIS cohort with the kinetics in the CAPRISA cohort (Figure 2). In the HISIS and CAPRISA cohorts, 14 of 16 participants (88%) and 12 of 14 participants (86%), respectively, developed responses with ID50 titers greater than 1:100 at 20 weeks after infection. At 12 months, 4 HISIS participants had titers greater than 1:10 000 (Figure 2A). No CAPRISA participant reached titers of 10 000 (Figure 2B). Median autologous titers at 12 months were significantly higher in the HISIS cohort, compared with the CAPRISA cohort (P = .0327, by the Wilcoxon signed rank test), but there was no significant correlation between median titer and breadth within each cohort or in a combined analysis.
Figure 2.
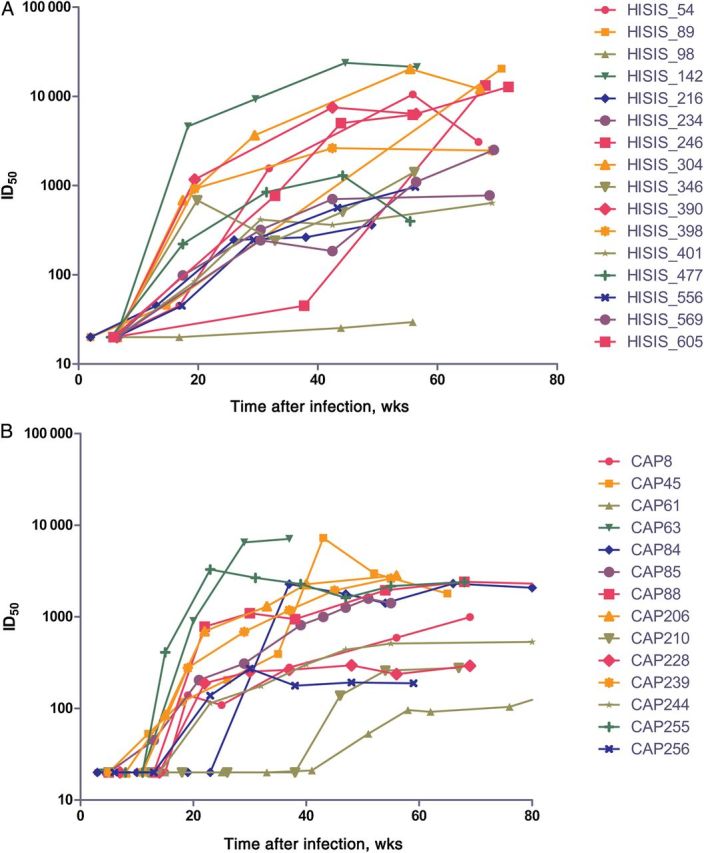
The development of autologous neutralizing antibody responses in the HISIS cohort (A) and the CAPRISA cohort (B). A functional envelope clone representing the transmitted/founder or an early virus for each of 16 HISIS and 14 CAPRISA participants was tested against longitudinal samples of autologous plasma in an Env-pseudotyped virus neutralization assay. Results are shown as neutralization titers (displayed as the median infectious dose [ID50]). Abbreviations: CAPRISA, Centre for the AIDS Programme of Research in South Africa; HISIS, HIV Superinfection Study.
Development of NAb Breadth in HISIS_605
Plasma specimens from one participant in the HISIS cohort, HISIS_605, who was infected with an A/C recombinant, neutralized 83% of the virus panel and fulfilled the definition of an elite neutralizer. Breadth developed rapidly, and her plasma neutralized 72% of the panel viruses 15 months after infection (Supplementary Figure 3A).
To identify the Ab specificity, plasma was screened against a panel of mutants introduced into a consensus C backbone to determine their ability to abrogate recognition by bNAbs. Mutations in the V1V2 and V3/C3 glycan epitopes did not affect neutralization titers. The CD4 binding loop spans 364–374, and mutations in or near this region (K360V, E362N, L369P, and S375M) reduced neutralization titers (<75% reduction in titer; Supplementary Figure 3B). Only position 369 has been shown to be a contact site for CD4 mAbs (CH103 and b12) [13], and we confirmed that the L369P mutation affected neutralization by VRC01. However, because of a lack of sample availability, we were unable to confirm whether this individual's potent responses targeted the CD4-bs.
DISCUSSION
We report different frequencies of bNAb responses in 2 female cohorts in Africa, with the HISIS participants from Tanzania having more bNAb responses at 2 years after infection, compared with the South African CAPRISA participants. More-potent autologous NAb responses were also found in the HISIS cohort at 12 months. Cumulative viral loads between 3 and 12 months were strongly predictive of breadth at 2 years and were significantly higher in HISIS participants.
Our study suggests that other factors may play a role, since the odds of developing breadth remained higher in the HISIS group even after correction for viral load. The role of subtype, dual infection, and other virological characteristics could not be established, because of the limited size of the study. Although subtype has been implicated in differences in viral load set point [14], other factors, including tropical coinfections such as malaria [15], which is absent in South Africa, may contribute to these differences in viral load. Other unknown environmental or genetic factors, such as the host genetic variation observed within Africa, may also influence levels of viral replication in different populations.
One HISIS individual (HISIS_605) developed bNAbs within 1 year of infection and was an elite neutralizer by 2 years after infection. This is rare [2] and was not observed in CAPRISA. Preliminary mapping data suggest that bNAbs produced by HISIS_605 may target the CD4 binding site. Further studies of this individual could help us understand the pathway to developing bNAbs more rapidly and inform vaccine design.
In conclusion, the CAPRISA and HISIS longitudinal cohorts, which recruited women recently infected with HIV, enabled us to directly compare the development of breadth in NAb responses in populations with different local epidemics. Viral load was the major factor associated with these differences, and further work is needed to understand whether the presence of coinfections influenced viral load and the development of bNAb responses. This and other cofactors, including virological characteristics, need to be considered because they may have implications for the continued efforts to understand how to elicit these responses through vaccination.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all of the study participants and the clinical and support staff of HIV Superinfection Study (HISIS) and Centre for the AIDS Programme of Research in South Africa (CAPRISA), for their participation and dedication; G. Shaw, for providing the HEK293 T cells; and the AIDS Research and Reference Reagent Program, for providing many of the reagents.
G. P. B. is the first author, was involved in manuscript writing, and performed single-genome amplification (SGA), sequencing, pseudovirion assays, data collection, sequence analysis, and statistical analysis. P. L. M. was involved in study design, performed pseudovirion assays, and assisted with manuscript writing. L. W. performed statistical analysis. E. S. G. performed pseudovirion assays. D. J. S. performed SGA, sequencing, sequence analysis, and statistical analysis. M. M. performed pseudovirion assays. A. N. performed SGA, sequencing, and pseudovirus cloning. R. T. performed SGA, sequencing, and pseudovirus cloning. J. C. M. performed SGA, sequencing, and pseudovirus cloning. L. Maboko is the clinical site project director. S. S. A. K. was involved in the CAPRISA study design. M. H. was involved in the HISIS study design. L. Morris was involved in manuscript writing. C. W. is the principal investigator and corresponding author and was involved in manuscript writing.
Financial support. This work was supported by the Bill and Melinda Gates Foundation's Collaboration for AIDS Vaccine Discovery/Comprehensive Antibody Vaccine Immune Monitoring Consortium (grant 1032144); the Poliomyelitis Research Foundation (grant 12/28); the Carnegie Corporation (research award to G. P. B.); the National Institute of Allergy and Infectious Diseases, National Institutes for Health, US Department of Health and Human Services (grant U19 AI51794 to CAPRISA); the European Commission Directorate General for Research (grants ICA4-CT-1999-10007 and ICA4-CT-2002-10048 to the HISIS cohort study); and the Wellcome Trust (Intermediate Fellowship in Public Health and Tropical Medicine; grant 089933/Z/09/Z to P. L. M.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 2014; 28:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray ES, Madiga MC, Hermanus T, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T Cell decline and high viral load during acute infection. J Virol 2011; 85:4828–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreja H, O'Sullivan E, Pade C, et al. Neutralization activity in a geographically diverse East London cohort of human immunodeficiency virus type 1-infected patients: clade C infection results in a stronger and broader humoral immune response than clade B infection. J Gen Virol 2010; 91:2794–803. [DOI] [PubMed] [Google Scholar]
- 4.Piantadosi A, Panteleeff D, Blish CA, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol 2009; 83:10269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortez V, Odem-Davis K, McClelland RS, Jaoko W, Overbaugh J. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS Pathog 2012; 8:e1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nofemela A, Bandawe G, Thebus R, et al. Defining the human immunodeficiency virus type 1 transmission genetic bottleneck in a region with multiple circulating subtypes and recombinant forms. Virology 2011; 415:107–13. [DOI] [PubMed] [Google Scholar]
- 7.Riedner G, Rusizoka M, Hoffmann O, et al. Baseline survey of sexually transmitted infections in a cohort of female bar workers in Mbeya Region, Tanzania. Sex Transm Infect 2003; 79:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Loggerenberg F, Mlisana K, Williamson C, et al. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 2008; 3:e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrahams M-R, Treurnicht FK, Ngandu NK, et al. Rapid, complex adaptation of transmitted HIV-1 full-length genomes in subtype C-infected individuals with differing disease progression. AIDS 2013; 27:507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blish CA, Dogan OC, Derby NR, et al. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J Virol 2008; 82:12094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saathoff E, Pritsch M, Geldmacher C, et al. Viral and host factors associated with the HIV-1 viral load setpoint in adults from Mbeya Region, Tanzania. J Acquir Immune Defic Syndr 2010; 54:324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodman Z, Mlisana K, Treurnicht F, et al. Short communication decreased incidence of dual infections in South African subtype C-infected women compared to a cohort ten years earlier. AIDS Res Hum Retroviruses 2011; 27:1167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonsignori M, Wiehe K, Grimm SK, et al. An autoreactive antibody from an SLE/HIV-1 individual broadly neutralizes HIV-1. J Clin Invest 2014; 124:1835–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodcroft E, Hadfield JD, Fearnhill E, et al. The contribution of viral genotype to plasma viral set-point in HIV infection. PLoS Pathog 2014; 10:e1004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuadros DF, Branscum AJ, Crowley PH. HIV-malaria co-infection: effects of malaria on the prevalence of HIV in East sub-Saharan Africa. Int J Epidemiol 2011; 40:931–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


